7fa153b383bfd956a107d55406ab79ef.ppt
- Количество слайдов: 64
 Chapter 2 - Fundamental Technologies • Eukaryotic gene organization • Molecular cloning • Genomic and c. DNA libraries • Polymerase chain reaction (PCR) • Chemical synthesis of DNA • DNA sequencing technologies • Sequencing whole genomes • Genomics (and other –omics) • Genome engineering using CRISPR Technology
Chapter 2 - Fundamental Technologies • Eukaryotic gene organization • Molecular cloning • Genomic and c. DNA libraries • Polymerase chain reaction (PCR) • Chemical synthesis of DNA • DNA sequencing technologies • Sequencing whole genomes • Genomics (and other –omics) • Genome engineering using CRISPR Technology
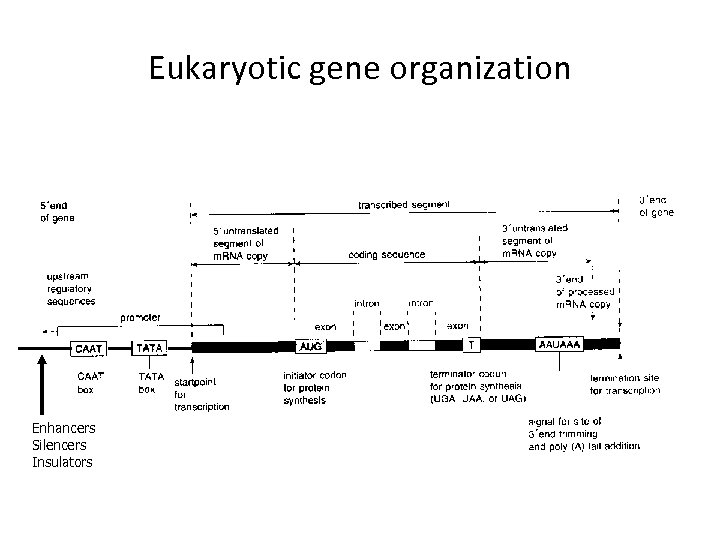 Eukaryotic gene organization Enhancers Silencers Insulators
Eukaryotic gene organization Enhancers Silencers Insulators
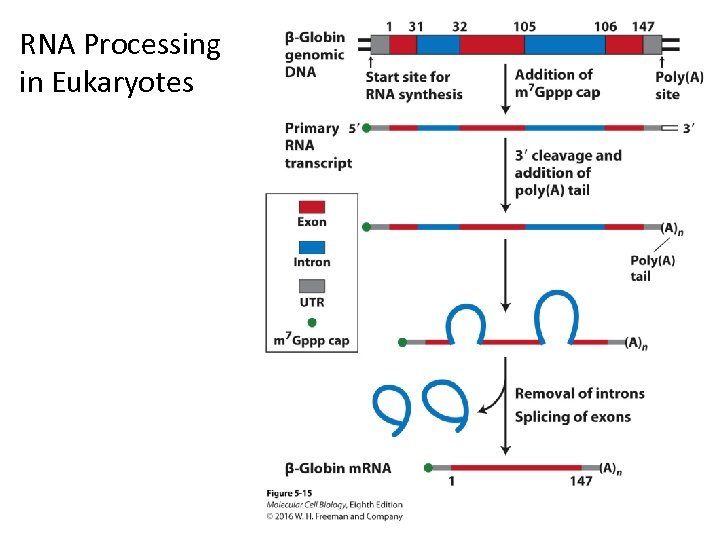 RNA Processing in Eukaryotes
RNA Processing in Eukaryotes
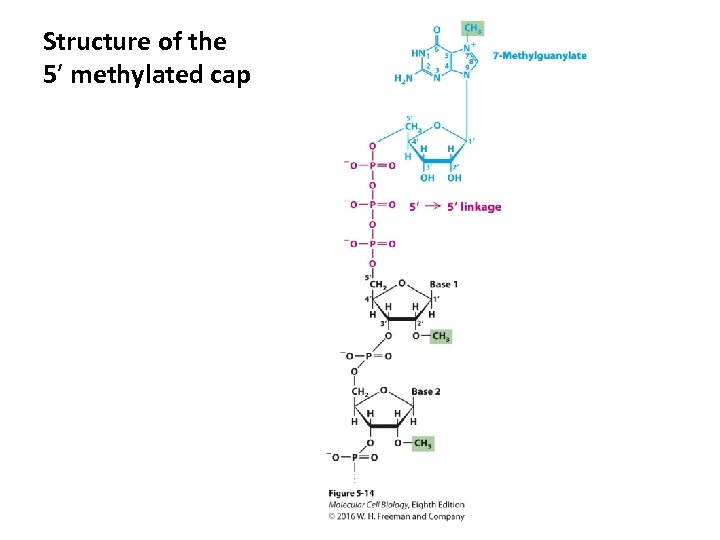 Structure of the 5′ methylated cap
Structure of the 5′ methylated cap
 Basic Transcriptional Mechanisms Life Cycle of m. RNA (https: //www. youtube. com/watch? v=h 7 FHqaet. MS 0) m. RNA splicing (https: //www. youtube. com/watch? v=FVu. Aw. BGw_p. Q)
Basic Transcriptional Mechanisms Life Cycle of m. RNA (https: //www. youtube. com/watch? v=h 7 FHqaet. MS 0) m. RNA splicing (https: //www. youtube. com/watch? v=FVu. Aw. BGw_p. Q)
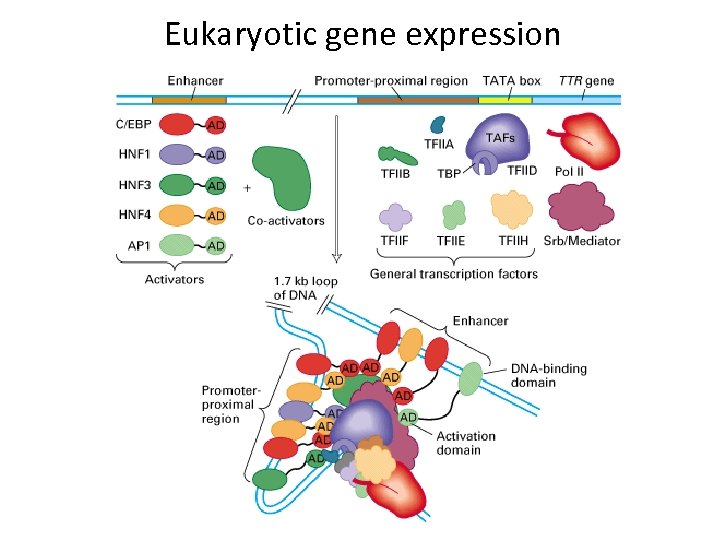 Eukaryotic gene expression
Eukaryotic gene expression
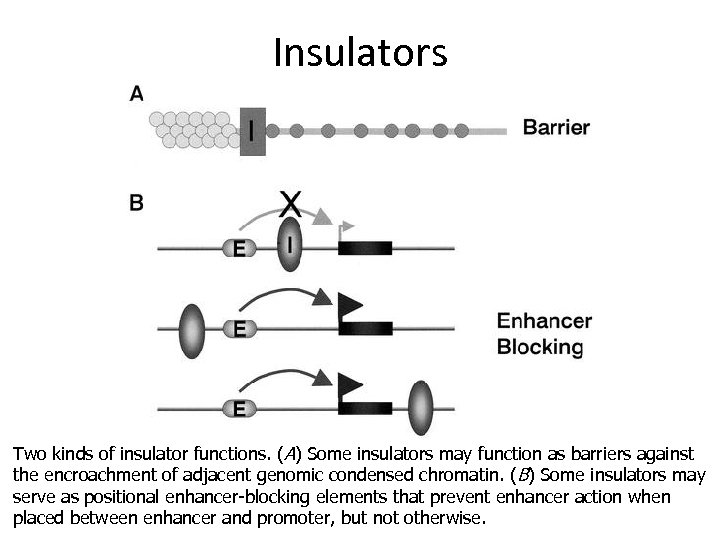 Insulators Two kinds of insulator functions. (A) Some insulators may function as barriers against the encroachment of adjacent genomic condensed chromatin. ( B) Some insulators may serve as positional enhancer-blocking elements that prevent enhancer action when placed between enhancer and promoter, but not otherwise.
