c1467cfdb06262c314f75a2705432cca.ppt
- Количество слайдов: 41
 Chapter 24 Nucleotides, Nucleic Acids, and Heredity
Chapter 24 Nucleotides, Nucleic Acids, and Heredity
 The Molecules of Heredity • Each cell has thousands of different proteins. • How do cells know which proteins to synthesize out of 100000 s possible amino acid sequences? • From the end of the 19 th century, biologists suspected that the transmission of hereditary information took place in the nucleus, more specifically in structures called chromosomes. • The hereditary information was thought to reside in genes within the chromosomes. • Chemical analysis of nuclei showed chromosomes are made up largely of proteins called histones and nucleic acids
The Molecules of Heredity • Each cell has thousands of different proteins. • How do cells know which proteins to synthesize out of 100000 s possible amino acid sequences? • From the end of the 19 th century, biologists suspected that the transmission of hereditary information took place in the nucleus, more specifically in structures called chromosomes. • The hereditary information was thought to reside in genes within the chromosomes. • Chemical analysis of nuclei showed chromosomes are made up largely of proteins called histones and nucleic acids
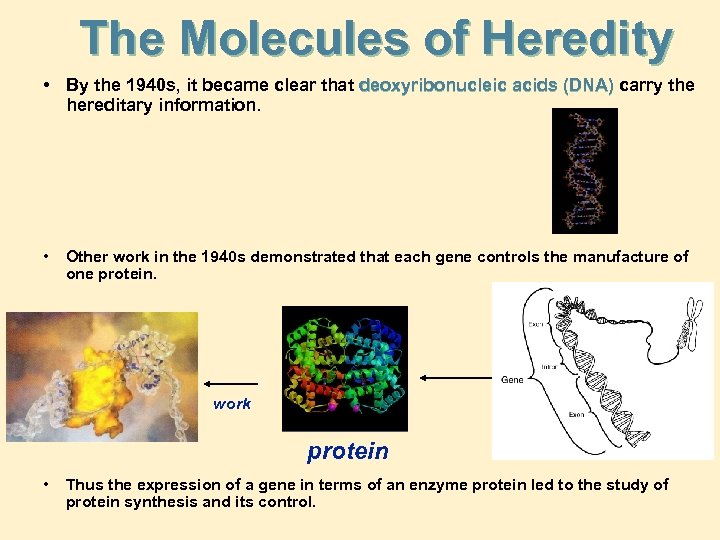 The Molecules of Heredity • By the 1940 s, it became clear that deoxyribonucleic acids (DNA) carry the hereditary information. • Other work in the 1940 s demonstrated that each gene controls the manufacture of one protein. work protein • Thus the expression of a gene in terms of an enzyme protein led to the study of protein synthesis and its control.
The Molecules of Heredity • By the 1940 s, it became clear that deoxyribonucleic acids (DNA) carry the hereditary information. • Other work in the 1940 s demonstrated that each gene controls the manufacture of one protein. work protein • Thus the expression of a gene in terms of an enzyme protein led to the study of protein synthesis and its control.
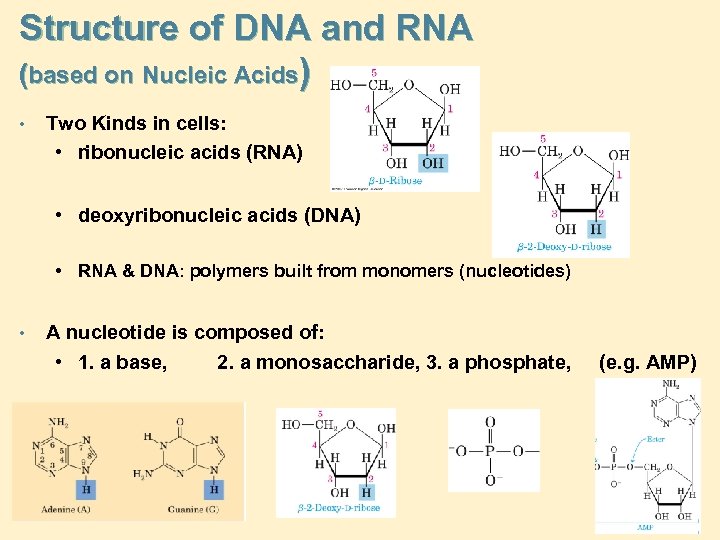 Structure of DNA and RNA (based on Nucleic Acids) • Two Kinds in cells: • ribonucleic acids (RNA) • deoxyribonucleic acids (DNA) • RNA & DNA: polymers built from monomers (nucleotides) • A nucleotide is composed of: • 1. a base, 2. a monosaccharide, 3. a phosphate, (e. g. AMP)
Structure of DNA and RNA (based on Nucleic Acids) • Two Kinds in cells: • ribonucleic acids (RNA) • deoxyribonucleic acids (DNA) • RNA & DNA: polymers built from monomers (nucleotides) • A nucleotide is composed of: • 1. a base, 2. a monosaccharide, 3. a phosphate, (e. g. AMP)
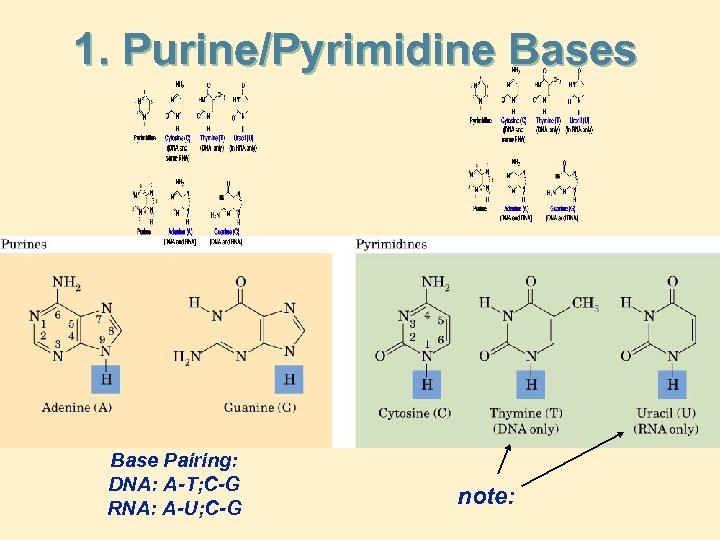 1. Purine/Pyrimidine Bases Base Pairing: DNA: A-T; C-G RNA: A-U; C-G note:
1. Purine/Pyrimidine Bases Base Pairing: DNA: A-T; C-G RNA: A-U; C-G note:
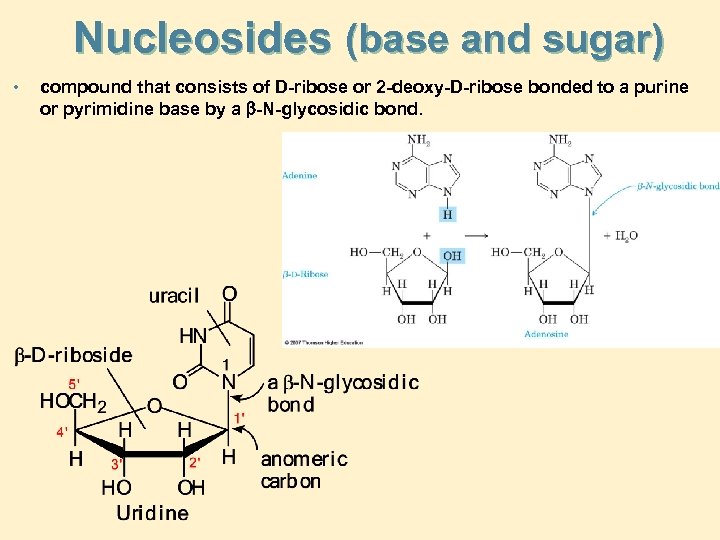 Nucleosides (base and sugar) • compound that consists of D-ribose or 2 -deoxy-D-ribose bonded to a purine or pyrimidine base by a -N-glycosidic bond.
