bba6322462dd9a88b832101c9737b2da.ppt
- Количество слайдов: 56
 Chapter 9 Genomics Mapping and characterizing whole genomes 16 and 20 February, 2004
Chapter 9 Genomics Mapping and characterizing whole genomes 16 and 20 February, 2004
 Overview • Genomics is the molecular mapping and characterization of whole genomes and whole sets of gene products. • Consecutive high-resolution genetic and physical maps culminate in the complete DNA sequence. • Sequencing strategies depend upon the size of the genome and the distribution of its repetitive sequences. • Assembly of sequences is done clone by clone or by whole genome assembly, or both. • Computational analysis is used to describe encoded information whereas functional genomics explores function and interaction of gene products.
Overview • Genomics is the molecular mapping and characterization of whole genomes and whole sets of gene products. • Consecutive high-resolution genetic and physical maps culminate in the complete DNA sequence. • Sequencing strategies depend upon the size of the genome and the distribution of its repetitive sequences. • Assembly of sequences is done clone by clone or by whole genome assembly, or both. • Computational analysis is used to describe encoded information whereas functional genomics explores function and interaction of gene products.
 Genomics • Focuses on the entire genome • Made possible by advances in technology – automated cloning and sequencing (robotics) allowing high throughput – computerized tracking and analysis of sequences • Insights into global organization, expression, regulation and evolution – enumeration of genes – identification regulatory and functional motifs • Functional genomics to determine actual function of genetic material