Insulators Two kinds of insulator functions. (A) Some insulators may function as barriers against the encroachment of adjacent genomic condensed chromatin. ( B) Some insulators may serve as positional enhancer-blocking elements that prevent enhancer action when placed between enhancer and promoter, but not otherwise.
 Molecular Cloning: Recombinant DNA cloning procedure • Requires a DNA fragment and a cloning vector • Plasmid Cloning (https: //www. youtube. com/watch? v=2 vrt 87 wj. OBg)
Molecular Cloning: Recombinant DNA cloning procedure • Requires a DNA fragment and a cloning vector • Plasmid Cloning (https: //www. youtube. com/watch? v=2 vrt 87 wj. OBg)
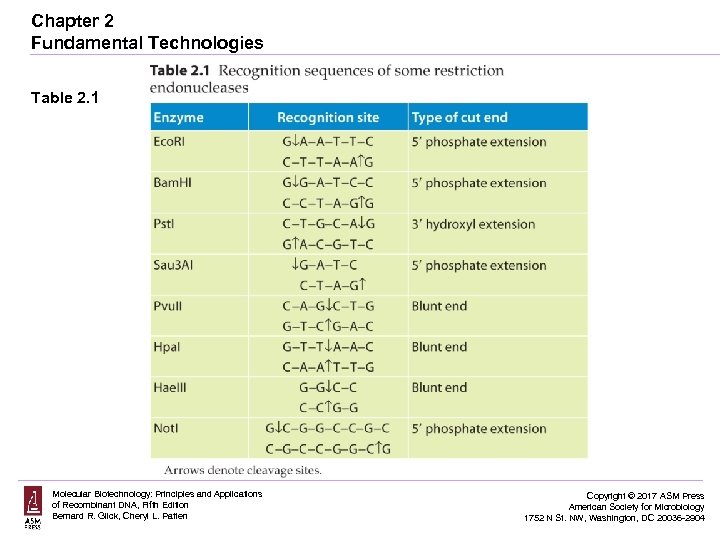 Chapter 2 Fundamental Technologies Table 2. 1 Molecular Biotechnology: Principles and Applications of Recombinant DNA, Fifth Edition Bernard R. Glick, Cheryl L. Patten Copyright © 2017 ASM Press American Society for Microbiology 1752 N St. NW, Washington, DC 20036 -2904
Chapter 2 Fundamental Technologies Table 2. 1 Molecular Biotechnology: Principles and Applications of Recombinant DNA, Fifth Edition Bernard R. Glick, Cheryl L. Patten Copyright © 2017 ASM Press American Society for Microbiology 1752 N St. NW, Washington, DC 20036 -2904
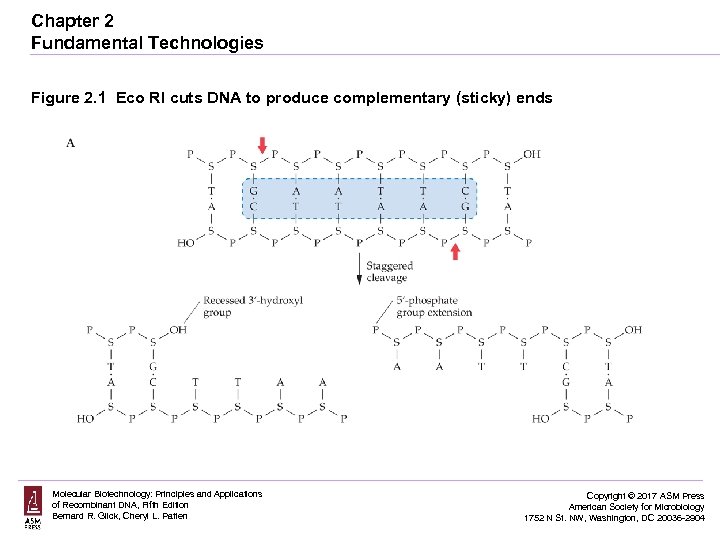 Chapter 2 Fundamental Technologies Figure 2. 1 Eco RI cuts DNA to produce complementary (sticky) ends Molecular Biotechnology: Principles and Applications of Recombinant DNA, Fifth Edition Bernard R. Glick, Cheryl L. Patten Copyright © 2017 ASM Press American Society for Microbiology 1752 N St. NW, Washington, DC 20036 -2904
Chapter 2 Fundamental Technologies Figure 2. 1 Eco RI cuts DNA to produce complementary (sticky) ends Molecular Biotechnology: Principles and Applications of Recombinant DNA, Fifth Edition Bernard R. Glick, Cheryl L. Patten Copyright © 2017 ASM Press American Society for Microbiology 1752 N St. NW, Washington, DC 20036 -2904
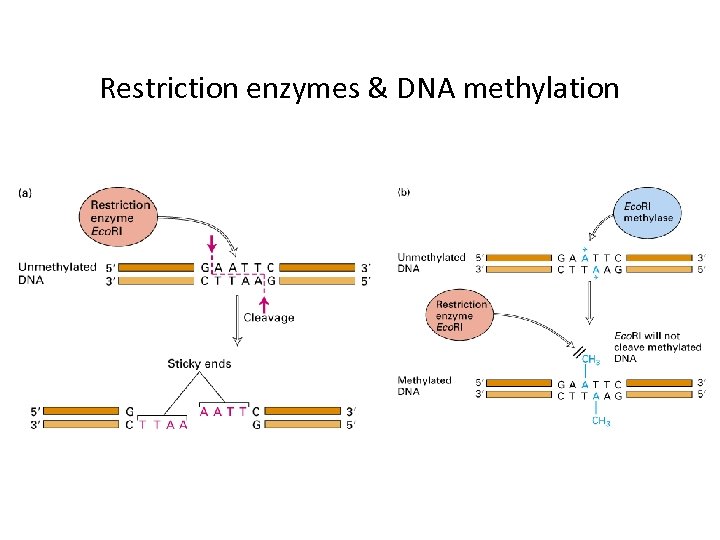 Restriction enzymes & DNA methylation
Restriction enzymes & DNA methylation
