c0f69d3ba4dc36bb9e34fe1d82ee1d5e.ppt
- Количество слайдов: 23

Chapter 3: Cells • Plasma membrane: structure • Plasma membrane: transport • Resting membrane potential • Cell-environment interactions • Cytoplasm • Nucleus • Cell growth & reproduction • Extracellular materials, developmental aspects Department of Kinesiology and Applied Physiology WCR
Cytoplasm • Located between plasma membrane and nucleus • Cytosol: water with solutes (protein, salts, sugars, etc. ) • Cytoplasmic organelles: metabolic machinery of cell • Mitochondria, ER, Golgi, lysosomes, etc. • Cytoskeleton, centrioles, ribosomes, etc. • Inclusions • Granules (glycogen, pigments), lipid droplets, etc. Copyright © 2010 Pearson Education, Inc.
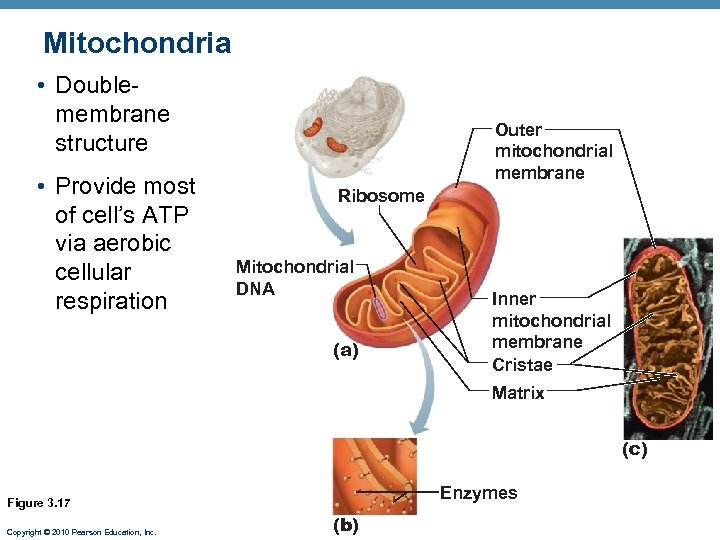
Mitochondria • Doublemembrane structure • Provide most of cell’s ATP via aerobic cellular respiration Outer mitochondrial membrane Ribosome Mitochondrial DNA (a) Inner mitochondrial membrane Cristae Matrix (c) Enzymes Figure 3. 17 Copyright © 2010 Pearson Education, Inc. (b)
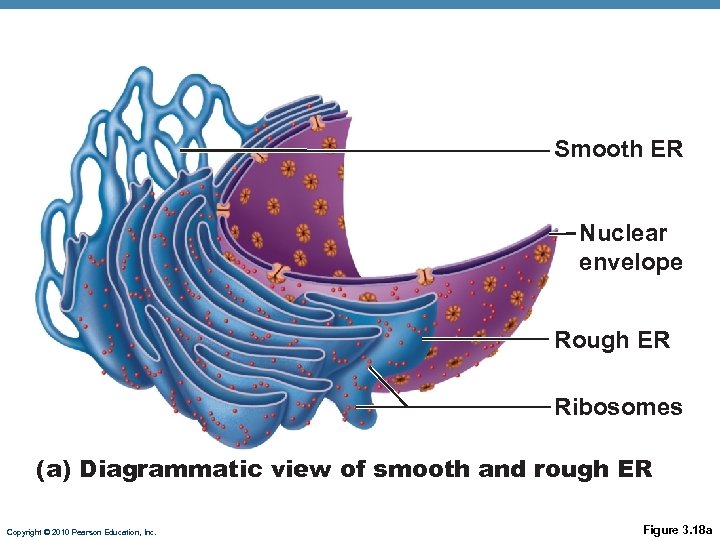
Smooth ER Nuclear envelope Rough ER Ribosomes (a) Diagrammatic view of smooth and rough ER Copyright © 2010 Pearson Education, Inc. Figure 3. 18 a
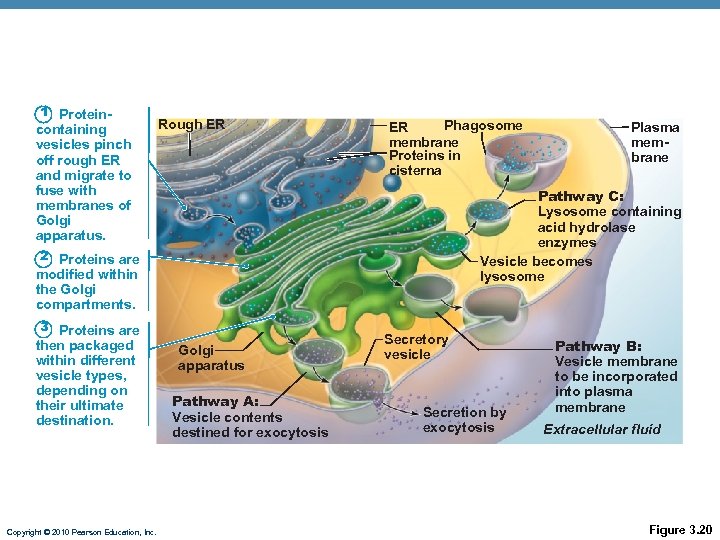
1 Protein- containing vesicles pinch off rough ER and migrate to fuse with membranes of Golgi apparatus. Rough ER Phagosome ER membrane Proteins in cisterna Pathway C: Lysosome containing acid hydrolase enzymes Vesicle becomes lysosome 2 Proteins are modified within the Golgi compartments. 3 Proteins are then packaged within different vesicle types, depending on their ultimate destination. Copyright © 2010 Pearson Education, Inc. Plasma membrane Golgi apparatus Pathway A: Vesicle contents destined for exocytosis Secretory vesicle Secretion by exocytosis Pathway B: Vesicle membrane to be incorporated into plasma membrane Extracellular fluid Figure 3. 20
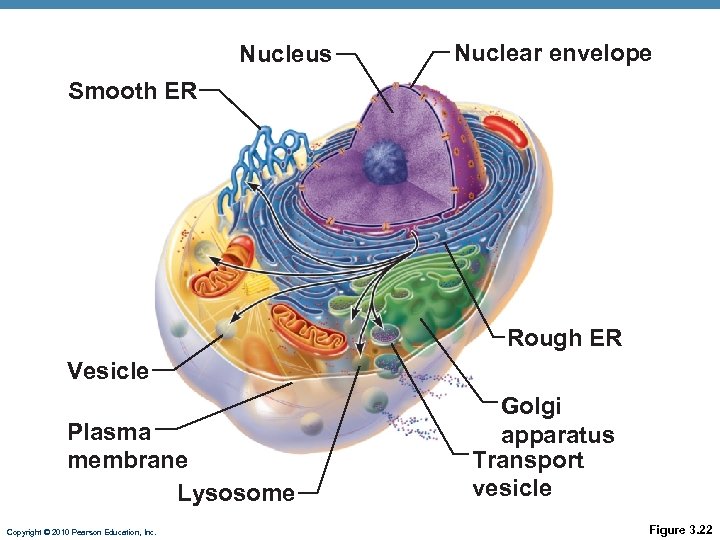