Nucleosides (base and sugar) • compound that consists of D-ribose or 2 -deoxy-D-ribose bonded to a purine or pyrimidine base by a -N-glycosidic bond.
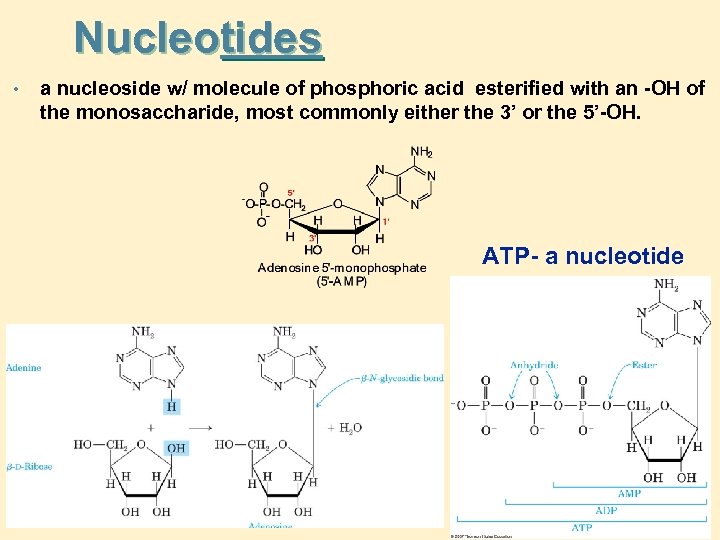 Nucleotides • a nucleoside w/ molecule of phosphoric acid esterified with an -OH of the monosaccharide, most commonly either the 3’ or the 5’-OH. ATP- a nucleotide
Nucleotides • a nucleoside w/ molecule of phosphoric acid esterified with an -OH of the monosaccharide, most commonly either the 3’ or the 5’-OH. ATP- a nucleotide
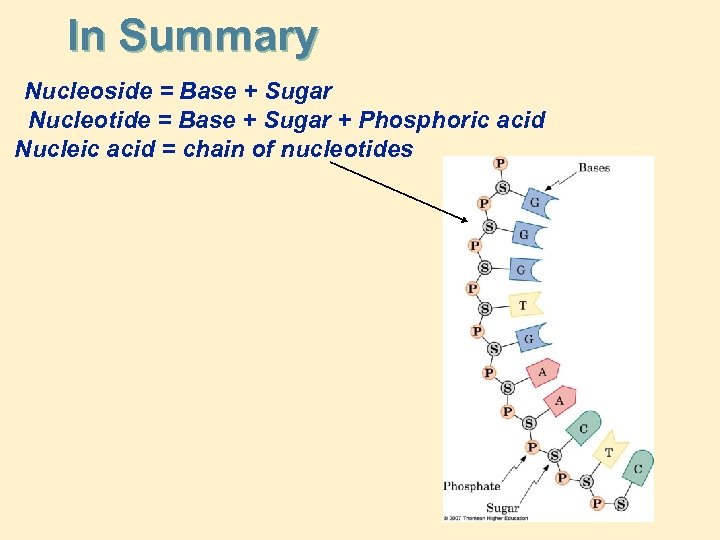 In Summary Nucleoside = Base + Sugar Nucleotide = Base + Sugar + Phosphoric acid Nucleic acid = chain of nucleotides
In Summary Nucleoside = Base + Sugar Nucleotide = Base + Sugar + Phosphoric acid Nucleic acid = chain of nucleotides
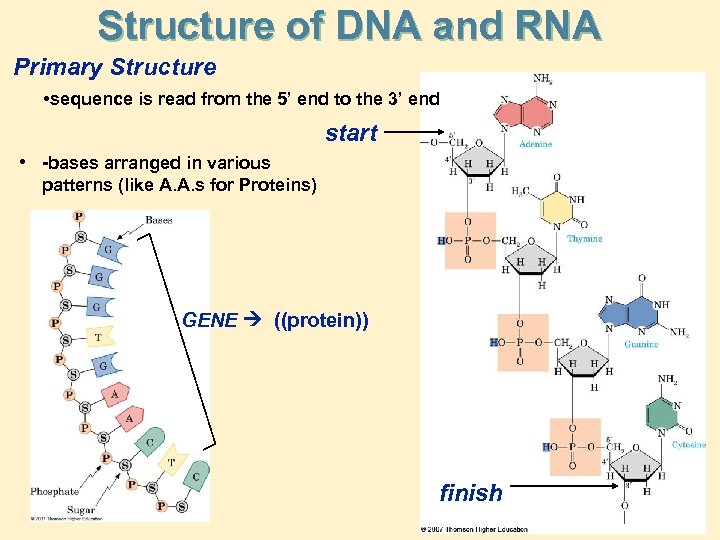 Structure of DNA and RNA Primary Structure • sequence is read from the 5’ end to the 3’ end start • -bases arranged in various patterns (like A. A. s for Proteins) GENE ((protein)) finish
Structure of DNA and RNA Primary Structure • sequence is read from the 5’ end to the 3’ end start • -bases arranged in various patterns (like A. A. s for Proteins) GENE ((protein)) finish
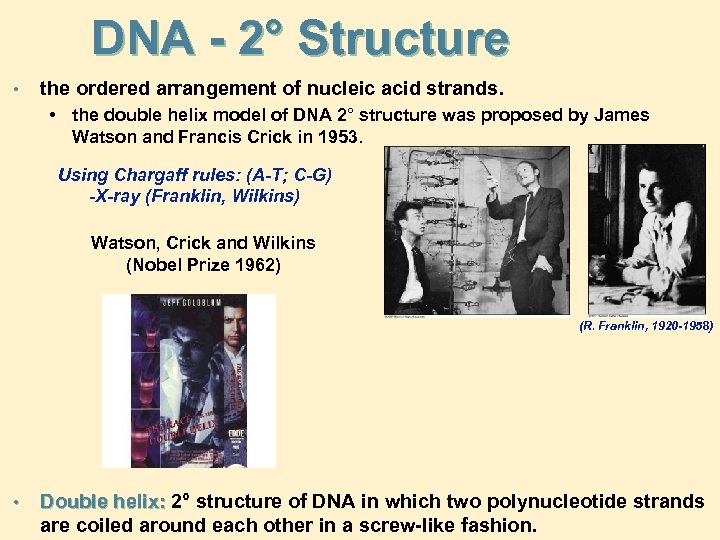 DNA - 2° Structure • the ordered arrangement of nucleic acid strands. • the double helix model of DNA 2° structure was proposed by James Watson and Francis Crick in 1953. Using Chargaff rules: (A-T; C-G) -X-ray (Franklin, Wilkins) Watson, Crick and Wilkins (Nobel Prize 1962) (R. Franklin, 1920 -1958) • Double helix: 2° structure of DNA in which two polynucleotide strands are coiled around each other in a screw-like fashion.
DNA - 2° Structure • the ordered arrangement of nucleic acid strands. • the double helix model of DNA 2° structure was proposed by James Watson and Francis Crick in 1953. Using Chargaff rules: (A-T; C-G) -X-ray (Franklin, Wilkins) Watson, Crick and Wilkins (Nobel Prize 1962) (R. Franklin, 1920 -1958) • Double helix: 2° structure of DNA in which two polynucleotide strands are coiled around each other in a screw-like fashion.