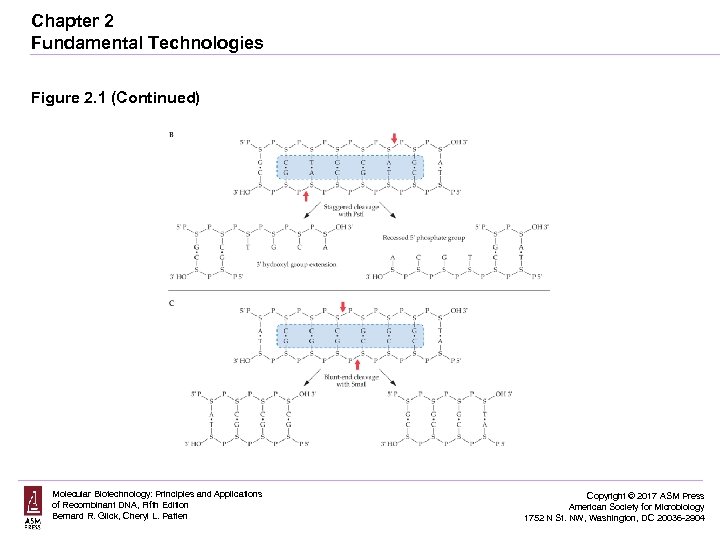 Chapter 2 Fundamental Technologies Figure 2. 1 (Continued) Molecular Biotechnology: Principles and Applications of Recombinant DNA, Fifth Edition Bernard R. Glick, Cheryl L. Patten Copyright © 2017 ASM Press American Society for Microbiology 1752 N St. NW, Washington, DC 20036 -2904
Chapter 2 Fundamental Technologies Figure 2. 1 (Continued) Molecular Biotechnology: Principles and Applications of Recombinant DNA, Fifth Edition Bernard R. Glick, Cheryl L. Patten Copyright © 2017 ASM Press American Society for Microbiology 1752 N St. NW, Washington, DC 20036 -2904
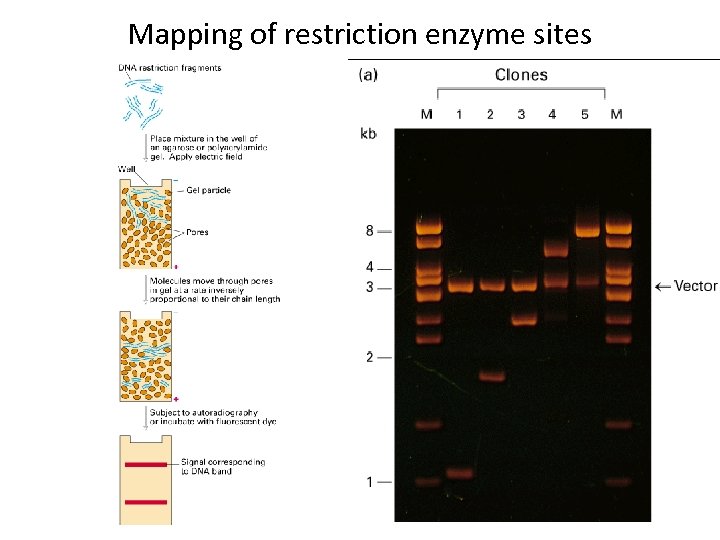 Mapping of restriction enzyme sites
Mapping of restriction enzyme sites
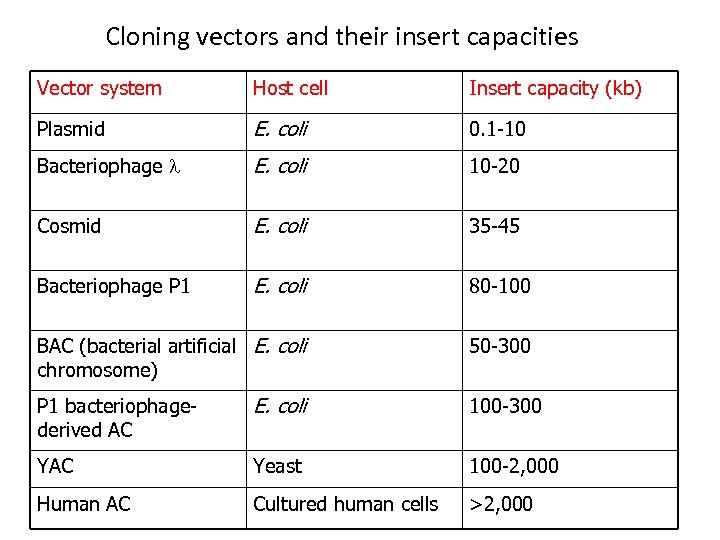 Cloning vectors and their insert capacities Vector system Host cell Insert capacity (kb) Plasmid E. coli 0. 1 -10 Bacteriophage l E. coli 10 -20 Cosmid E. coli 35 -45 Bacteriophage P 1 E. coli 80 -100 BAC (bacterial artificial E. coli chromosome) 50 -300 P 1 bacteriophagederived AC E. coli 100 -300 YAC Yeast 100 -2, 000 Human AC Cultured human cells >2, 000
Cloning vectors and their insert capacities Vector system Host cell Insert capacity (kb) Plasmid E. coli 0. 1 -10 Bacteriophage l E. coli 10 -20 Cosmid E. coli 35 -45 Bacteriophage P 1 E. coli 80 -100 BAC (bacterial artificial E. coli chromosome) 50 -300 P 1 bacteriophagederived AC E. coli 100 -300 YAC Yeast 100 -2, 000 Human AC Cultured human cells >2, 000
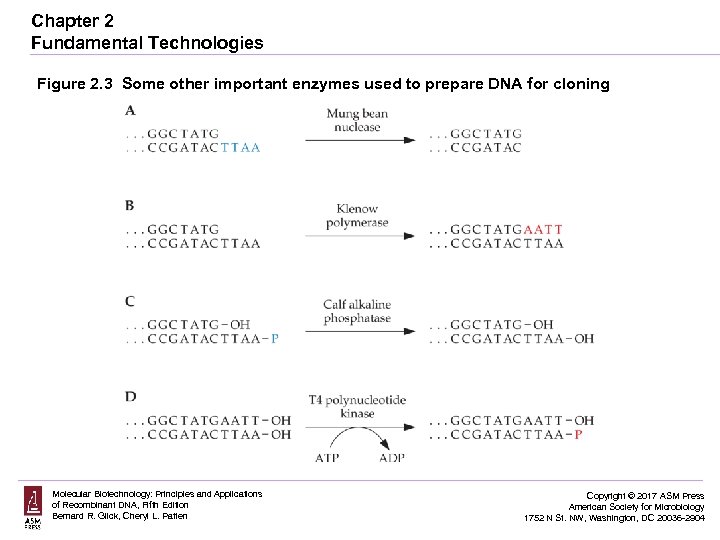 Chapter 2 Fundamental Technologies Figure 2. 3 Some other important enzymes used to prepare DNA for cloning Molecular Biotechnology: Principles and Applications of Recombinant DNA, Fifth Edition Bernard R. Glick, Cheryl L. Patten Copyright © 2017 ASM Press American Society for Microbiology 1752 N St. NW, Washington, DC 20036 -2904