Nucleus Nuclear envelope Smooth ER Rough ER Vesicle Plasma membrane Lysosome Copyright © 2010 Pearson Education, Inc. Golgi apparatus Transport vesicle Figure 3. 22

Exercise-induced BCL 2 -regulated autophagy is required for muscle glucose homeostasis He et al. , Nature (2012) doi: 10. 1038/nature 10758 Exercise has beneficial effects on human health, including protection against metabolic disorders such as diabetes 1. However, the cellular mechanisms underlying these effects are incompletely understood. The lysosomal degradation pathway, autophagy, is an intracellular recycling system that functions during basal conditions in organelle and protein quality control 2. During stress, increased levels of autophagy permit cells to adapt to changing nutritional and energy demands through protein catabolism 3. Moreover, in animal models, autophagy protects against diseases such as cancer, neurodegenerative disorders, infections, inflammatory diseases, ageing and insulin resistance 4, 5, 6. Here we show that acute exercise induces autophagy in skeletal and cardiac muscle of fed mice. To investigate the role of exercise-mediated autophagy in vivo, we generated mutant mice that show normal levels of basal autophagy but are deficient in stimulus (exercise- or starvation)-induced autophagy. These mice (termed BCL 2 AAA mice) contain knock-in mutations in BCL 2 phosphorylation sites (Thr 69 Ala, Ser 70 Ala and Ser 84 Ala) that prevent stimulus-induced disruption of the BCL 2–beclin-1 complex and autophagy activation. BCL 2 AAA mice show decreased endurance and altered glucose metabolism during acute exercise, as well as impaired chronic exercise-mediated protection against high-fat-diet-induced glucose intolerance. Thus, exercise induces autophagy, BCL 2 is a crucial regulator of exercise- (and starvation)-induced autophagy in vivo, and autophagy induction may contribute to the beneficial metabolic effects of exercise.