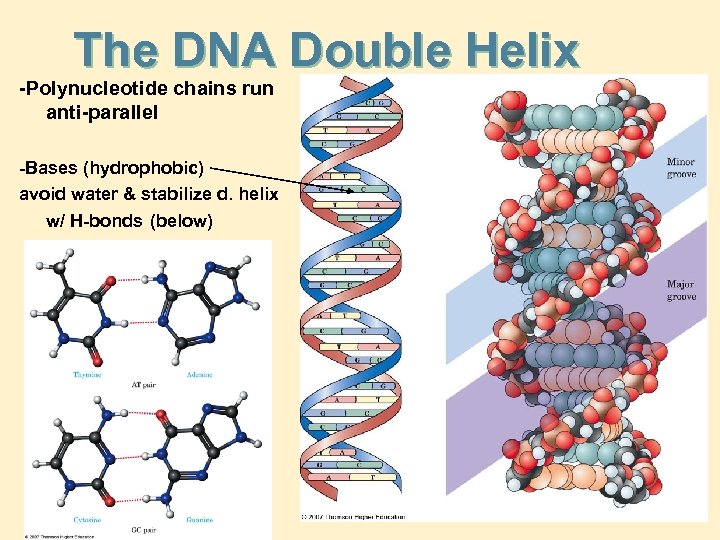 The DNA Double Helix -Polynucleotide chains run anti-parallel -Bases (hydrophobic) avoid water & stabilize d. helix w/ H-bonds (below)
The DNA Double Helix -Polynucleotide chains run anti-parallel -Bases (hydrophobic) avoid water & stabilize d. helix w/ H-bonds (below)
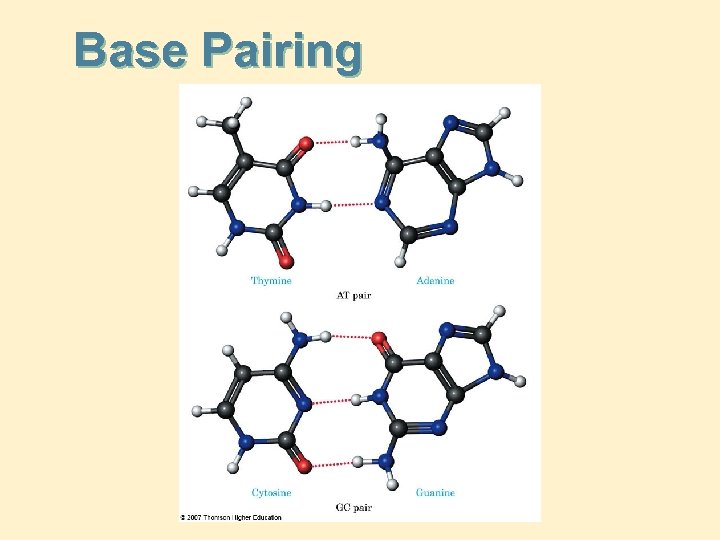 Base Pairing
Base Pairing
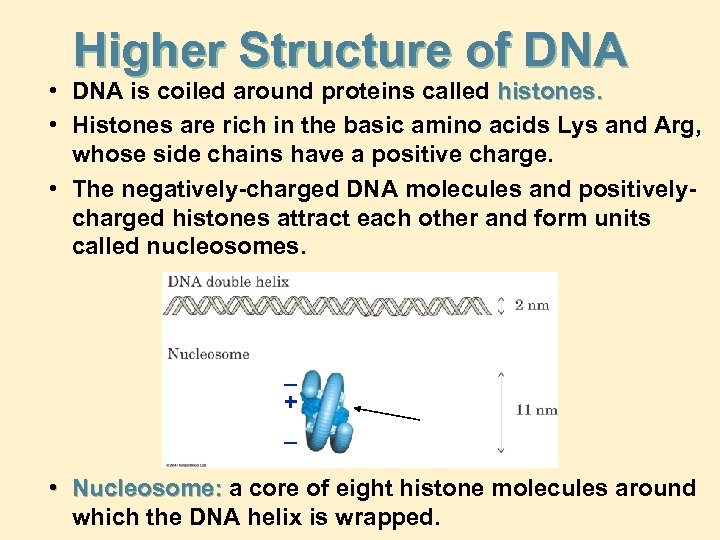 Higher Structure of DNA • DNA is coiled around proteins called histones. • Histones are rich in the basic amino acids Lys and Arg, whose side chains have a positive charge. • The negatively-charged DNA molecules and positivelycharged histones attract each other and form units called nucleosomes. _ + _ • Nucleosome: a core of eight histone molecules around which the DNA helix is wrapped.
Higher Structure of DNA • DNA is coiled around proteins called histones. • Histones are rich in the basic amino acids Lys and Arg, whose side chains have a positive charge. • The negatively-charged DNA molecules and positivelycharged histones attract each other and form units called nucleosomes. _ + _ • Nucleosome: a core of eight histone molecules around which the DNA helix is wrapped.
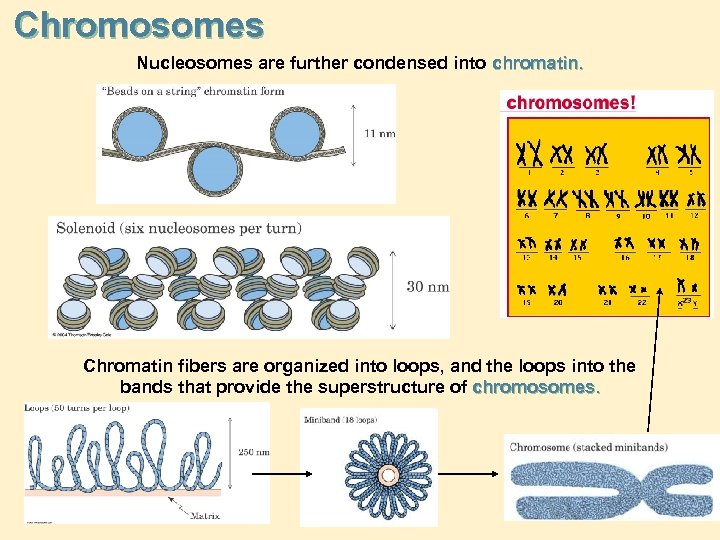 Chromosomes Nucleosomes are further condensed into chromatin. Chromatin fibers are organized into loops, and the loops into the bands that provide the superstructure of chromosomes.
Chromosomes Nucleosomes are further condensed into chromatin. Chromatin fibers are organized into loops, and the loops into the bands that provide the superstructure of chromosomes.
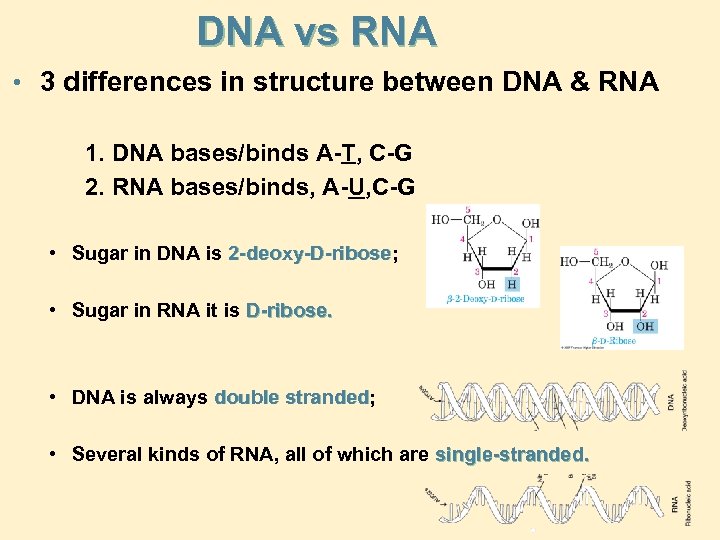 DNA vs RNA • 3 differences in structure between DNA & RNA 1. DNA bases/binds A-T, C-G 2. RNA bases/binds, A-U, C-G • Sugar in DNA is 2 -deoxy-D-ribose; 2 -deoxy-D-ribose • Sugar in RNA it is D-ribose. • DNA is always double stranded; stranded • Several kinds of RNA, all of which are single-stranded.
DNA vs RNA • 3 differences in structure between DNA & RNA 1. DNA bases/binds A-T, C-G 2. RNA bases/binds, A-U, C-G • Sugar in DNA is 2 -deoxy-D-ribose; 2 -deoxy-D-ribose • Sugar in RNA it is D-ribose. • DNA is always double stranded; stranded • Several kinds of RNA, all of which are single-stranded.