Chapter 2 Fundamental Technologies Figure 2. 3 Some other important enzymes used to prepare DNA for cloning Molecular Biotechnology: Principles and Applications of Recombinant DNA, Fifth Edition Bernard R. Glick, Cheryl L. Patten Copyright © 2017 ASM Press American Society for Microbiology 1752 N St. NW, Washington, DC 20036 -2904
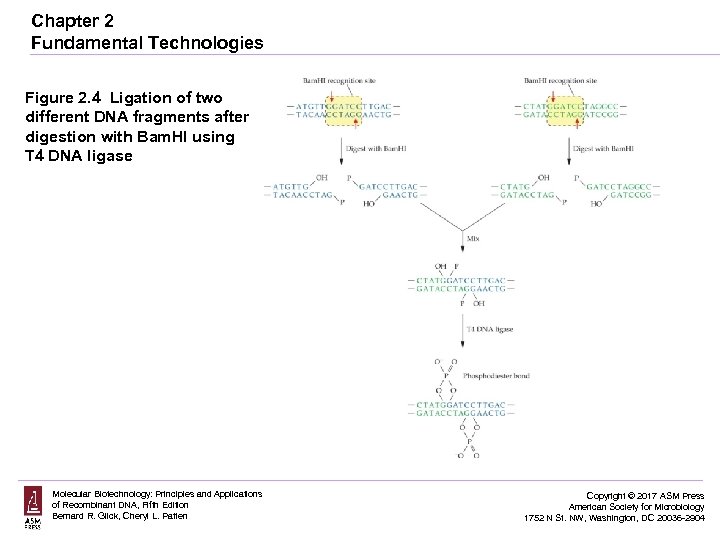 Chapter 2 Fundamental Technologies Figure 2. 4 Ligation of two different DNA fragments after digestion with Bam. HI using T 4 DNA ligase Molecular Biotechnology: Principles and Applications of Recombinant DNA, Fifth Edition Bernard R. Glick, Cheryl L. Patten Copyright © 2017 ASM Press American Society for Microbiology 1752 N St. NW, Washington, DC 20036 -2904
Chapter 2 Fundamental Technologies Figure 2. 4 Ligation of two different DNA fragments after digestion with Bam. HI using T 4 DNA ligase Molecular Biotechnology: Principles and Applications of Recombinant DNA, Fifth Edition Bernard R. Glick, Cheryl L. Patten Copyright © 2017 ASM Press American Society for Microbiology 1752 N St. NW, Washington, DC 20036 -2904
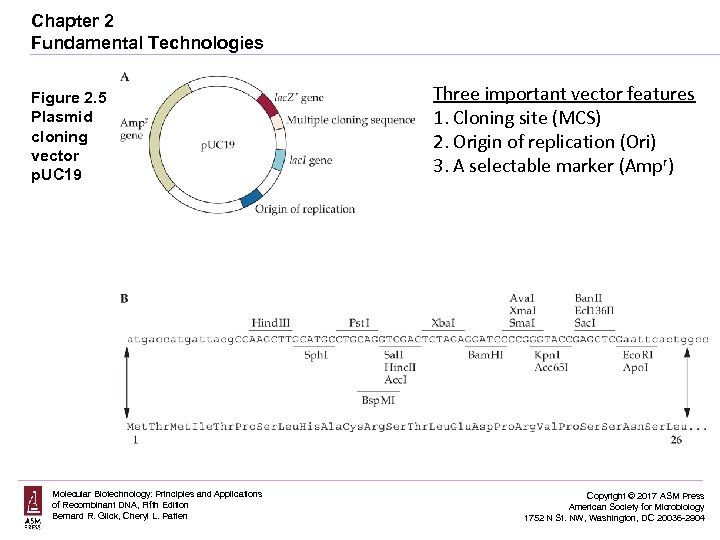 Chapter 2 Fundamental Technologies Figure 2. 5 Plasmid cloning vector p. UC 19 Molecular Biotechnology: Principles and Applications of Recombinant DNA, Fifth Edition Bernard R. Glick, Cheryl L. Patten Three important vector features 1. Cloning site (MCS) 2. Origin of replication (Ori) 3. A selectable marker (Ampr) Copyright © 2017 ASM Press American Society for Microbiology 1752 N St. NW, Washington, DC 20036 -2904
Chapter 2 Fundamental Technologies Figure 2. 5 Plasmid cloning vector p. UC 19 Molecular Biotechnology: Principles and Applications of Recombinant DNA, Fifth Edition Bernard R. Glick, Cheryl L. Patten Three important vector features 1. Cloning site (MCS) 2. Origin of replication (Ori) 3. A selectable marker (Ampr) Copyright © 2017 ASM Press American Society for Microbiology 1752 N St. NW, Washington, DC 20036 -2904