Genomics • Focuses on the entire genome • Made possible by advances in technology – automated cloning and sequencing (robotics) allowing high throughput – computerized tracking and analysis of sequences • Insights into global organization, expression, regulation and evolution – enumeration of genes – identification regulatory and functional motifs • Functional genomics to determine actual function of genetic material
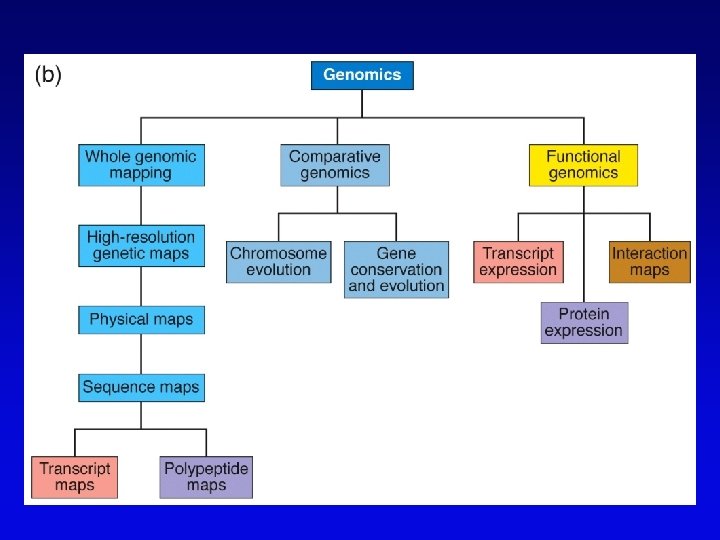
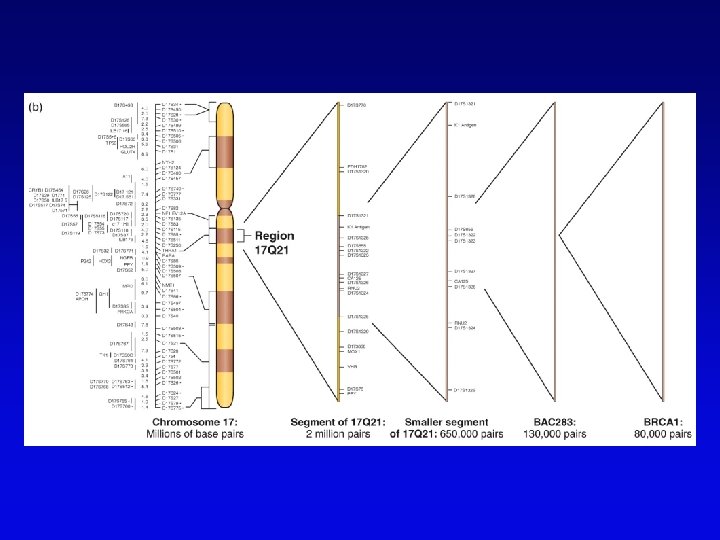
 Genome projects • Starts with high-resolution recombination and cytogenetic maps of each chromosome • Followed by physical characterization and positioning of cloned DNA fragments to anchor to high-resolution map • Followed by large-scale sequencing and analysis – clone-based sequencing – whole genome shotgun sequencing • Last step: functional genomics (the hard part)
Genome projects • Starts with high-resolution recombination and cytogenetic maps of each chromosome • Followed by physical characterization and positioning of cloned DNA fragments to anchor to high-resolution map • Followed by large-scale sequencing and analysis – clone-based sequencing – whole genome shotgun sequencing • Last step: functional genomics (the hard part)
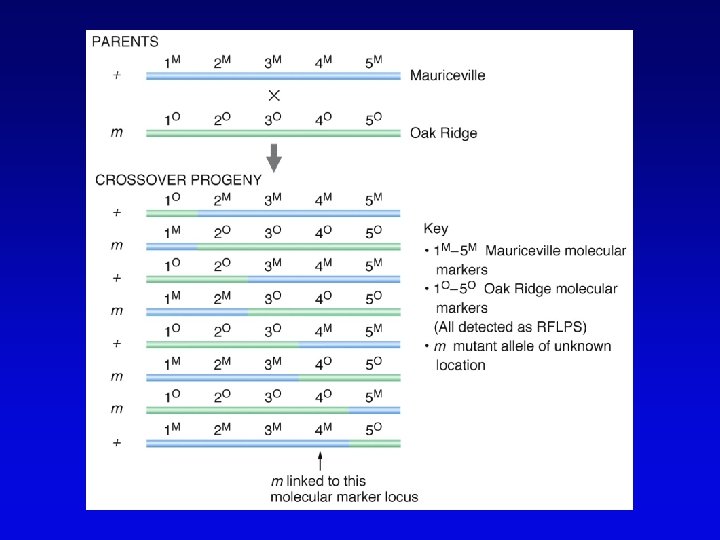
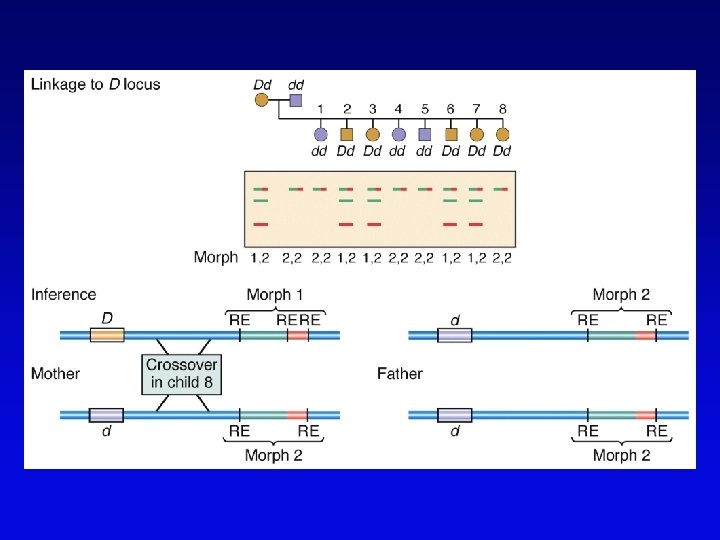
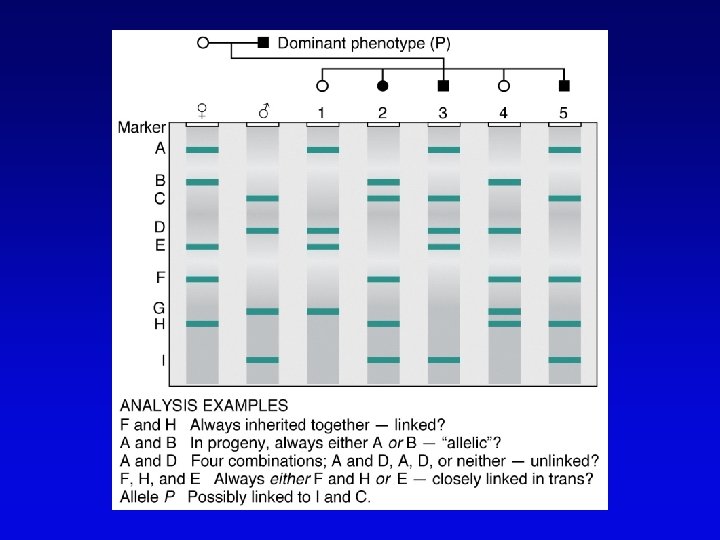
 High-resolution genetic maps • Start with low-resolution maps from existing recombination maps • Next, layer DNA polymorphisms onto map –e. g. , neutral DNA sequence variation not associated with phenotypic variation • phenotypic consequences, if any, irrelevant –such DNA markers behave as allelic gene pairs and can be detected by Southern blotting or PCR –mapped by recombination or cytogenetics
High-resolution genetic maps • Start with low-resolution maps from existing recombination maps • Next, layer DNA polymorphisms onto map –e. g. , neutral DNA sequence variation not associated with phenotypic variation • phenotypic consequences, if any, irrelevant –such DNA markers behave as allelic gene pairs and can be detected by Southern blotting or PCR –mapped by recombination or cytogenetics