Exercise-induced BCL 2 -regulated autophagy is required for muscle glucose homeostasis He et al. , Nature (2012) doi: 10. 1038/nature 10758 This study shows that exercise can activate cellular recycling (autophagy). Other researchers have already shown that autophagy protects against cancer, diabetes, aging, etc. The authors also made genetically altered mice with normal “resting” autophagy but which did not “ramp up” autophagy in response to exercise. These mice had lower endurance and were more likely to develop a diabetic condition. This study is a strong step forward in understanding how and why exercise is good for us.
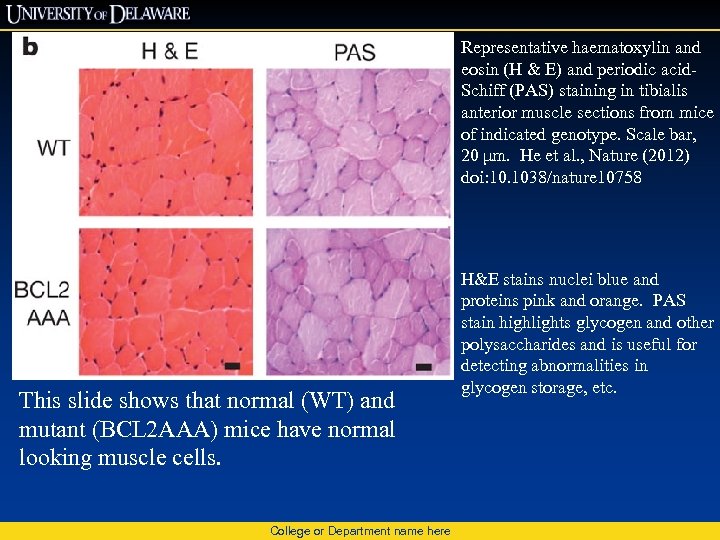
Representative haematoxylin and eosin (H & E) and periodic acid. Schiff (PAS) staining in tibialis anterior muscle sections from mice of indicated genotype. Scale bar, 20 μm. He et al. , Nature (2012) doi: 10. 1038/nature 10758 This slide shows that normal (WT) and mutant (BCL 2 AAA) mice have normal looking muscle cells. College or Department name here H&E stains nuclei blue and proteins pink and orange. PAS stain highlights glycogen and other polysaccharides and is useful for detecting abnormalities in glycogen storage, etc.
Endomembrane System PLAY Copyright © 2010 Pearson Education, Inc. Animation: Endomembrane System
Cytoskeleton Rods & struts throughout cytosol • Microtubules – hollow tubes; tubulin; radiate from centrosomes; help regulate cell motility & shape; position intracellular structures • Microfilaments – double helix; actin; help regulate cell motility & shape • Intermediate filaments – various types including keratins; resist force; attach to desmosomes Copyright © 2010 Pearson Education, Inc.
Motor Molecules • Protein complexes that generate a pulling force • Motility function: contraction, movement of organelles, etc. • Powered by ATP • Some types (substrate – what they “walk” on) (example) • Myosin (actin) (muscle) • Kinesin “walkers” (microtubules) (see video) • Dynein (microtubules) (sperm flagellum) Copyright © 2010 Pearson Education, Inc.