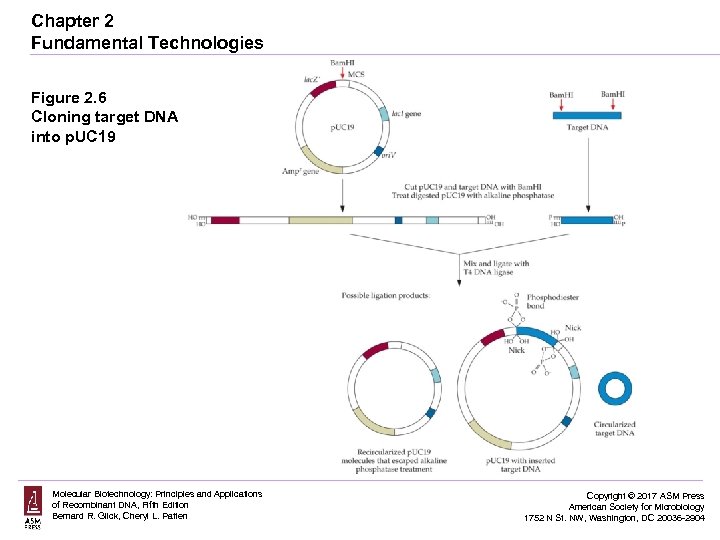 Chapter 2 Fundamental Technologies Figure 2. 6 Cloning target DNA into p. UC 19 Molecular Biotechnology: Principles and Applications of Recombinant DNA, Fifth Edition Bernard R. Glick, Cheryl L. Patten Copyright © 2017 ASM Press American Society for Microbiology 1752 N St. NW, Washington, DC 20036 -2904
Chapter 2 Fundamental Technologies Figure 2. 6 Cloning target DNA into p. UC 19 Molecular Biotechnology: Principles and Applications of Recombinant DNA, Fifth Edition Bernard R. Glick, Cheryl L. Patten Copyright © 2017 ASM Press American Society for Microbiology 1752 N St. NW, Washington, DC 20036 -2904
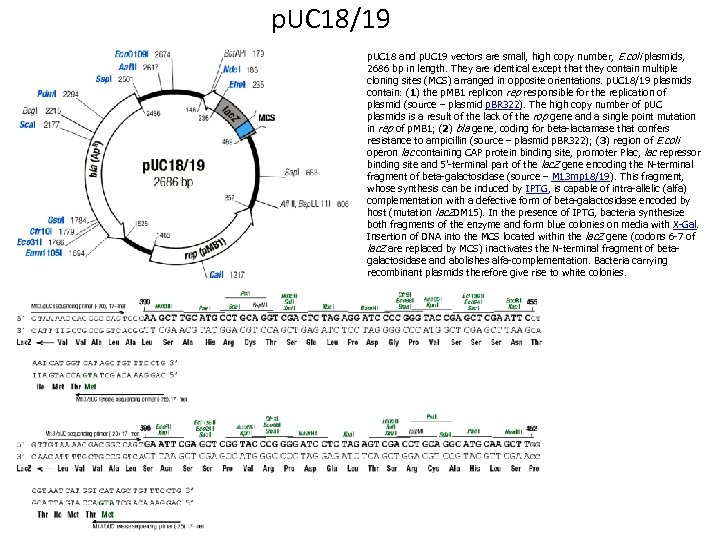 p. UC 18/19 p. UC 18 and p. UC 19 vectors are small, high copy number, E. coli plasmids, 2686 bp in length. They are identical except that they contain multiple cloning sites (MCS) arranged in opposite orientations. p. UC 18/19 plasmids contain: (1) the p. MB 1 replicon rep responsible for the replication of plasmid (source – plasmid p. BR 322). The high copy number of p. UC plasmids is a result of the lack of the rop gene and a single point mutation in rep of p. MB 1; (2) bla gene, coding for beta-lactamase that confers resistance to ampicillin (source – plasmid p. BR 322); (3) region of E. coli operon lac containing CAP protein binding site, promoter Plac, lac repressor binding site and 5’-terminal part of the lac. Z gene encoding the N-terminal fragment of beta-galactosidase (source – M 13 mp 18/19). This fragment, whose synthesis can be induced by IPTG, is capable of intra-allelic (alfa) complementation with a defective form of beta-galactosidase encoded by host (mutation lac. ZDM 15). In the presence of IPTG, bacteria synthesize both fragments of the enzyme and form blue colonies on media with X-Gal. Insertion of DNA into the MCS located within the lac. Z gene (codons 6 -7 of lac. Z are replaced by MCS) inactivates the N-terminal fragment of betagalactosidase and abolishes alfa-complementation. Bacteria carrying recombinant plasmids therefore give rise to white colonies.