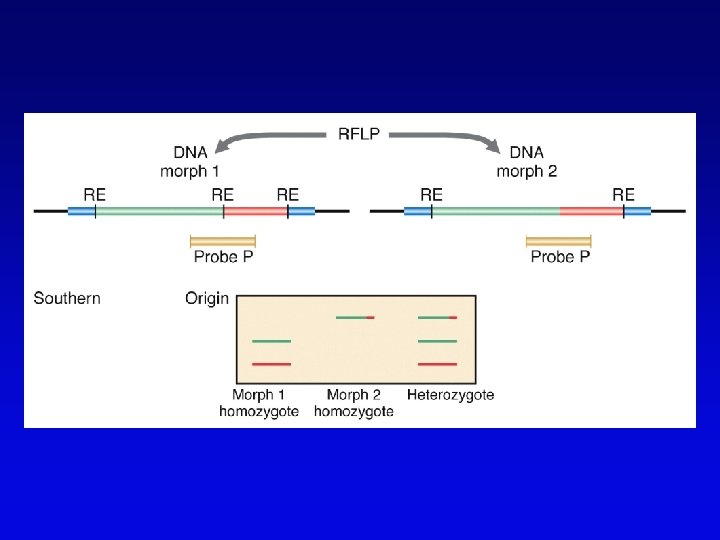
 RFLP • • Restriction fragment length polymorphism May or may not be neutral Detected by Southern blotting or PCR Multiple RFLPs can be mapped by classical segregation analysis • Example: – restriction digestion of PCR products yields one fragment for allele A and two fragments for a – note that homozygotes and heterozygote have different restriction patterns, permitting identification of carrier
RFLP • • Restriction fragment length polymorphism May or may not be neutral Detected by Southern blotting or PCR Multiple RFLPs can be mapped by classical segregation analysis • Example: – restriction digestion of PCR products yields one fragment for allele A and two fragments for a – note that homozygotes and heterozygote have different restriction patterns, permitting identification of carrier
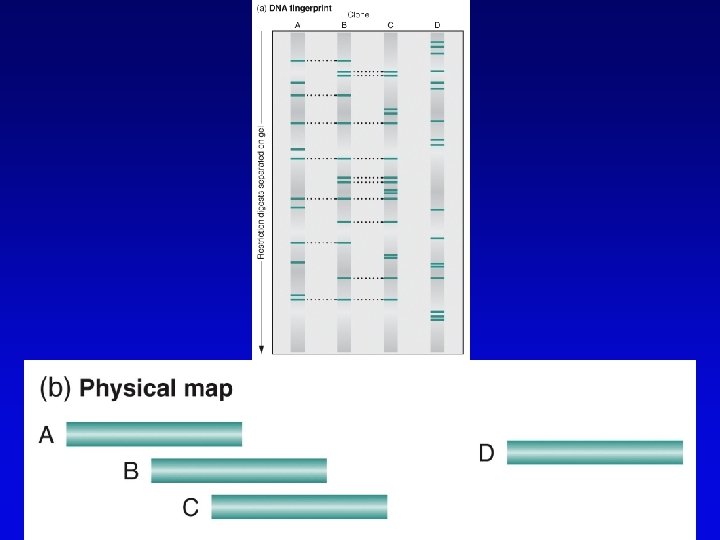
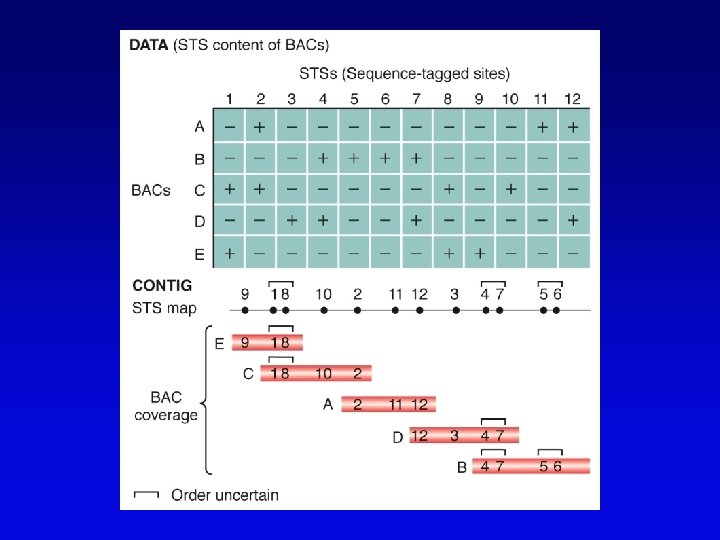
 Physical maps • Maps of physically isolated pieces of genome, i. e. , cloned DNA – previously cloned DNA can be localized to map by Southern blotting or PCR to measure location and distance – useful in assembly of sequences • Vectors with large inserts are most useful • Overlapping clones are assembled into contigs, ideally, one per chromosome – mapping of restriction sites – sequence-tagged sites (STSs)
Physical maps • Maps of physically isolated pieces of genome, i. e. , cloned DNA – previously cloned DNA can be localized to map by Southern blotting or PCR to measure location and distance – useful in assembly of sequences • Vectors with large inserts are most useful • Overlapping clones are assembled into contigs, ideally, one per chromosome – mapping of restriction sites – sequence-tagged sites (STSs)
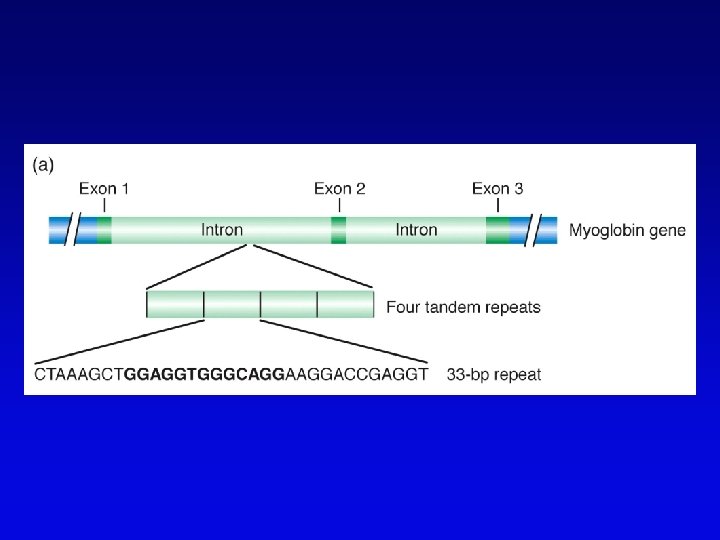
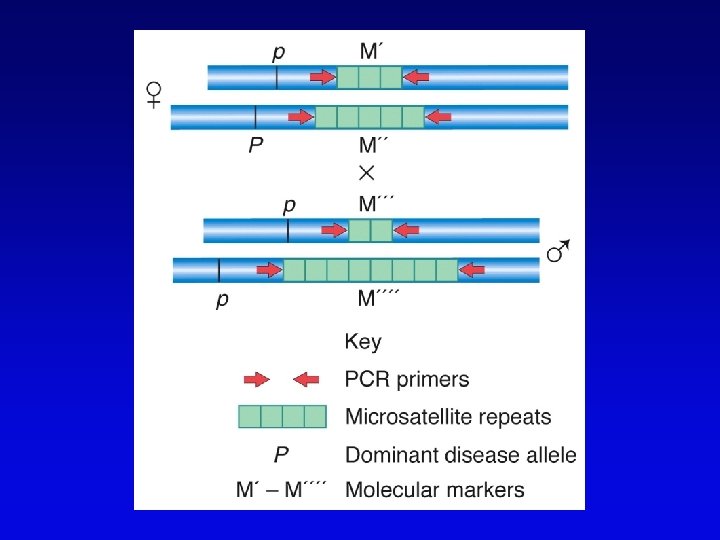
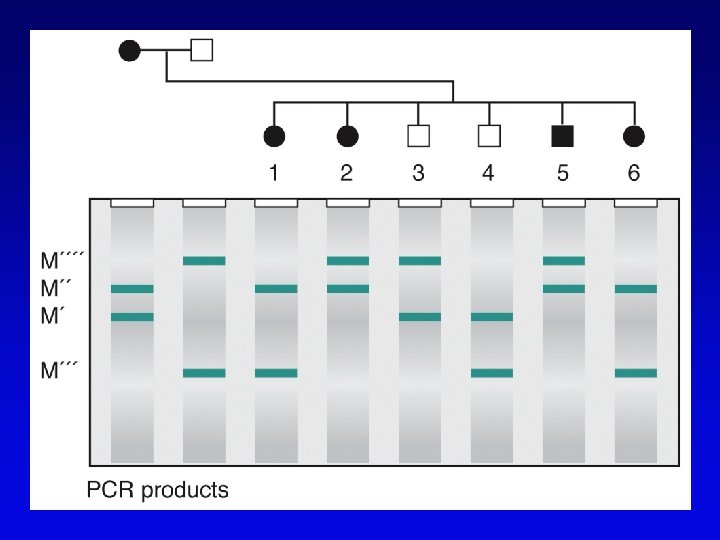