 RNA- 4 types: 1. Messenger RNA (m. RNA) -Carries genetic info f/ DNA to ribosome -Acts as template for protein synthesis 2. Transfer RNA (t. RNA) -RNA that transports amino acids to site of protein synthesis (ribosomes) 3. Ribosomal RNA (RNA) -RNA complexed with proteins in ribosomes 4. Ribozymes -Catalytic RNA, with special enzyme functions (e. g. splicing)
RNA- 4 types: 1. Messenger RNA (m. RNA) -Carries genetic info f/ DNA to ribosome -Acts as template for protein synthesis 2. Transfer RNA (t. RNA) -RNA that transports amino acids to site of protein synthesis (ribosomes) 3. Ribosomal RNA (RNA) -RNA complexed with proteins in ribosomes 4. Ribozymes -Catalytic RNA, with special enzyme functions (e. g. splicing)
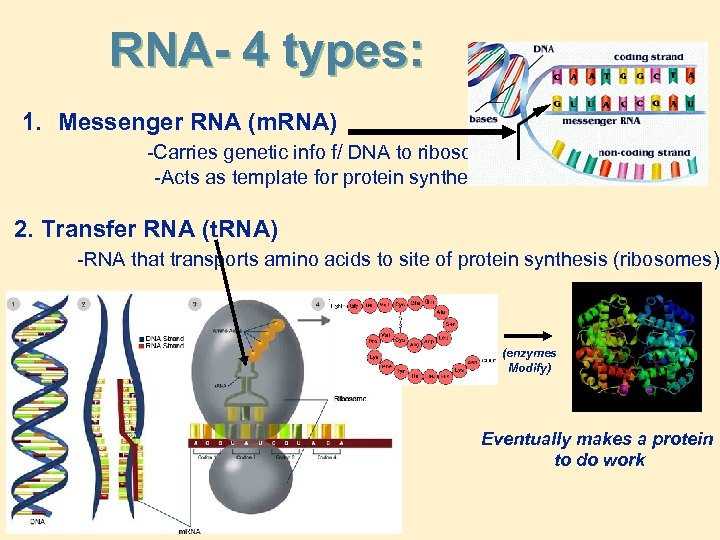 RNA- 4 types: 1. Messenger RNA (m. RNA) -Carries genetic info f/ DNA to ribosome -Acts as template for protein synthesis 2. Transfer RNA (t. RNA) -RNA that transports amino acids to site of protein synthesis (ribosomes) (enzymes Modify) Eventually makes a protein to do work
RNA- 4 types: 1. Messenger RNA (m. RNA) -Carries genetic info f/ DNA to ribosome -Acts as template for protein synthesis 2. Transfer RNA (t. RNA) -RNA that transports amino acids to site of protein synthesis (ribosomes) (enzymes Modify) Eventually makes a protein to do work
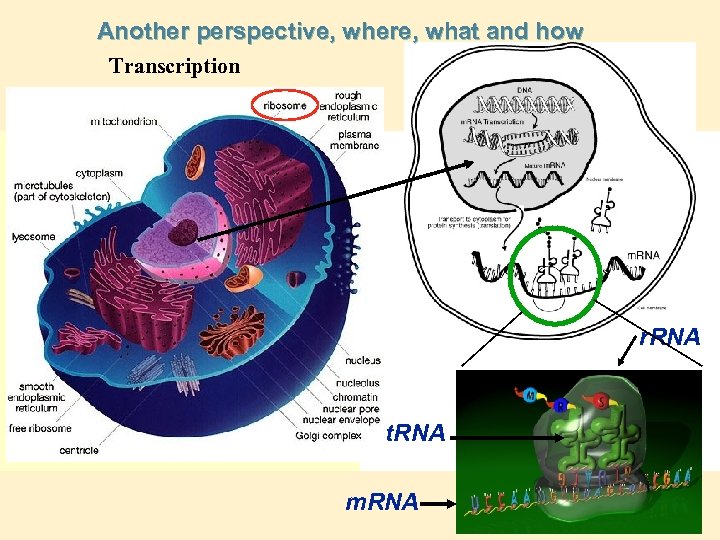 Another perspective, where, what and how Transcription r. RNA t. RNA m. RNA
Another perspective, where, what and how Transcription r. RNA t. RNA m. RNA
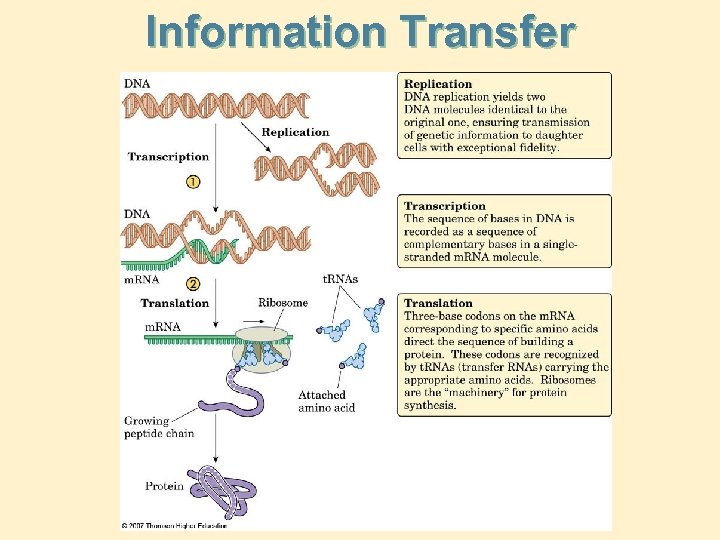 Information Transfer
Information Transfer
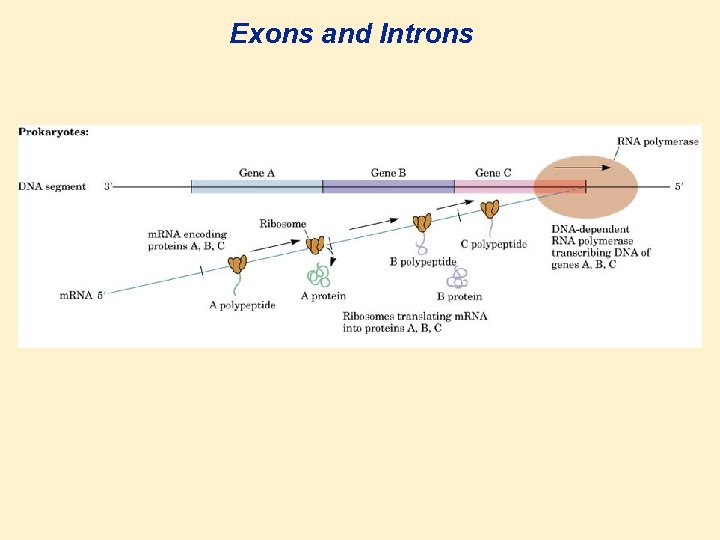
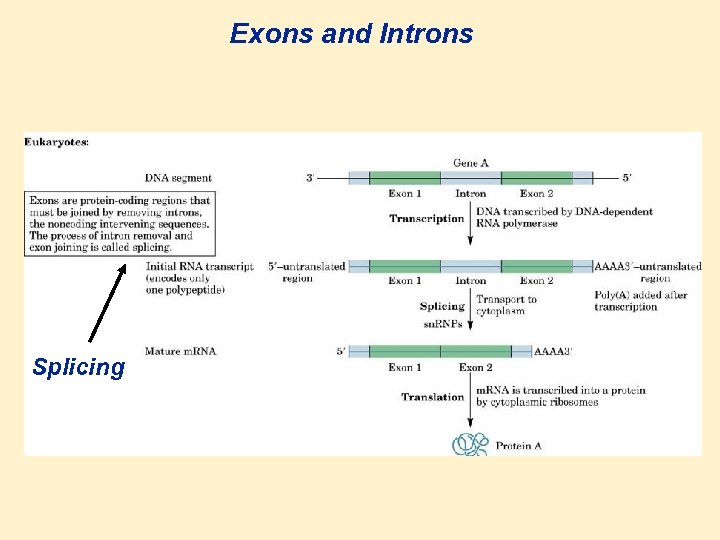
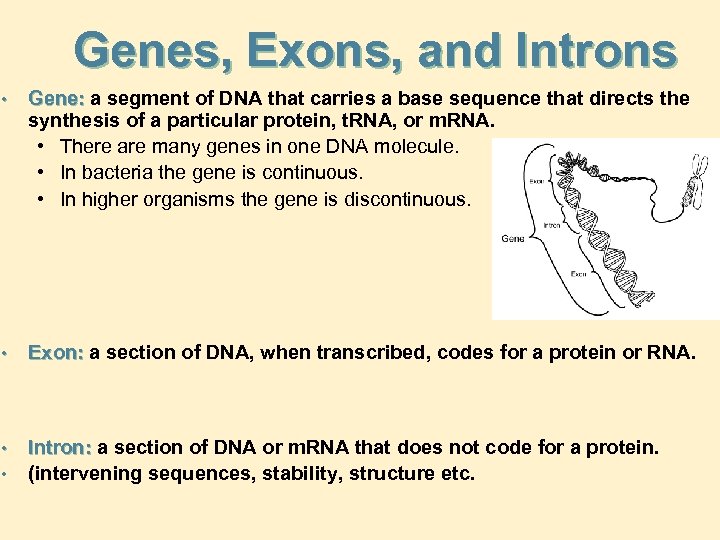 Genes, Exons, and Introns • Gene: a segment of DNA that carries a base sequence that directs the synthesis of a particular protein, t. RNA, or m. RNA. • There are many genes in one DNA molecule. • In bacteria the gene is continuous. • In higher organisms the gene is discontinuous. • Exon: a section of DNA, when transcribed, codes for a protein or RNA. • • Intron: a section of DNA or m. RNA that does not code for a protein. (intervening sequences, stability, structure etc.