p. UC 18/19 p. UC 18 and p. UC 19 vectors are small, high copy number, E. coli plasmids, 2686 bp in length. They are identical except that they contain multiple cloning sites (MCS) arranged in opposite orientations. p. UC 18/19 plasmids contain: (1) the p. MB 1 replicon rep responsible for the replication of plasmid (source – plasmid p. BR 322). The high copy number of p. UC plasmids is a result of the lack of the rop gene and a single point mutation in rep of p. MB 1; (2) bla gene, coding for beta-lactamase that confers resistance to ampicillin (source – plasmid p. BR 322); (3) region of E. coli operon lac containing CAP protein binding site, promoter Plac, lac repressor binding site and 5’-terminal part of the lac. Z gene encoding the N-terminal fragment of beta-galactosidase (source – M 13 mp 18/19). This fragment, whose synthesis can be induced by IPTG, is capable of intra-allelic (alfa) complementation with a defective form of beta-galactosidase encoded by host (mutation lac. ZDM 15). In the presence of IPTG, bacteria synthesize both fragments of the enzyme and form blue colonies on media with X-Gal. Insertion of DNA into the MCS located within the lac. Z gene (codons 6 -7 of lac. Z are replaced by MCS) inactivates the N-terminal fragment of betagalactosidase and abolishes alfa-complementation. Bacteria carrying recombinant plasmids therefore give rise to white colonies.
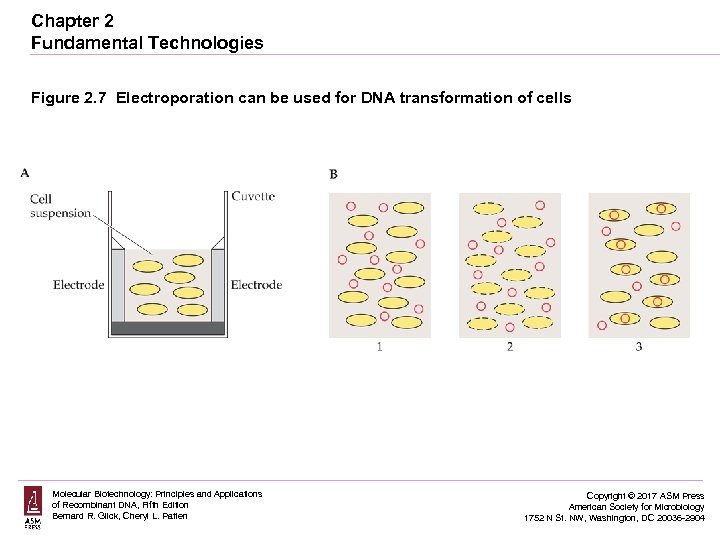 Chapter 2 Fundamental Technologies Figure 2. 7 Electroporation can be used for DNA transformation of cells Molecular Biotechnology: Principles and Applications of Recombinant DNA, Fifth Edition Bernard R. Glick, Cheryl L. Patten Copyright © 2017 ASM Press American Society for Microbiology 1752 N St. NW, Washington, DC 20036 -2904
Chapter 2 Fundamental Technologies Figure 2. 7 Electroporation can be used for DNA transformation of cells Molecular Biotechnology: Principles and Applications of Recombinant DNA, Fifth Edition Bernard R. Glick, Cheryl L. Patten Copyright © 2017 ASM Press American Society for Microbiology 1752 N St. NW, Washington, DC 20036 -2904