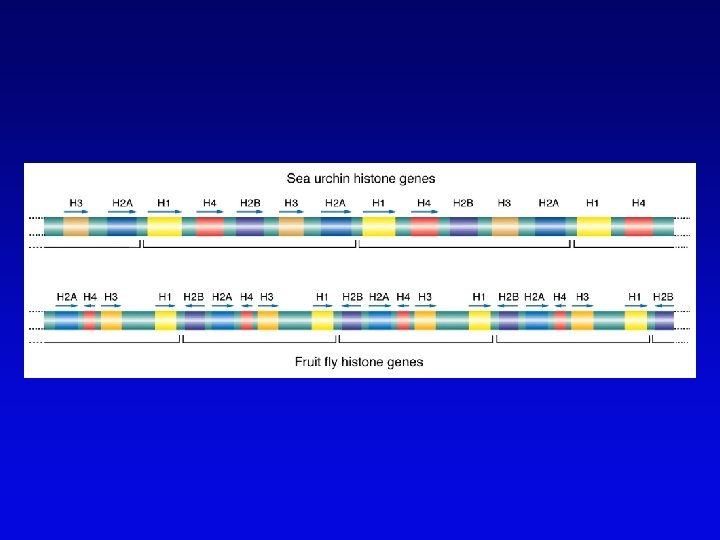
 Short-sequence repeat markers • Tandemly repeated • Variable numbers of repeats, give different size restriction fragments detected on Southern blots • Single sequence length polymorphisms (SSLPs) – e. g. , TGACGTATGACGTA – mutations give rise to large number of alleles – higher proportion of heterozygotes – two types in genomics • minisatellite (VNTRs) • microsatellite
Short-sequence repeat markers • Tandemly repeated • Variable numbers of repeats, give different size restriction fragments detected on Southern blots • Single sequence length polymorphisms (SSLPs) – e. g. , TGACGTATGACGTA – mutations give rise to large number of alleles – higher proportion of heterozygotes – two types in genomics • minisatellite (VNTRs) • microsatellite
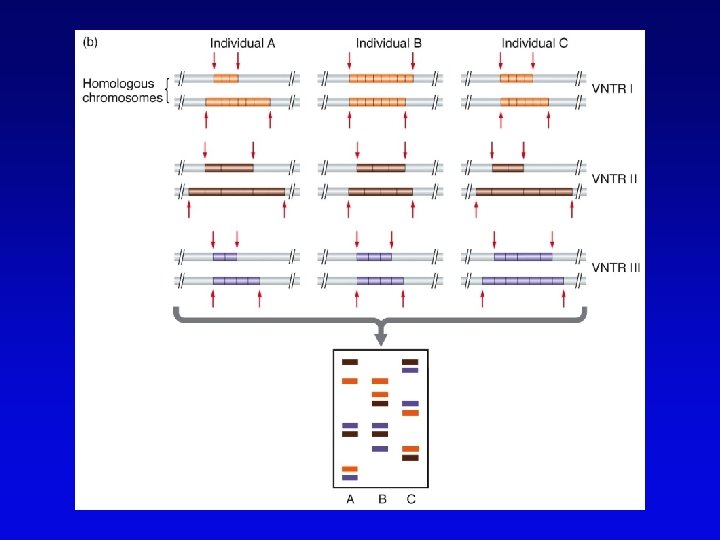
 Minisatellites and microsatellites • Minisatellites – based on variation of number of tandem repeats (VNTRs) which segregate as alleles – in humans, repeat unit is 15 -100 nucleotides, for total of 1 -5 kb – if number of repeats is variable, Southern blot will show numerous bands – basis of DNA fingerprinting and can be used in mapping • Microsatellites – sequences dispersed throughout the genome – variable numbers of dinucleotide repeats – detected by PCR
Minisatellites and microsatellites • Minisatellites – based on variation of number of tandem repeats (VNTRs) which segregate as alleles – in humans, repeat unit is 15 -100 nucleotides, for total of 1 -5 kb – if number of repeats is variable, Southern blot will show numerous bands – basis of DNA fingerprinting and can be used in mapping • Microsatellites – sequences dispersed throughout the genome – variable numbers of dinucleotide repeats – detected by PCR
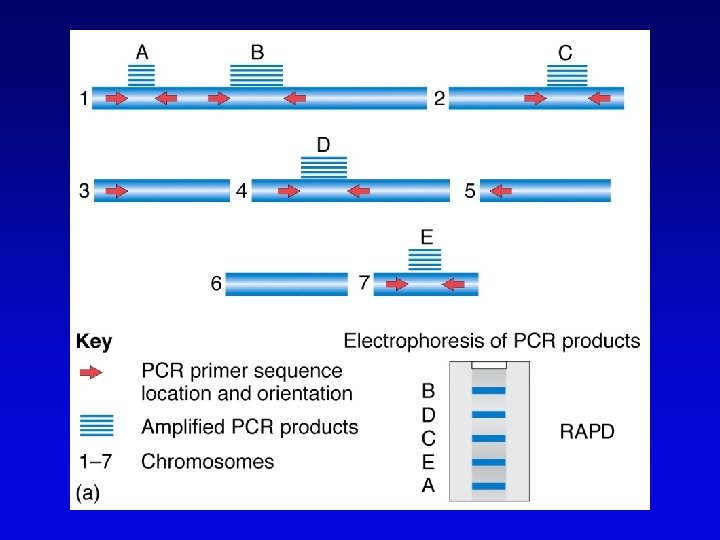