Genes, Exons, and Introns • Gene: a segment of DNA that carries a base sequence that directs the synthesis of a particular protein, t. RNA, or m. RNA. • There are many genes in one DNA molecule. • In bacteria the gene is continuous. • In higher organisms the gene is discontinuous. • Exon: a section of DNA, when transcribed, codes for a protein or RNA. • • Intron: a section of DNA or m. RNA that does not code for a protein. (intervening sequences, stability, structure etc.
 Exons and Introns
Exons and Introns
 Exons and Introns Splicing
Exons and Introns Splicing
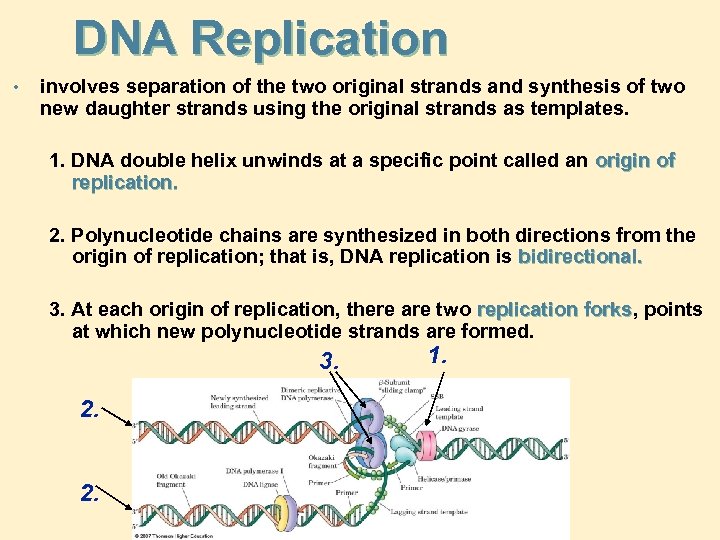 DNA Replication • involves separation of the two original strands and synthesis of two new daughter strands using the original strands as templates. 1. DNA double helix unwinds at a specific point called an origin of replication. 2. Polynucleotide chains are synthesized in both directions from the origin of replication; that is, DNA replication is bidirectional. 3. At each origin of replication, there are two replication forks, points forks at which new polynucleotide strands are formed. 3. 2. 2. 1.
DNA Replication • involves separation of the two original strands and synthesis of two new daughter strands using the original strands as templates. 1. DNA double helix unwinds at a specific point called an origin of replication. 2. Polynucleotide chains are synthesized in both directions from the origin of replication; that is, DNA replication is bidirectional. 3. At each origin of replication, there are two replication forks, points forks at which new polynucleotide strands are formed. 3. 2. 2. 1.
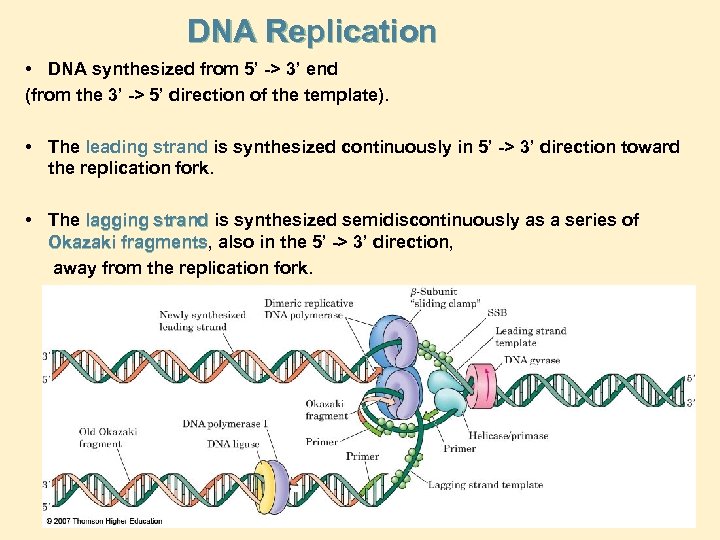 DNA Replication • DNA synthesized from 5’ -> 3’ end (from the 3’ -> 5’ direction of the template). • The leading strand is synthesized continuously in 5’ -> 3’ direction toward the replication fork. • The lagging strand is synthesized semidiscontinuously as a series of Okazaki fragments, also in the 5’ -> 3’ direction, fragments away from the replication fork.
DNA Replication • DNA synthesized from 5’ -> 3’ end (from the 3’ -> 5’ direction of the template). • The leading strand is synthesized continuously in 5’ -> 3’ direction toward the replication fork. • The lagging strand is synthesized semidiscontinuously as a series of Okazaki fragments, also in the 5’ -> 3’ direction, fragments away from the replication fork.
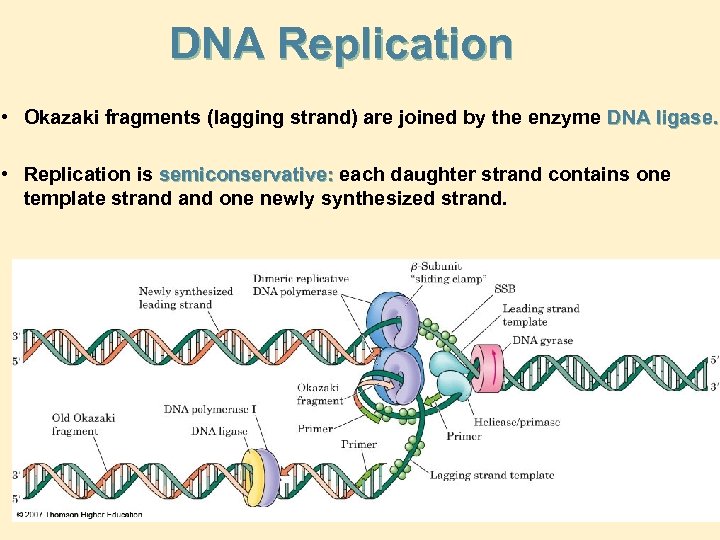 DNA Replication • Okazaki fragments (lagging strand) are joined by the enzyme DNA ligase. • Replication is semiconservative: each daughter strand contains one template strand one newly synthesized strand.