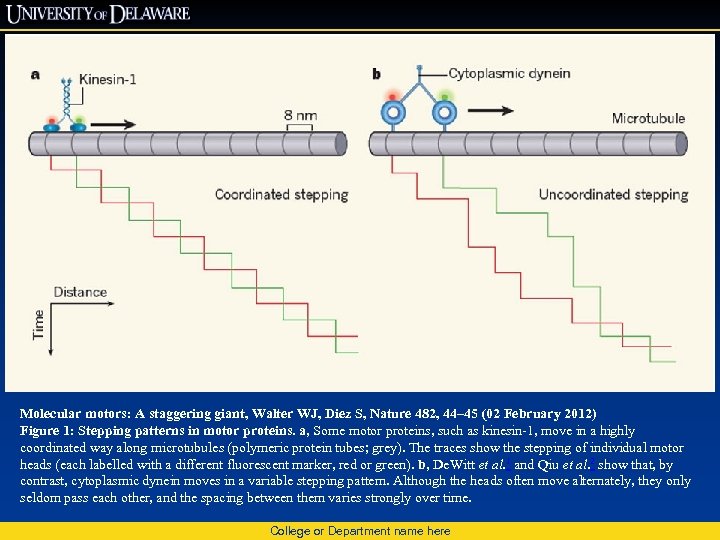
Molecular motors: A staggering giant, Walter WJ, Diez S, Nature 482, 44– 45 (02 February 2012) Figure 1: Stepping patterns in motor proteins. a, Some motor proteins, such as kinesin-1, move in a highly coordinated way along microtubules (polymeric protein tubes; grey). The traces show the stepping of individual motor heads (each labelled with a different fluorescent marker, red or green). b, De. Witt et al. 1 and Qiu et al. 2 show that, by contrast, cytoplasmic dynein moves in a variable stepping pattern. Although the heads often move alternately, they only seldom pass each other, and the spacing between them varies strongly over time. College or Department name here
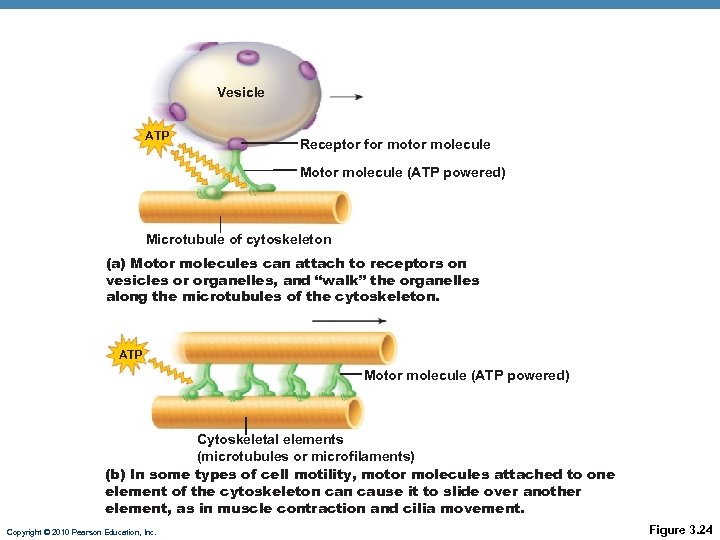
Vesicle ATP Receptor for motor molecule Motor molecule (ATP powered) Microtubule of cytoskeleton (a) Motor molecules can attach to receptors on vesicles or organelles, and “walk” the organelles along the microtubules of the cytoskeleton. ATP Motor molecule (ATP powered) Cytoskeletal elements (microtubules or microfilaments) (b) In some types of cell motility, motor molecules attached to one element of the cytoskeleton cause it to slide over another element, as in muscle contraction and cilia movement. Copyright © 2010 Pearson Education, Inc. Figure 3. 24
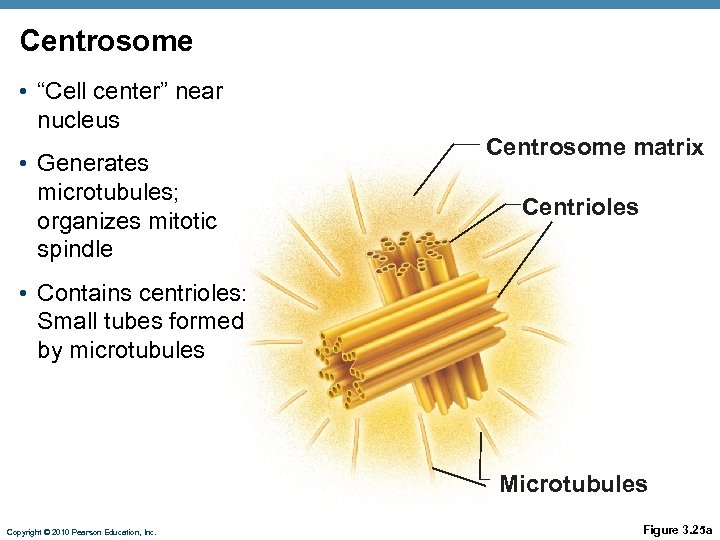