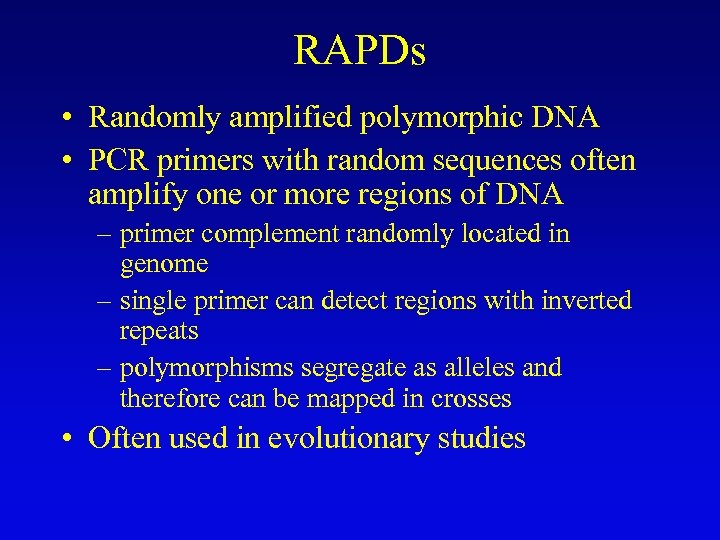 RAPDs • Randomly amplified polymorphic DNA • PCR primers with random sequences often amplify one or more regions of DNA – primer complement randomly located in genome – single primer can detect regions with inverted repeats – polymorphisms segregate as alleles and therefore can be mapped in crosses • Often used in evolutionary studies
RAPDs • Randomly amplified polymorphic DNA • PCR primers with random sequences often amplify one or more regions of DNA – primer complement randomly located in genome – single primer can detect regions with inverted repeats – polymorphisms segregate as alleles and therefore can be mapped in crosses • Often used in evolutionary studies
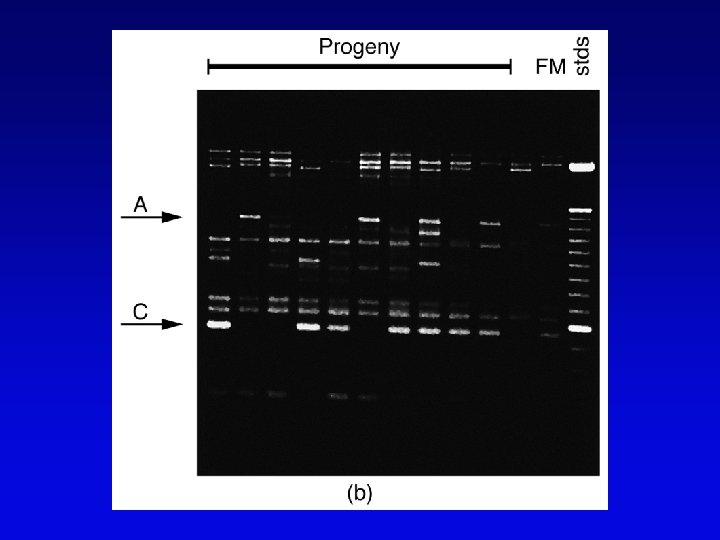
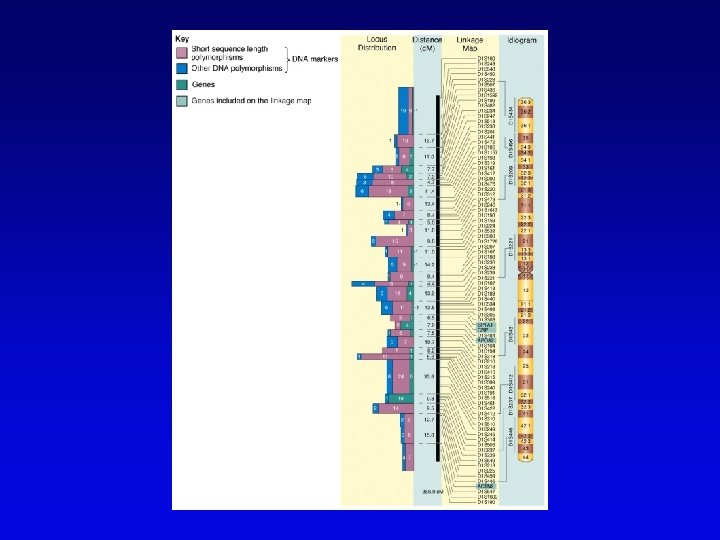
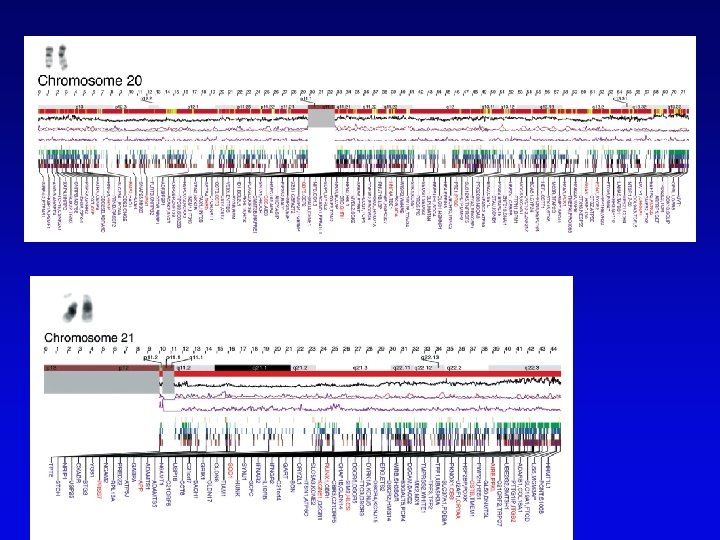
 Human high-resolution map • RFLP, SSLP, and RAPD markers have been mapped to 1 c. M density • Provide landmarks for anchoring sequence information • 1 c. M of human DNA is ~ 1 Mb of DNA, still a large amount • Single nucleotide polymorphisms (SNPs) estimated number about 3 million between any two genomes
Human high-resolution map • RFLP, SSLP, and RAPD markers have been mapped to 1 c. M density • Provide landmarks for anchoring sequence information • 1 c. M of human DNA is ~ 1 Mb of DNA, still a large amount • Single nucleotide polymorphisms (SNPs) estimated number about 3 million between any two genomes
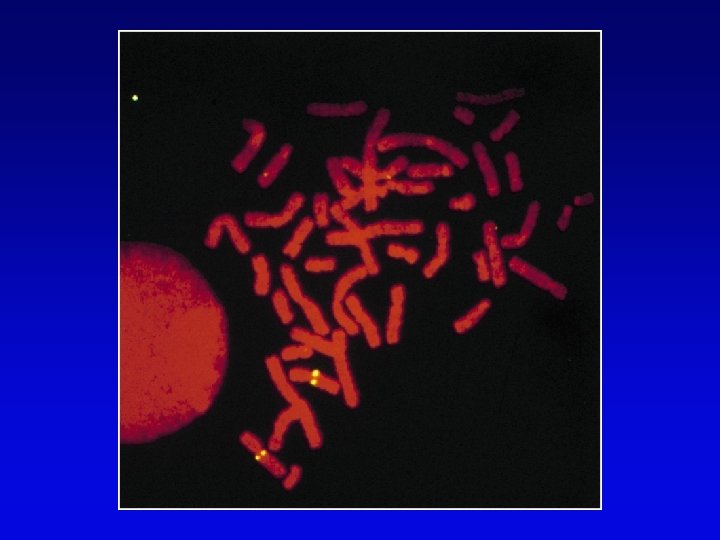
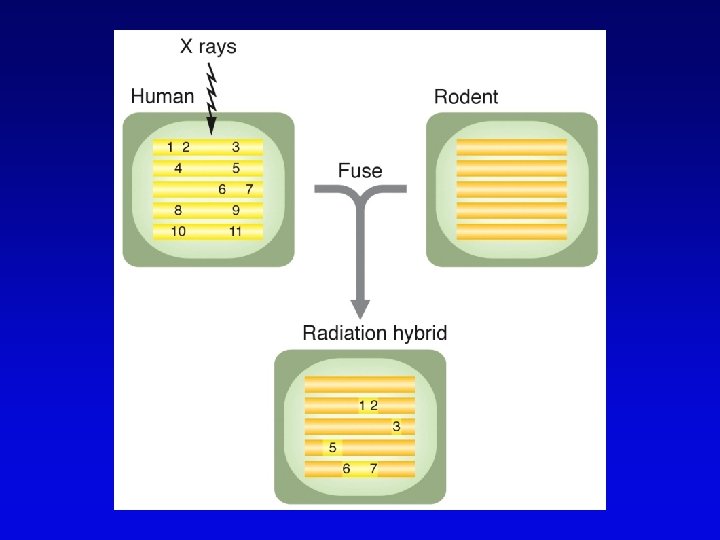
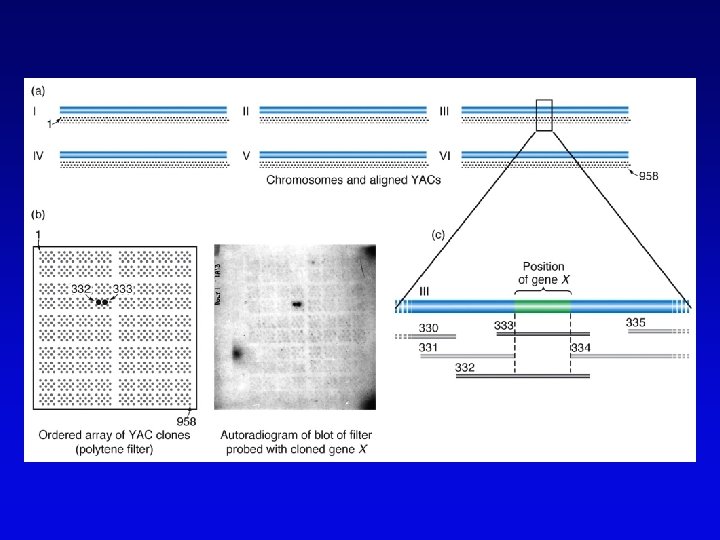
 High-resolution cytogenetic maps • Relates markers to chromosome bands, puffs or disruptions • In situ hybridization – cloned DNA labeled with radioactivity or fluorescent dye (FISH) – hybridized to denatured metaphase or polytene chromosomes – indicates approximate locations – FISH extension allows chromosome painting • Rearrangement breakpoint mapping – detected by Southern blotting