DNA Replication • Okazaki fragments (lagging strand) are joined by the enzyme DNA ligase. • Replication is semiconservative: each daughter strand contains one template strand one newly synthesized strand.
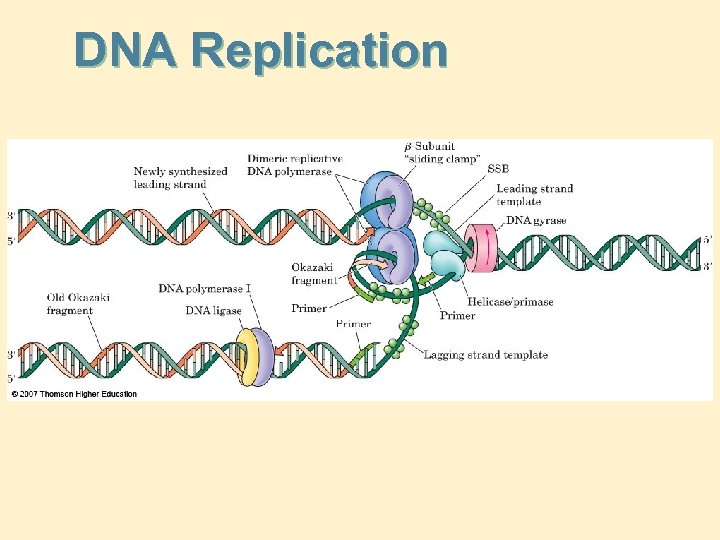 DNA Replication
DNA Replication
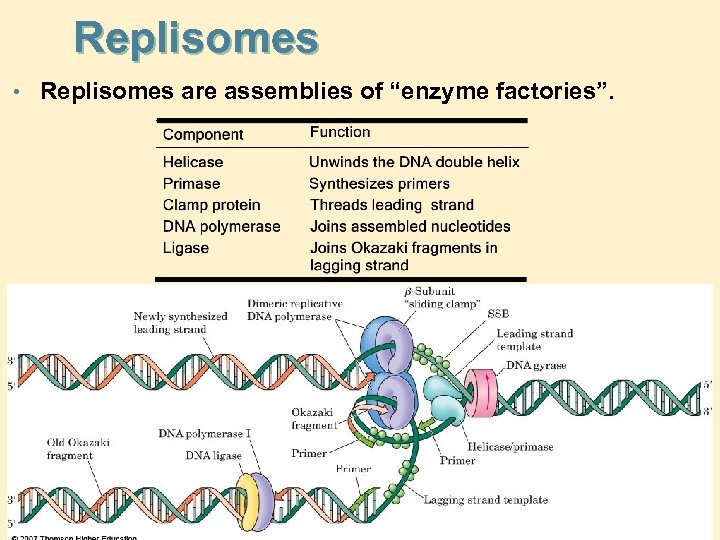 Replisomes • Replisomes are assemblies of “enzyme factories”.
Replisomes • Replisomes are assemblies of “enzyme factories”.
 Telomers: TTAGGG Somatic cells vs Stem Cells, fetal cells and cancer cells (immortal) Stem cells have telomerase enzyme confers immortality to cells Possible organ regeneration etc.
Telomers: TTAGGG Somatic cells vs Stem Cells, fetal cells and cancer cells (immortal) Stem cells have telomerase enzyme confers immortality to cells Possible organ regeneration etc.
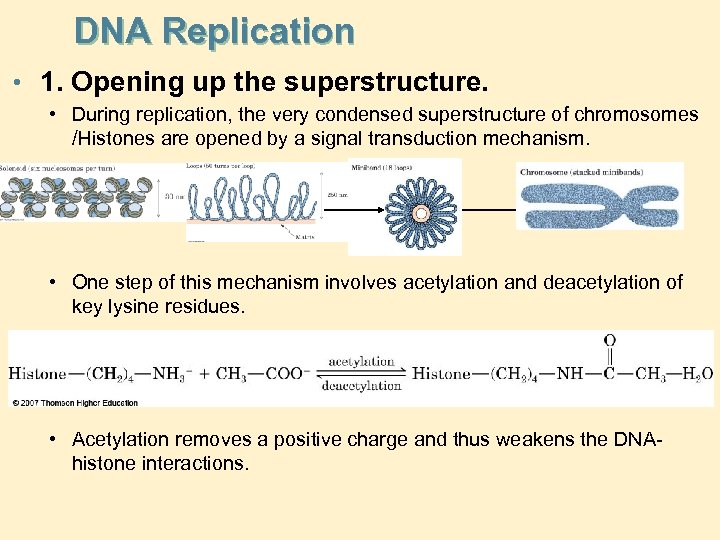 DNA Replication • 1. Opening up the superstructure. • During replication, the very condensed superstructure of chromosomes /Histones are opened by a signal transduction mechanism. • One step of this mechanism involves acetylation and deacetylation of key lysine residues. • Acetylation removes a positive charge and thus weakens the DNAhistone interactions.
DNA Replication • 1. Opening up the superstructure. • During replication, the very condensed superstructure of chromosomes /Histones are opened by a signal transduction mechanism. • One step of this mechanism involves acetylation and deacetylation of key lysine residues. • Acetylation removes a positive charge and thus weakens the DNAhistone interactions.
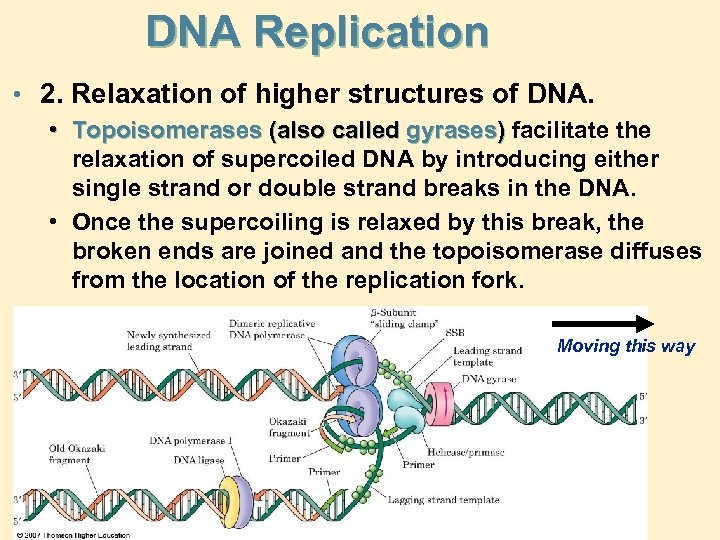 DNA Replication • 2. Relaxation of higher structures of DNA. • Topoisomerases (also called gyrases) facilitate the relaxation of supercoiled DNA by introducing either single strand or double strand breaks in the DNA. • Once the supercoiling is relaxed by this break, the broken ends are joined and the topoisomerase diffuses from the location of the replication fork. Moving this way
DNA Replication • 2. Relaxation of higher structures of DNA. • Topoisomerases (also called gyrases) facilitate the relaxation of supercoiled DNA by introducing either single strand or double strand breaks in the DNA. • Once the supercoiling is relaxed by this break, the broken ends are joined and the topoisomerase diffuses from the location of the replication fork. Moving this way
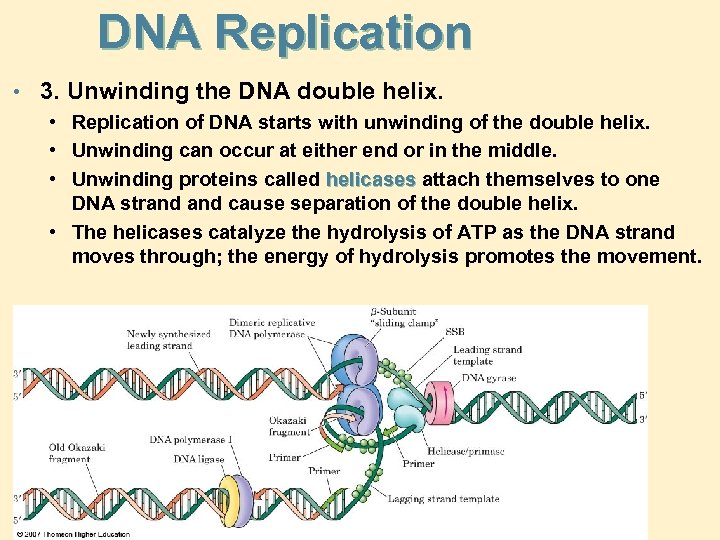 DNA Replication • 3. • • • Unwinding the DNA double helix. Replication of DNA starts with unwinding of the double helix. Unwinding can occur at either end or in the middle. Unwinding proteins called helicases attach themselves to one DNA strand cause separation of the double helix. • The helicases catalyze the hydrolysis of ATP as the DNA strand moves through; the energy of hydrolysis promotes the movement.