Centrosome • “Cell center” near nucleus • Generates microtubules; organizes mitotic spindle Centrosome matrix Centrioles • Contains centrioles: Small tubes formed by microtubules Microtubules Copyright © 2010 Pearson Education, Inc. Figure 3. 25 a
Cellular Extensions • Cilia and flagella • Whiplike, motile extensions on surfaces of certain cells • Contain microtubules and motor molecules • Cilia move substances across cell surfaces • Longer flagella propel whole cells (tail of sperm) • Microvilli • Fingerlike extensions of plasma membrane • Increase surface area for absorption • Core of actin filaments for stiffening PLAY Copyright © 2010 Pearson Education, Inc. Animation: Cilia and Flagella
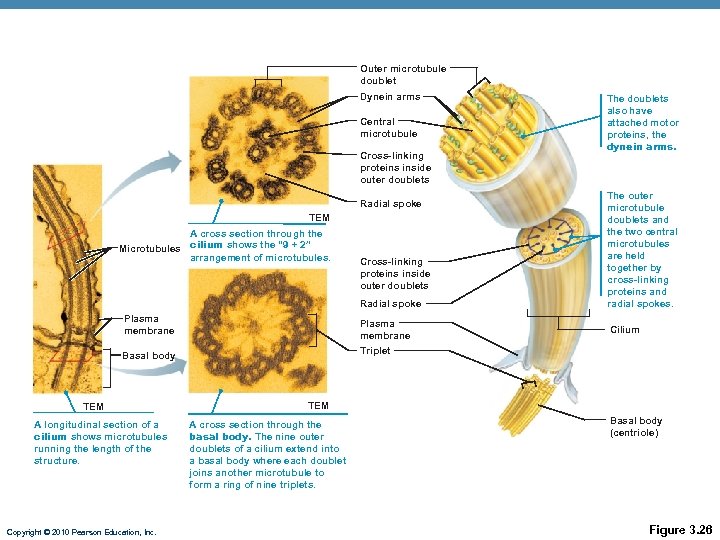
Outer microtubule doublet Dynein arms Central microtubule Cross-linking proteins inside outer doublets Radial spoke TEM A cross section through the Microtubules cilium shows the “ 9 + 2” arrangement of microtubules. Cross-linking proteins inside outer doublets Radial spoke Plasma membrane Basal body The doublets also have attached motor proteins, the dynein arms. The outer microtubule doublets and the two central microtubules are held together by cross-linking proteins and radial spokes. Triplet TEM A longitudinal section of a cilium shows microtubules running the length of the