High-resolution cytogenetic maps • Relates markers to chromosome bands, puffs or disruptions • In situ hybridization – cloned DNA labeled with radioactivity or fluorescent dye (FISH) – hybridized to denatured metaphase or polytene chromosomes – indicates approximate locations – FISH extension allows chromosome painting • Rearrangement breakpoint mapping – detected by Southern blotting
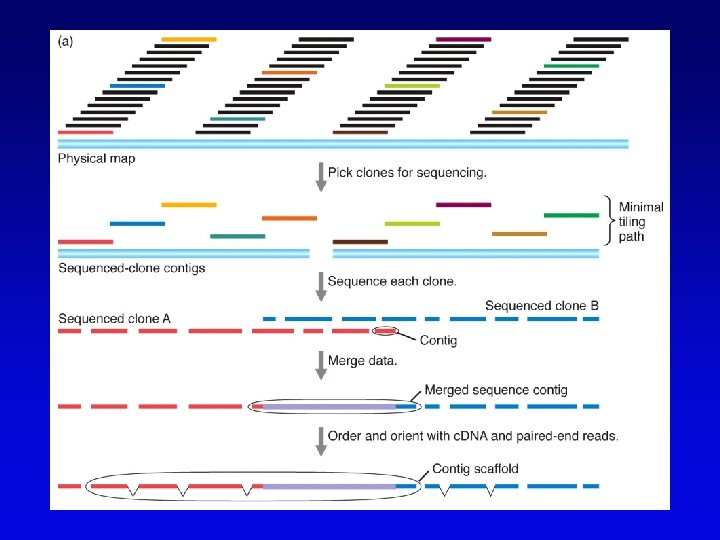
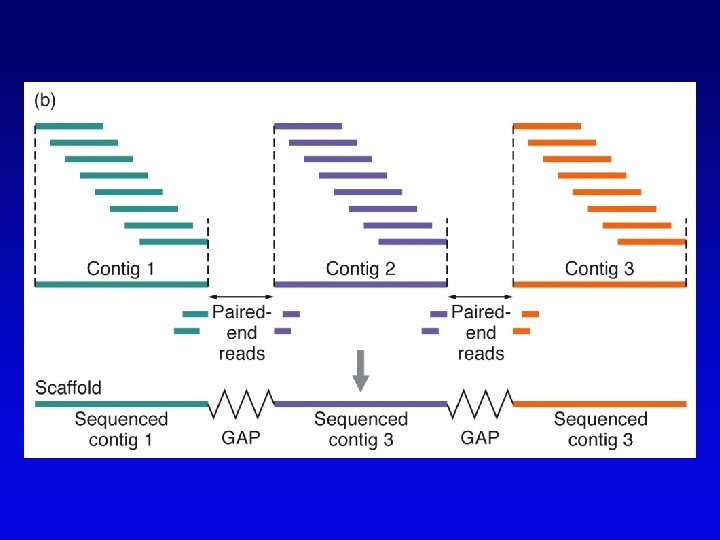
 Assembling genomes with repetitive sequences • Use of ordered clones – e. g. , C. elegans – large mapped, cosmids with minimum overlap (minimum tiling path) subcloned into sequencing vectors – inserts sequenced by automated methods – sequence assembled by computer based on map • Whole genome shotgun – e. g. , D. melanogaster – three libraries (2 -kb, 100 -kb, 150 -kb) of genomic clones, each sequenced from both ends – sequences aligned by homologous sequence overlap and by use of paired-end sequences to produce scaffolds of contigs
Assembling genomes with repetitive sequences • Use of ordered clones – e. g. , C. elegans – large mapped, cosmids with minimum overlap (minimum tiling path) subcloned into sequencing vectors – inserts sequenced by automated methods – sequence assembled by computer based on map • Whole genome shotgun – e. g. , D. melanogaster – three libraries (2 -kb, 100 -kb, 150 -kb) of genomic clones, each sequenced from both ends – sequences aligned by homologous sequence overlap and by use of paired-end sequences to produce scaffolds of contigs
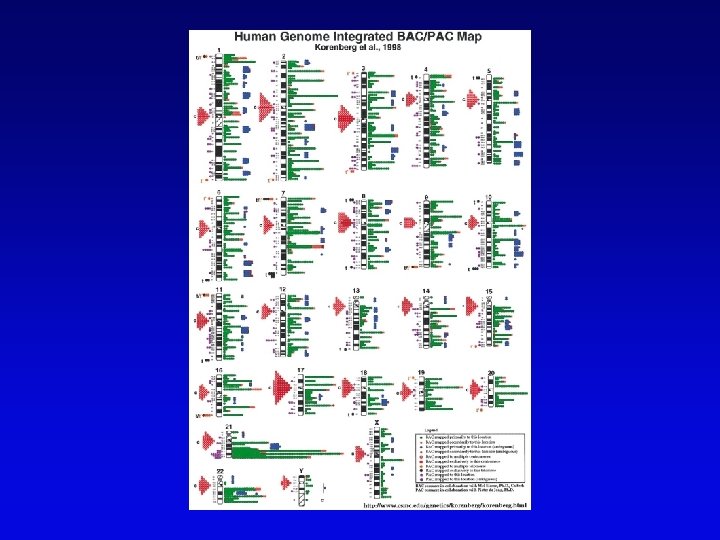
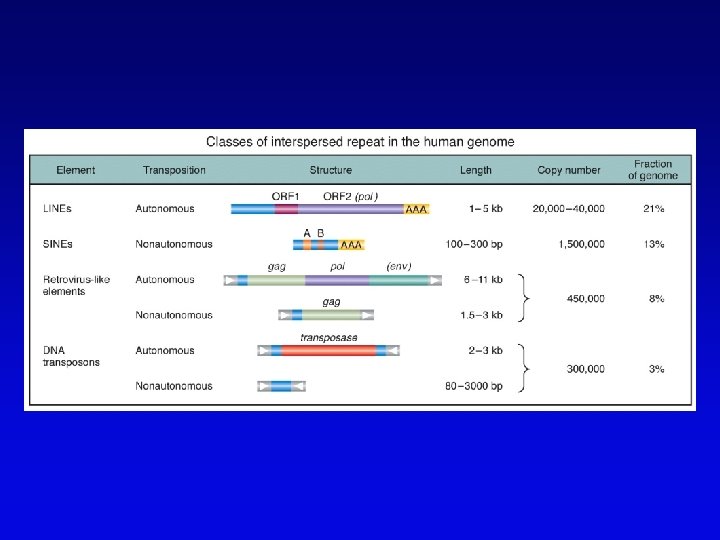
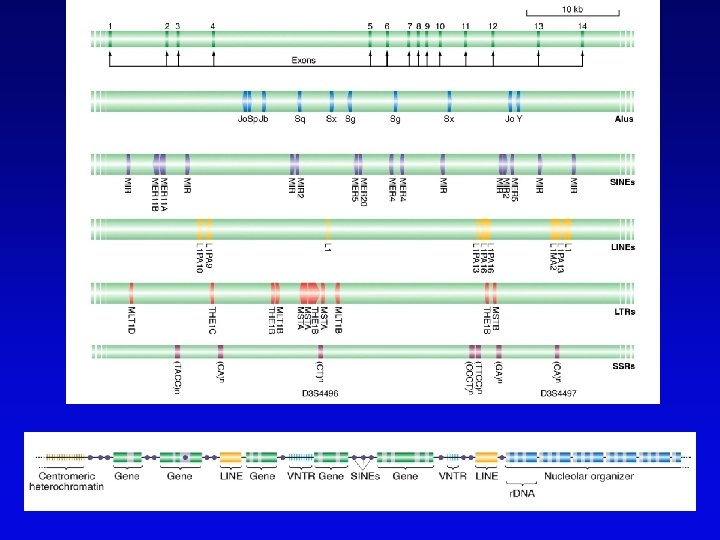
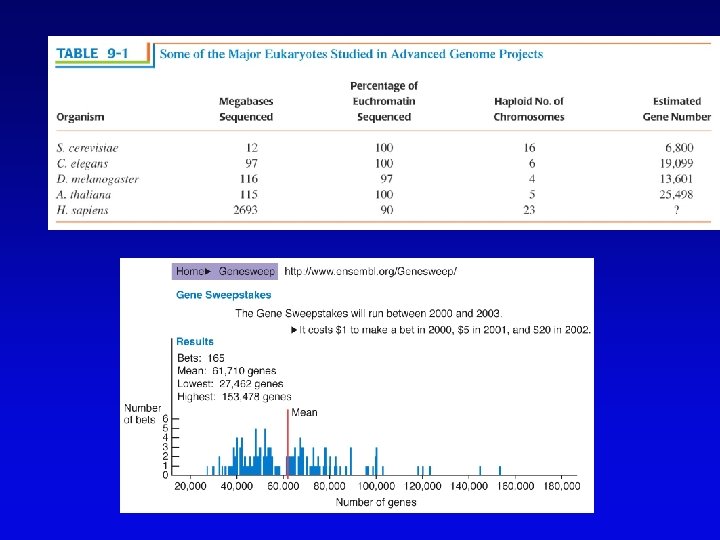
 Assembling genomes • If genome is rich in repetitive elements, contigs may be short • Gaps usually occur, regardless of technique – short gaps filled by PCR – long gaps require additional cloning, sometimes in different host • Sequenced eukaryotic genomes include: Saccharomyces cerevisiae, Caenorhabditis elegans, Drosophila. melanogaster, Arabidopsis thaliania, Mus musculus, Danio rerio, Homo sapiens