DNA Replication • 3. • • • Unwinding the DNA double helix. Replication of DNA starts with unwinding of the double helix. Unwinding can occur at either end or in the middle. Unwinding proteins called helicases attach themselves to one DNA strand cause separation of the double helix. • The helicases catalyze the hydrolysis of ATP as the DNA strand moves through; the energy of hydrolysis promotes the movement.
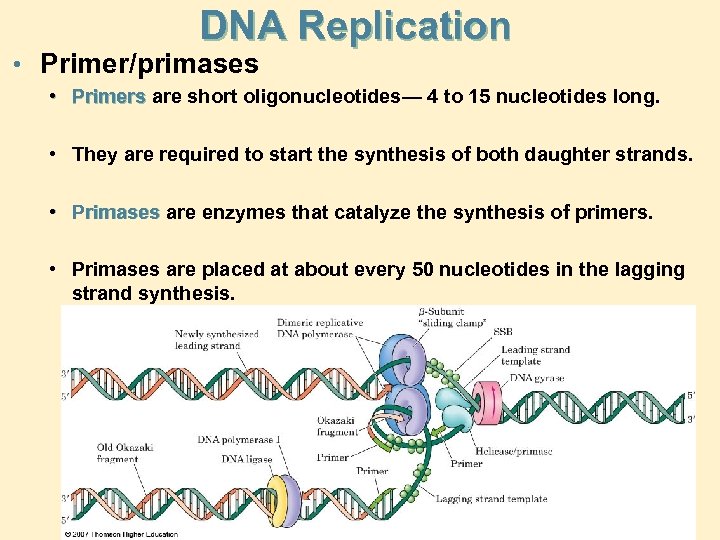 DNA Replication • Primer/primases • Primers are short oligonucleotides— 4 to 15 nucleotides long. • They are required to start the synthesis of both daughter strands. • Primases are enzymes that catalyze the synthesis of primers. • Primases are placed at about every 50 nucleotides in the lagging strand synthesis.
DNA Replication • Primer/primases • Primers are short oligonucleotides— 4 to 15 nucleotides long. • They are required to start the synthesis of both daughter strands. • Primases are enzymes that catalyze the synthesis of primers. • Primases are placed at about every 50 nucleotides in the lagging strand synthesis.
 DNA Replication • DNA polymerases are key enzymes in replication. • Once the two strands have separated at the replication fork, the nucleotides must be lined up in proper order for DNA synthesis. • In the absence of DNA polymerase, alignment is slow. • DNA polymerase provides the speed and specificity of alignment. • Along lagging (3’ -> 5’) strand, polymerases can synthesize only short fragments, because these enzymes only work from 5’ -> 3’. • These short fragments are called Okazaki fragments. • Joining the Okazaki fragments and any remaining nicks is catalyzed by DNA ligase.
DNA Replication • DNA polymerases are key enzymes in replication. • Once the two strands have separated at the replication fork, the nucleotides must be lined up in proper order for DNA synthesis. • In the absence of DNA polymerase, alignment is slow. • DNA polymerase provides the speed and specificity of alignment. • Along lagging (3’ -> 5’) strand, polymerases can synthesize only short fragments, because these enzymes only work from 5’ -> 3’. • These short fragments are called Okazaki fragments. • Joining the Okazaki fragments and any remaining nicks is catalyzed by DNA ligase.
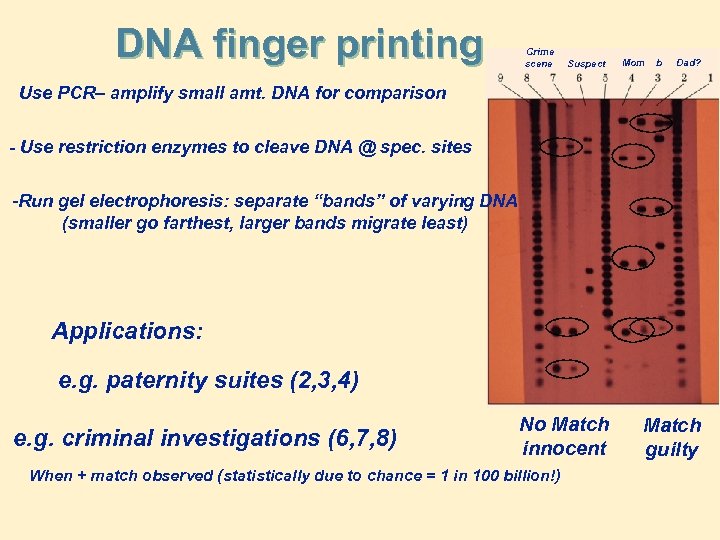 DNA finger printing Crime scene Suspect Mom b Dad? Use PCR– amplify small amt. DNA for comparison - Use restriction enzymes to cleave DNA @ spec. sites -Run gel electrophoresis: separate “bands” of varying DNA (smaller go farthest, larger bands migrate least) Applications: e. g. paternity suites (2, 3, 4) e. g. criminal investigations (6, 7, 8) No Match innocent When + match observed (statistically due to chance = 1 in 100 billion!) Match guilty
DNA finger printing Crime scene Suspect Mom b Dad? Use PCR– amplify small amt. DNA for comparison - Use restriction enzymes to cleave DNA @ spec. sites -Run gel electrophoresis: separate “bands” of varying DNA (smaller go farthest, larger bands migrate least) Applications: e. g. paternity suites (2, 3, 4) e. g. criminal investigations (6, 7, 8) No Match innocent When + match observed (statistically due to chance = 1 in 100 billion!) Match guilty
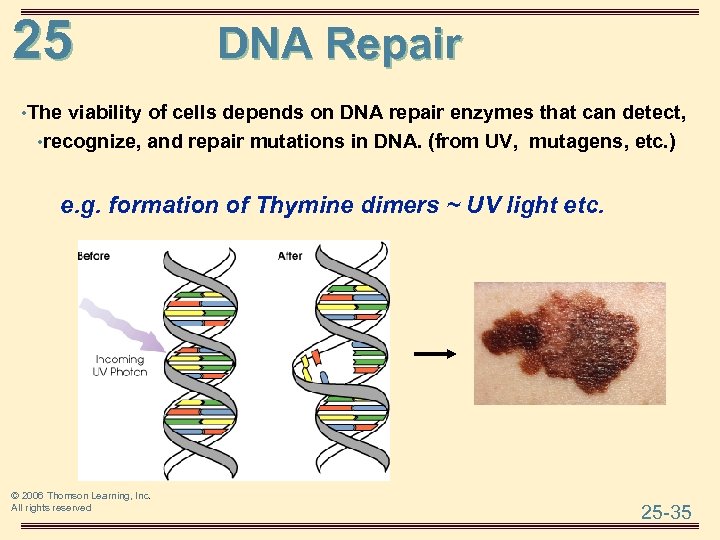 25 DNA Repair • The viability of cells depends on DNA repair enzymes that can detect, • recognize, and repair mutations in DNA. (from UV, mutagens, etc. ) e. g. formation of Thymine dimers ~ UV light etc. © 2006 Thomson Learning, Inc. All rights reserved 25 -35