structure. Copyright © 2010 Pearson Education, Inc. Cilium TEM A cross section through the basal body. The nine outer doublets of a cilium extend into a basal body where each doublet joins another microtubule to form a ring of nine triplets. Basal body (centriole) Figure 3. 26
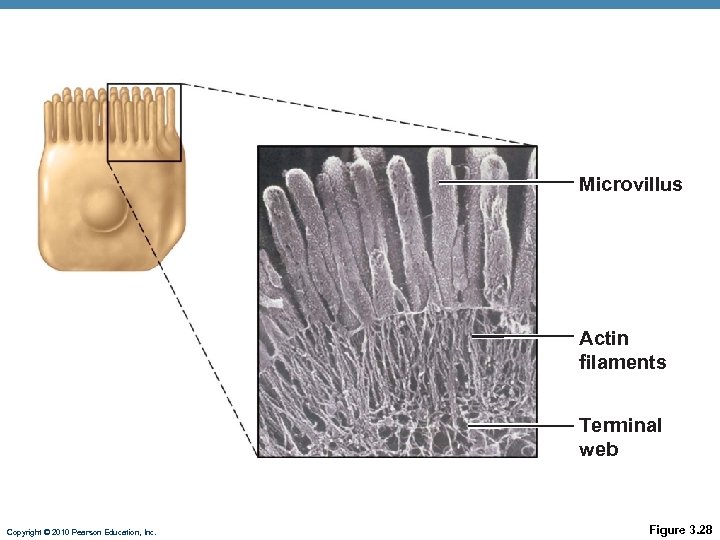
Microvillus Actin filaments Terminal web Copyright © 2010 Pearson Education, Inc. Figure 3. 28
Nucleus • Genetic library with blueprints for nearly all cellular proteins • Responds to signals; dictates kinds and amounts of proteins to be synthesized • Most cells: 1 • RBCs: 0 • Skeletal muscle cells, osteoclasts, some hepatocytes: >1. • Double-membrane barrier containing pores which regulate transport of large molecules in and out Copyright © 2010 Pearson Education, Inc.
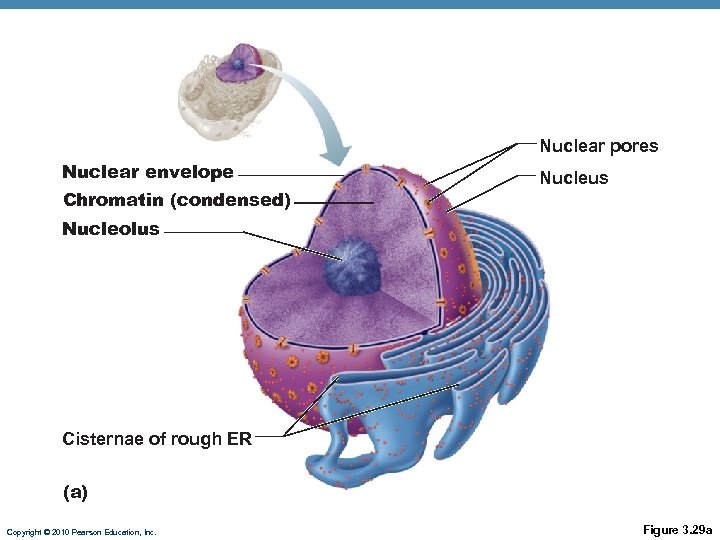
Nuclear pores Nuclear envelope Chromatin (condensed) Nucleus Nucleolus Cisternae of rough ER (a) Copyright © 2010 Pearson Education, Inc. Figure 3. 29 a
Nucleoli • Dark-staining spherical bodies within nucleus • Involved in r. RNA synthesis, ribosome subunit assembly Chromatin • Threadlike strands of DNA (30%), histone proteins (60%), RNA (10%) • Arranged in fundamental units called nucleosomes (8 histones plus 2 wraps of DNA) • Condenses into chromosomes during cell division Copyright © 2010 Pearson Education, Inc.