Assembling genomes • If genome is rich in repetitive elements, contigs may be short • Gaps usually occur, regardless of technique – short gaps filled by PCR – long gaps require additional cloning, sometimes in different host • Sequenced eukaryotic genomes include: Saccharomyces cerevisiae, Caenorhabditis elegans, Drosophila. melanogaster, Arabidopsis thaliania, Mus musculus, Danio rerio, Homo sapiens
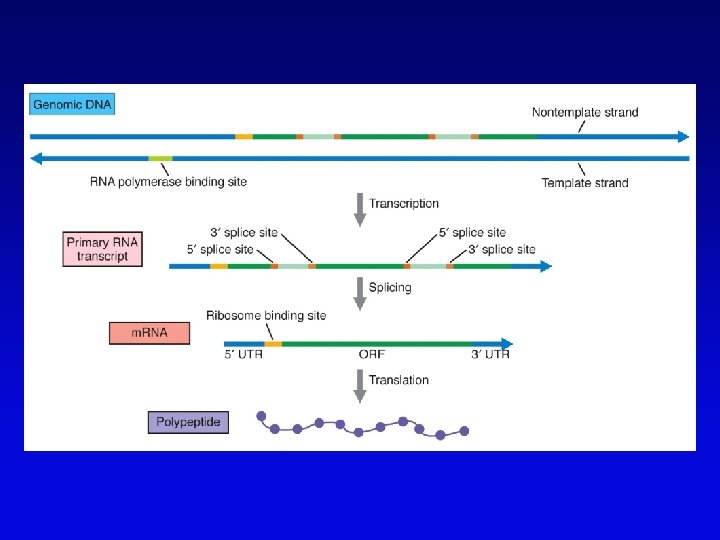
 Bioinformatics (1) • Not clear what all of nucleotide sequence of draft genome means • In addition to proteome (protein encoding sequences), genome contains additional information • Considerable ignorance due to the following: – docking (target) sequences of many DNA binding proteins are unknown – alternative splicing complicates ORF finding – some sequences have context-dependent meaning – some sequences have multiple uses • Bioinformatics and functional genomics attempt to decipher genome
Bioinformatics (1) • Not clear what all of nucleotide sequence of draft genome means • In addition to proteome (protein encoding sequences), genome contains additional information • Considerable ignorance due to the following: – docking (target) sequences of many DNA binding proteins are unknown – alternative splicing complicates ORF finding – some sequences have context-dependent meaning – some sequences have multiple uses • Bioinformatics and functional genomics attempt to decipher genome
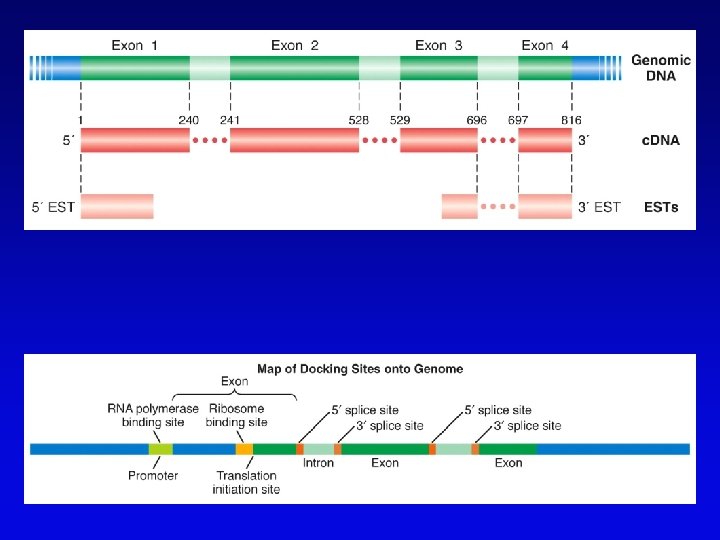
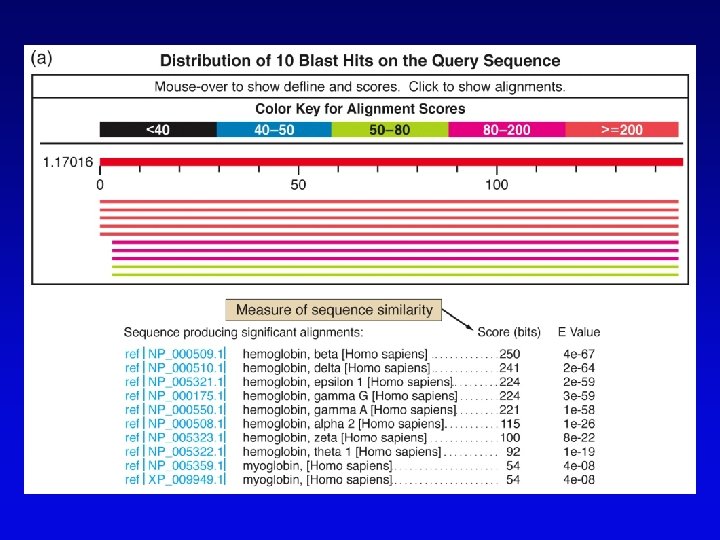
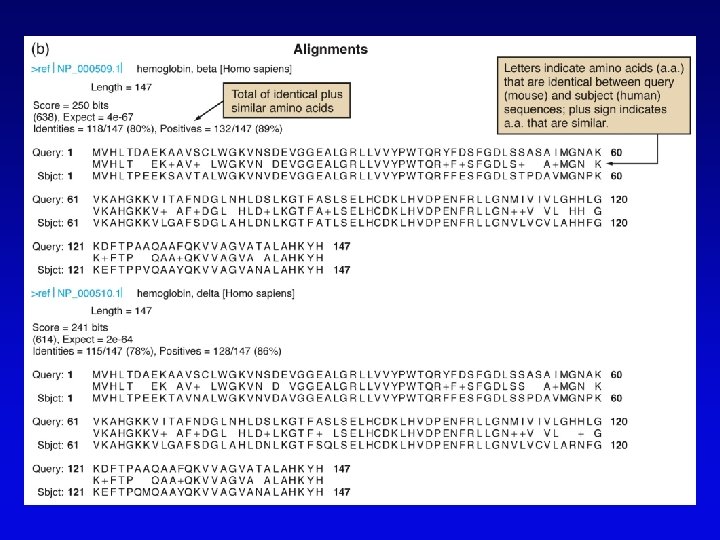
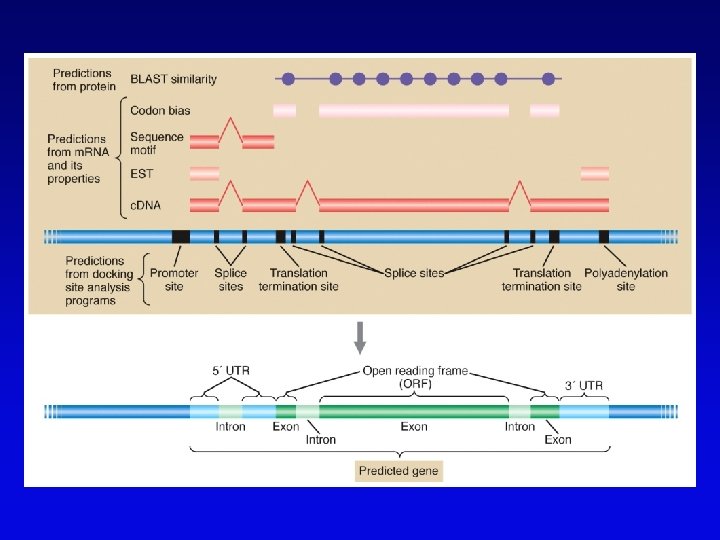
 Bioinformatics (2) • Uses available information (much of it available on the Web) to predict function of sequences • c. DNA evidence – motifs, e. g. , start codons, ORFs – expressed sequence tags (ESTs), from reverse transcribed m. RNA • m. RNA and ORF structure – gene and intron finding programs • Polypeptide similarity evidence – at level of >35% sequence identity, polypeptides likely have common function – often identified by BLASTp search • Codon bias information