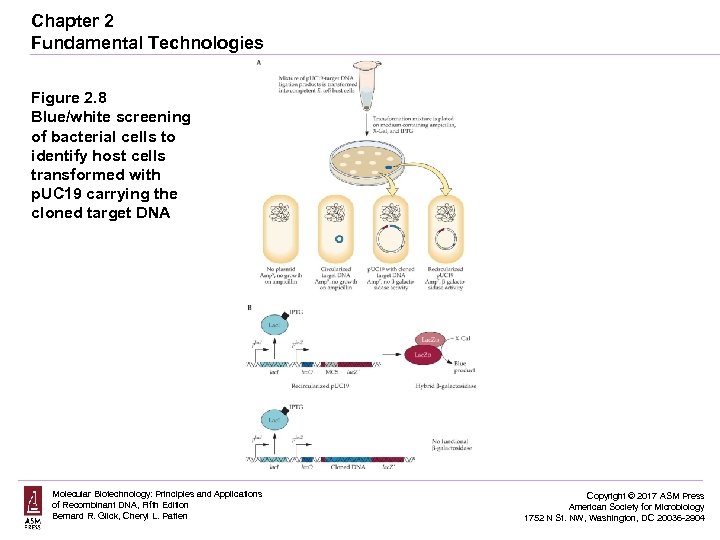 Chapter 2 Fundamental Technologies Figure 2. 8 Blue/white screening of bacterial cells to identify host cells transformed with p. UC 19 carrying the cloned target DNA Molecular Biotechnology: Principles and Applications of Recombinant DNA, Fifth Edition Bernard R. Glick, Cheryl L. Patten Copyright © 2017 ASM Press American Society for Microbiology 1752 N St. NW, Washington, DC 20036 -2904
Chapter 2 Fundamental Technologies Figure 2. 8 Blue/white screening of bacterial cells to identify host cells transformed with p. UC 19 carrying the cloned target DNA Molecular Biotechnology: Principles and Applications of Recombinant DNA, Fifth Edition Bernard R. Glick, Cheryl L. Patten Copyright © 2017 ASM Press American Society for Microbiology 1752 N St. NW, Washington, DC 20036 -2904
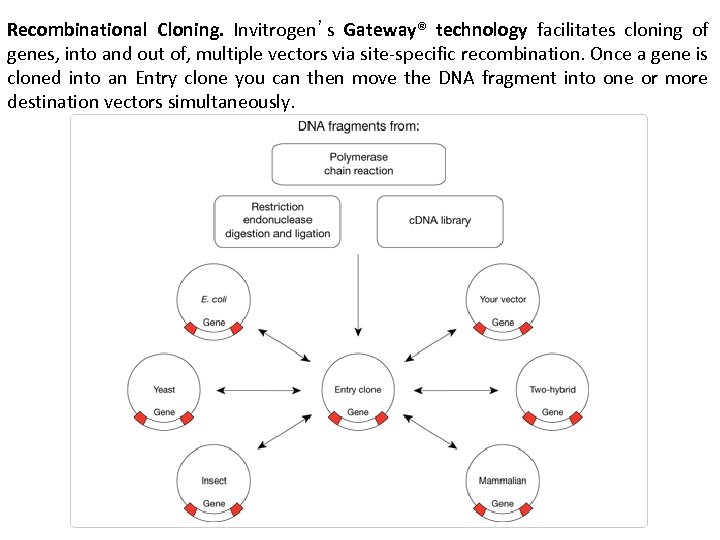 Recombinational Cloning. Invitrogen’s Gateway® technology facilitates cloning of genes, into and out of, multiple vectors via site-specific recombination. Once a gene is cloned into an Entry clone you can then move the DNA fragment into one or more destination vectors simultaneously.
Recombinational Cloning. Invitrogen’s Gateway® technology facilitates cloning of genes, into and out of, multiple vectors via site-specific recombination. Once a gene is cloned into an Entry clone you can then move the DNA fragment into one or more destination vectors simultaneously.
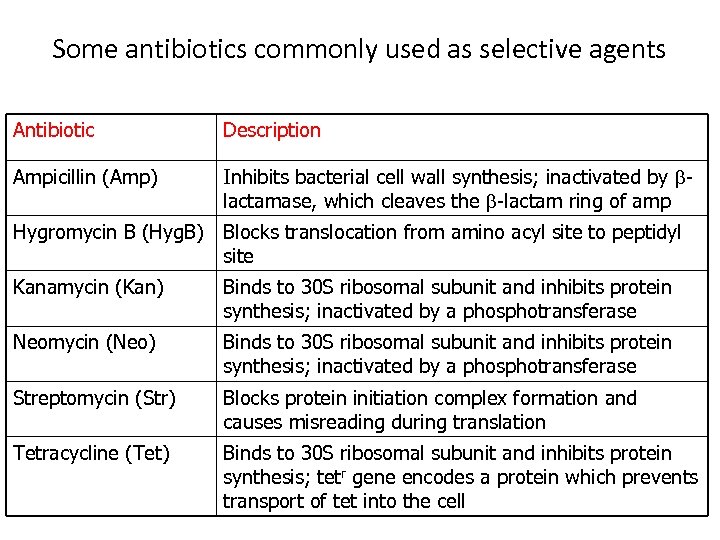 Some antibiotics commonly used as selective agents Antibiotic Description Ampicillin (Amp) Inhibits bacterial cell wall synthesis; inactivated by blactamase, which cleaves the b-lactam ring of amp Hygromycin B (Hyg. B) Blocks translocation from amino acyl site to peptidyl site Kanamycin (Kan) Binds to 30 S ribosomal subunit and inhibits protein synthesis; inactivated by a phosphotransferase Neomycin (Neo) Binds to 30 S ribosomal subunit and inhibits protein synthesis; inactivated by a phosphotransferase Streptomycin (Str) Blocks protein initiation complex formation and causes misreading during translation Tetracycline (Tet) Binds to 30 S ribosomal subunit and inhibits protein synthesis; tetr gene encodes a protein which prevents transport of tet into the cell