25 DNA Repair • The viability of cells depends on DNA repair enzymes that can detect, • recognize, and repair mutations in DNA. (from UV, mutagens, etc. ) e. g. formation of Thymine dimers ~ UV light etc. © 2006 Thomson Learning, Inc. All rights reserved 25 -35
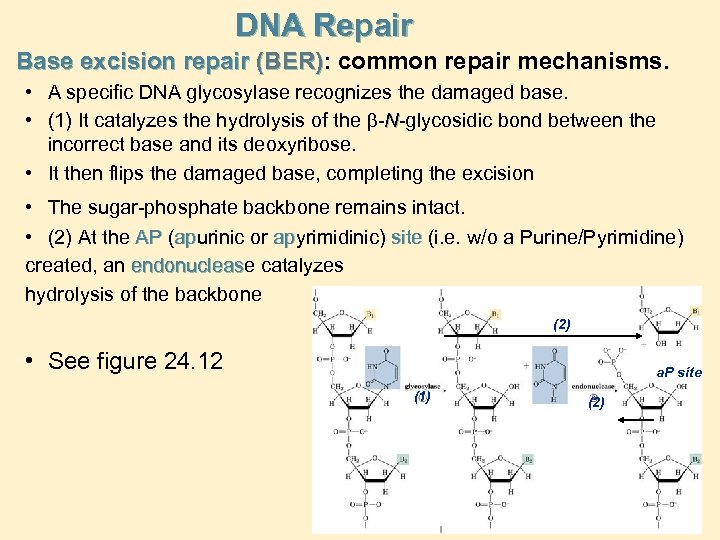 DNA Repair Base excision repair (BER): common repair mechanisms. (BER) • A specific DNA glycosylase recognizes the damaged base. • (1) It catalyzes the hydrolysis of the b-N-glycosidic bond between the incorrect base and its deoxyribose. • It then flips the damaged base, completing the excision • The sugar-phosphate backbone remains intact. • (2) At the AP (apurinic or apyrimidinic) site (i. e. w/o a Purine/Pyrimidine) ap ap created, an endonuclease catalyzes endonucleas hydrolysis of the backbone (2) • See figure 24. 12 a. P site (1) (2)
DNA Repair Base excision repair (BER): common repair mechanisms. (BER) • A specific DNA glycosylase recognizes the damaged base. • (1) It catalyzes the hydrolysis of the b-N-glycosidic bond between the incorrect base and its deoxyribose. • It then flips the damaged base, completing the excision • The sugar-phosphate backbone remains intact. • (2) At the AP (apurinic or apyrimidinic) site (i. e. w/o a Purine/Pyrimidine) ap ap created, an endonuclease catalyzes endonucleas hydrolysis of the backbone (2) • See figure 24. 12 a. P site (1) (2)
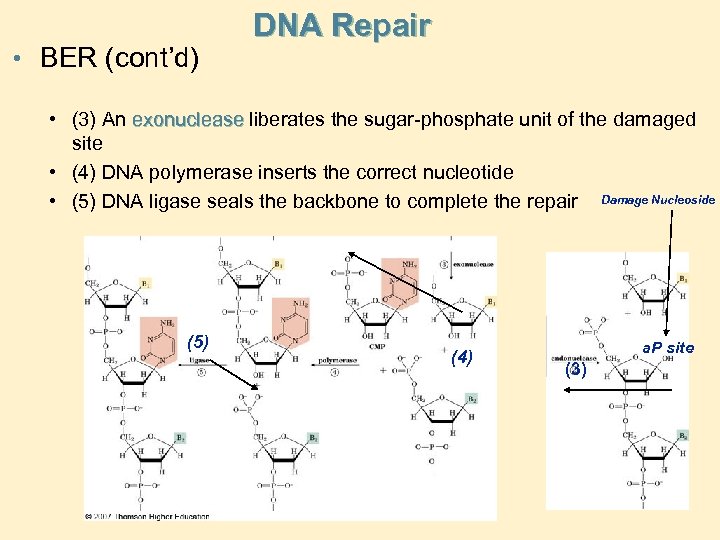 • BER (cont’d) DNA Repair • (3) An exonuclease liberates the sugar-phosphate unit of the damaged site • (4) DNA polymerase inserts the correct nucleotide • (5) DNA ligase seals the backbone to complete the repair Damage Nucleoside (5) (4) a. P site (3)
• BER (cont’d) DNA Repair • (3) An exonuclease liberates the sugar-phosphate unit of the damaged site • (4) DNA polymerase inserts the correct nucleotide • (5) DNA ligase seals the backbone to complete the repair Damage Nucleoside (5) (4) a. P site (3)
 25 DNA Repair (cont. ) NER (nucleotide excision repair) removes and repairs • up to 24 -32 units by a similar mechanism • involving a number of repair enzymes © 2006 Thomson Learning, Inc. All rights reserved 25 -38
25 DNA Repair (cont. ) NER (nucleotide excision repair) removes and repairs • up to 24 -32 units by a similar mechanism • involving a number of repair enzymes © 2006 Thomson Learning, Inc. All rights reserved 25 -38
 Cloning • Clone: a genetically identical population. • Cloning: a process whereby DNA is amplified by inserting it into a host and having the host replicate it along with the host’s own DNA. • Polymerase chain reaction (PCR): an automated technique for amplifying DNA using a heat-stable DNA polymerase from a thermophilic bacterium. Steps: • • 1. Heat (95*C), unwinds DNA, add primer, cool (70*C) replicates 1 st strand. . Etc. . Repeat step 1. • See next slide
Cloning • Clone: a genetically identical population. • Cloning: a process whereby DNA is amplified by inserting it into a host and having the host replicate it along with the host’s own DNA. • Polymerase chain reaction (PCR): an automated technique for amplifying DNA using a heat-stable DNA polymerase from a thermophilic bacterium. Steps: • • 1. Heat (95*C), unwinds DNA, add primer, cool (70*C) replicates 1 st strand. . Etc. . Repeat step 1. • See next slide
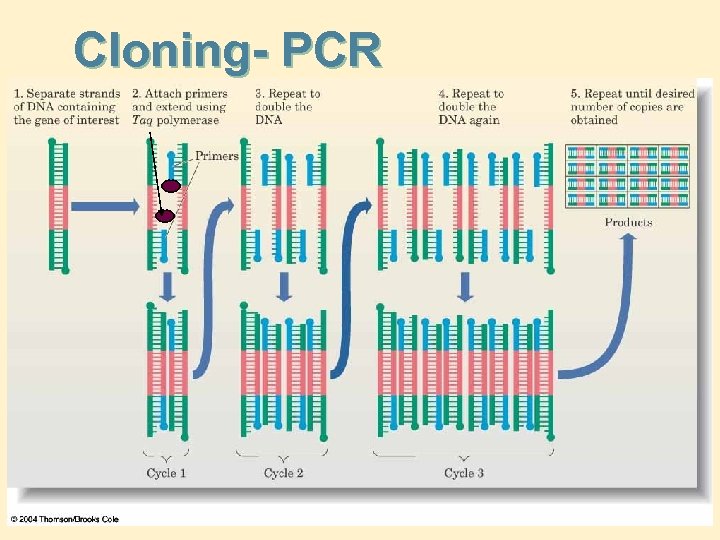 Cloning- PCR
Cloning- PCR
 from Nucleotides, and Nucleic Acids, to cloned genetics
from Nucleotides, and Nucleic Acids, to cloned genetics