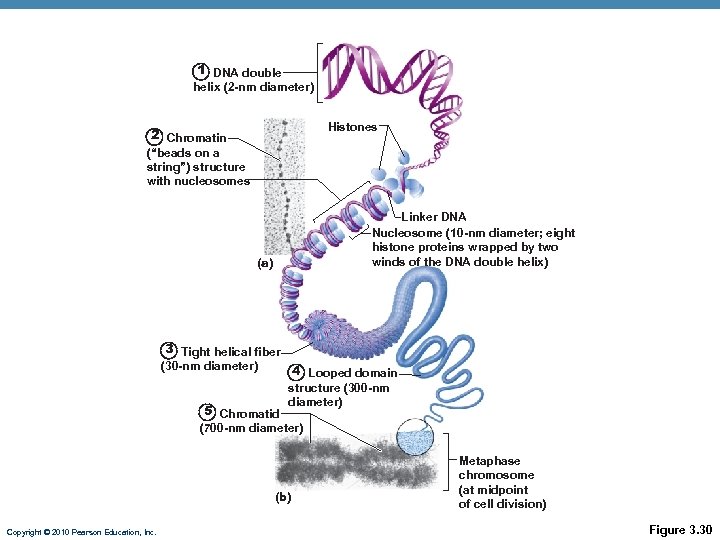
1 DNA double helix (2 -nm diameter) Histones 2 Chromatin (“beads on a string”) structure with nucleosomes Linker DNA Nucleosome (10 -nm diameter; eight histone proteins wrapped by two winds of the DNA double helix) (a) 3 Tight helical fiber (30 -nm diameter) 4 Looped domain 5 Chromatid structure (300 -nm diameter) (700 -nm diameter) (b) Copyright © 2010 Pearson Education, Inc. Metaphase chromosome (at midpoint of cell division) Figure 3. 30
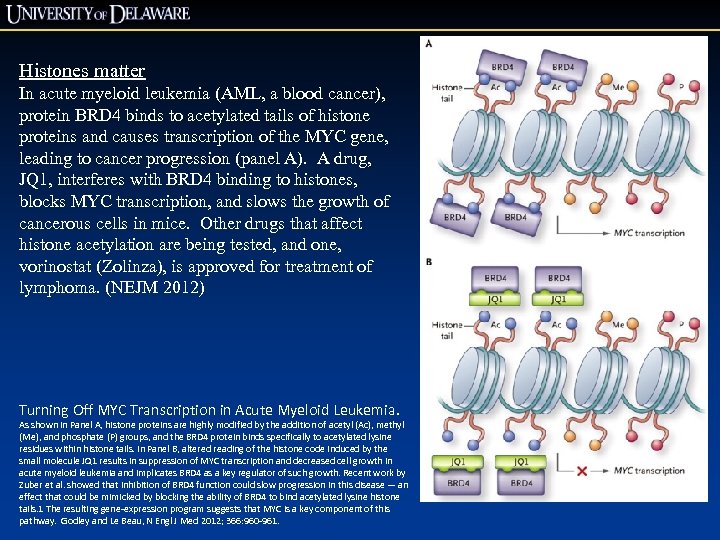
Histones matter In acute myeloid leukemia (AML, a blood cancer), protein BRD 4 binds to acetylated tails of histone proteins and causes transcription of the MYC gene, leading to cancer progression (panel A). A drug, JQ 1, interferes with BRD 4 binding to histones, blocks MYC transcription, and slows the growth of cancerous cells in mice. Other drugs that affect histone acetylation are being tested, and one, vorinostat (Zolinza), is approved for treatment of lymphoma. (NEJM 2012) Turning Off MYC Transcription in Acute Myeloid Leukemia. As shown in Panel A, histone proteins are highly modified by the addition of acetyl (Ac), methyl (Me), and phosphate (P) groups, and the BRD 4 protein binds specifically to acetylated lysine residues within histone tails. In Panel B, altered reading of the histone code induced by the small molecule JQ 1 results in suppression of MYC transcription and decreased cell growth in acute myeloid leukemia and implicates BRD 4 as a key regulator of such growth. Recent work by Zuber et al. showed that inhibition of BRD 4 function could slow progression in this disease — an effect that could be mimicked by blocking the ability of BRD 4 to bind acetylated lysine histone tails. 1 The resulting gene-expression program suggests that MYC is a key component of this pathway. Godley and Le Beau, N Engl J Med 2012; 366: 960 -961.
c0f69d3ba4dc36bb9e34fe1d82ee1d5e.ppt