Bioinformatics (2) • Uses available information (much of it available on the Web) to predict function of sequences • c. DNA evidence – motifs, e. g. , start codons, ORFs – expressed sequence tags (ESTs), from reverse transcribed m. RNA • m. RNA and ORF structure – gene and intron finding programs • Polypeptide similarity evidence – at level of >35% sequence identity, polypeptides likely have common function – often identified by BLASTp search • Codon bias information
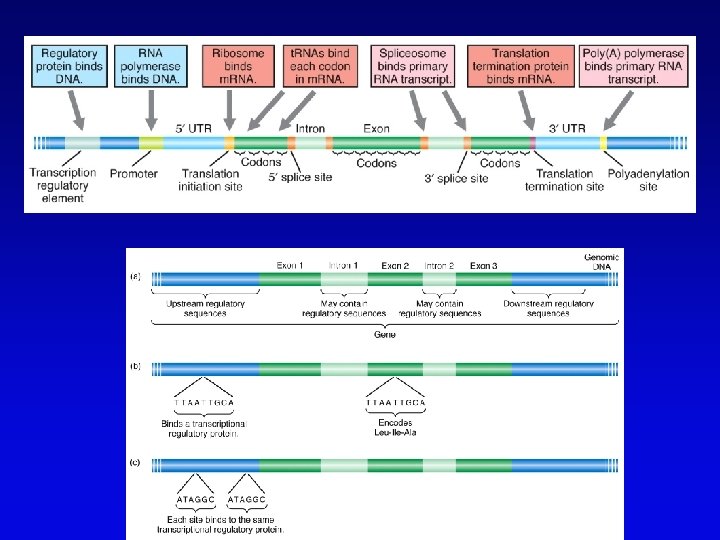
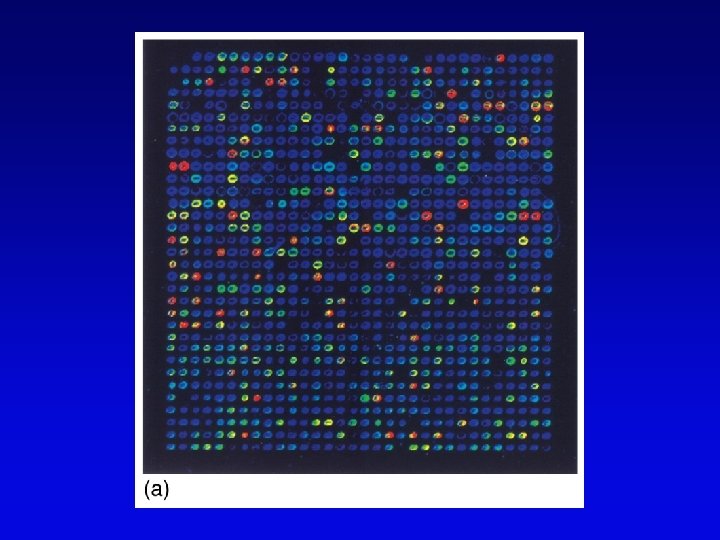
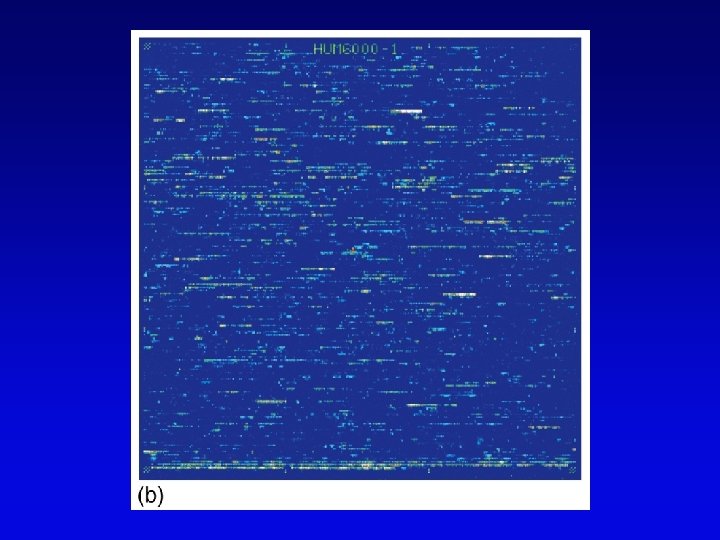
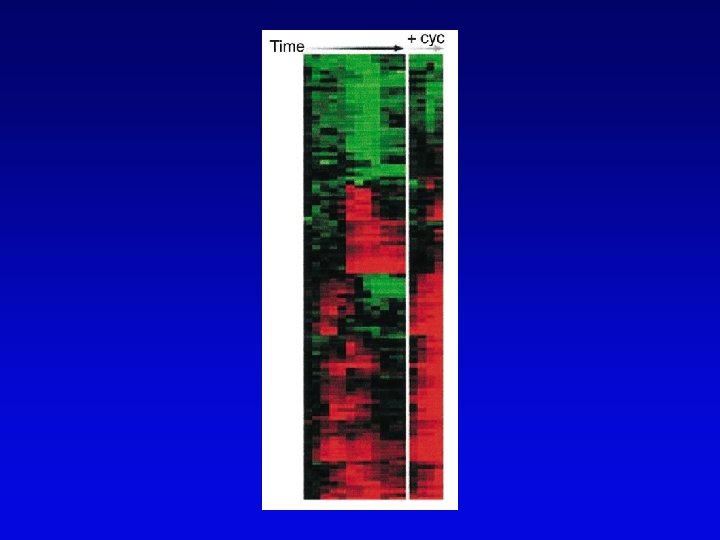
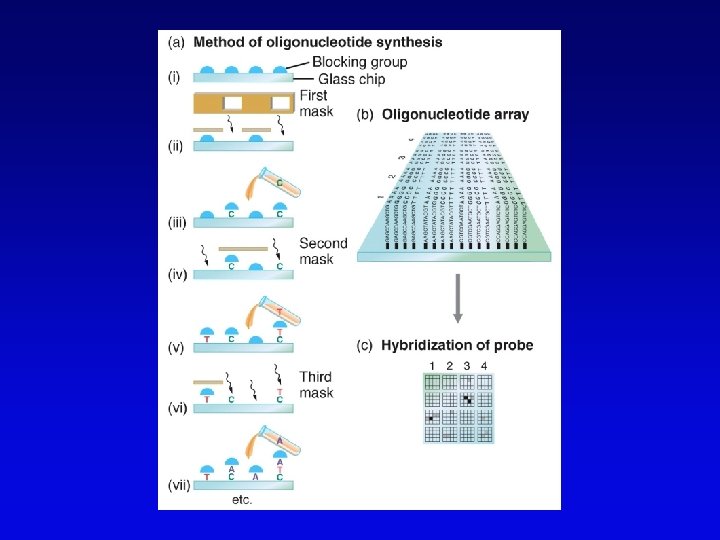
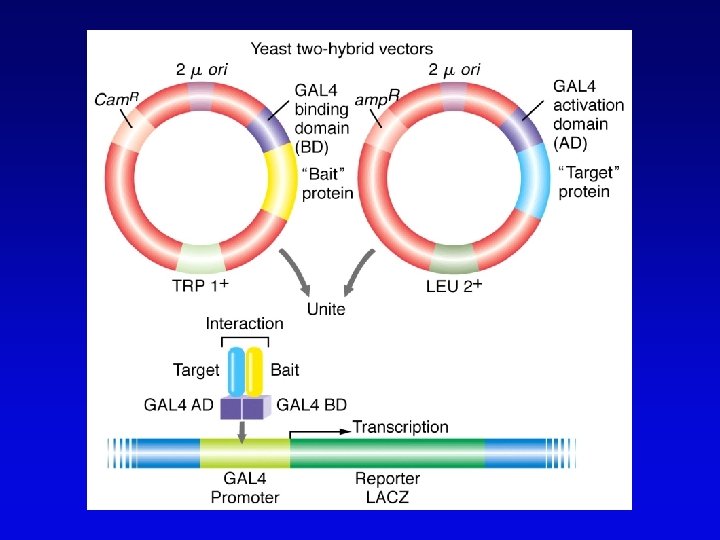
 Functional genomics • Study of expression and interaction of gene products • Requires new vocabulary and techniques – transcriptome: all DNA transcripts • may be monitored by use of DNA chips – proteome: all encoded proteins • complicated by alternative splicing – interactome: all interactions between all categories of molecules • detected by two-hybrid system and related procedures – phenome: phenotype of each gene knockout
Functional genomics • Study of expression and interaction of gene products • Requires new vocabulary and techniques – transcriptome: all DNA transcripts • may be monitored by use of DNA chips – proteome: all encoded proteins • complicated by alternative splicing – interactome: all interactions between all categories of molecules • detected by two-hybrid system and related procedures – phenome: phenotype of each gene knockout
 Assignment: Continue with * section of the Web tutorial.
Assignment: Continue with * section of the Web tutorial.