Some antibiotics commonly used as selective agents Antibiotic Description Ampicillin (Amp) Inhibits bacterial cell wall synthesis; inactivated by blactamase, which cleaves the b-lactam ring of amp Hygromycin B (Hyg. B) Blocks translocation from amino acyl site to peptidyl site Kanamycin (Kan) Binds to 30 S ribosomal subunit and inhibits protein synthesis; inactivated by a phosphotransferase Neomycin (Neo) Binds to 30 S ribosomal subunit and inhibits protein synthesis; inactivated by a phosphotransferase Streptomycin (Str) Blocks protein initiation complex formation and causes misreading during translation Tetracycline (Tet) Binds to 30 S ribosomal subunit and inhibits protein synthesis; tetr gene encodes a protein which prevents transport of tet into the cell
 Library Construction and Screening • Genomic (gene) libraries • c. DNA libraries • Library screening
Library Construction and Screening • Genomic (gene) libraries • c. DNA libraries • Library screening
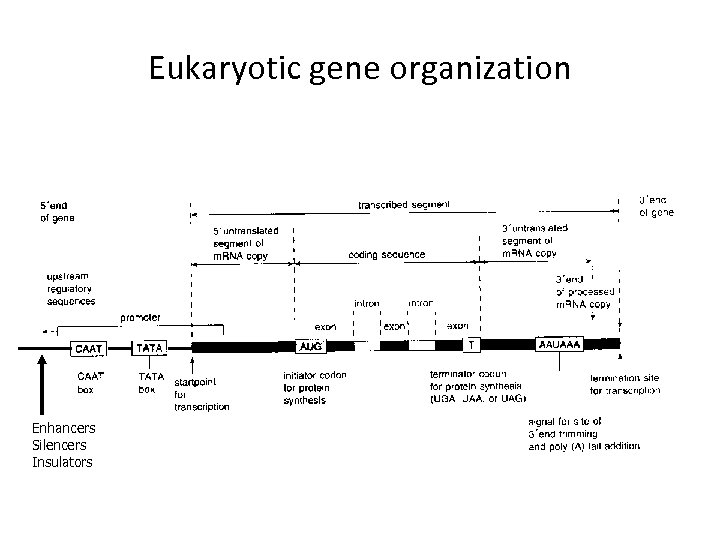 Eukaryotic gene organization Enhancers Silencers Insulators
Eukaryotic gene organization Enhancers Silencers Insulators
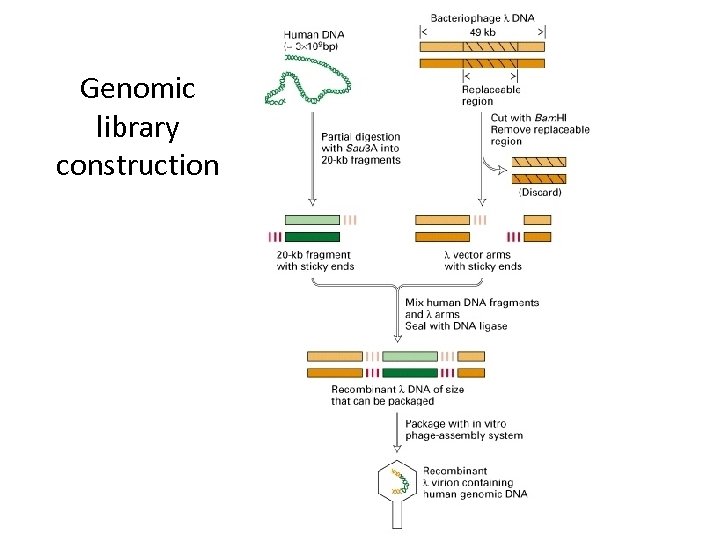 Genomic library construction
Genomic library construction
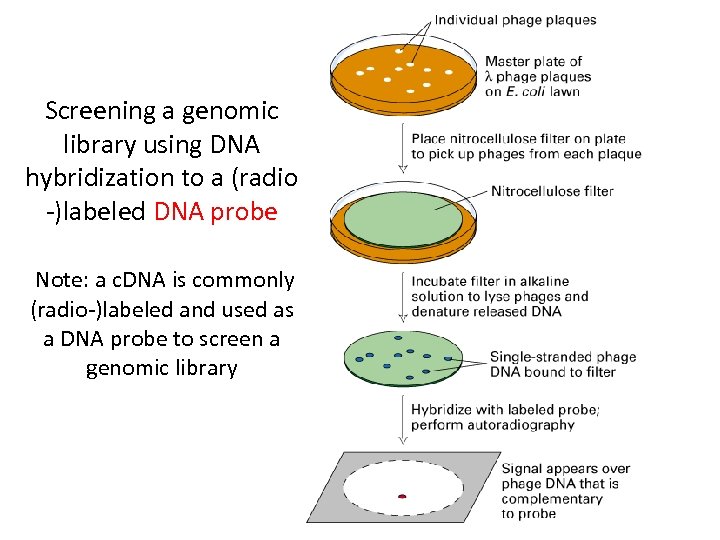 Screening a genomic library using DNA hybridization to a (radio -)labeled DNA probe Note: a c. DNA is commonly (radio-)labeled and used as a DNA probe to screen a genomic library
Screening a genomic library using DNA hybridization to a (radio -)labeled DNA probe Note: a c. DNA is commonly (radio-)labeled and used as a DNA probe to screen a genomic library
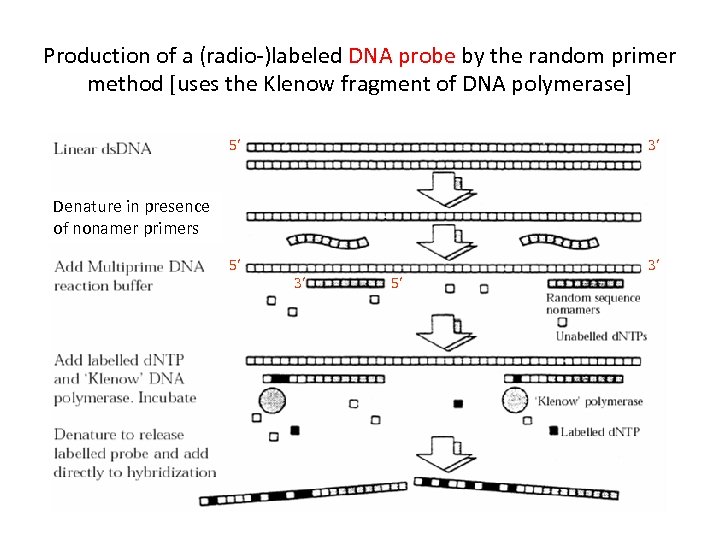 Production of a (radio-)labeled DNA probe by the random primer method [uses the Klenow fragment of DNA polymerase] 5’ 3’ Denature in presence of nonamer primers 3’ 5’
Production of a (radio-)labeled DNA probe by the random primer method [uses the Klenow fragment of DNA polymerase] 5’ 3’ Denature in presence of nonamer primers 3’ 5’
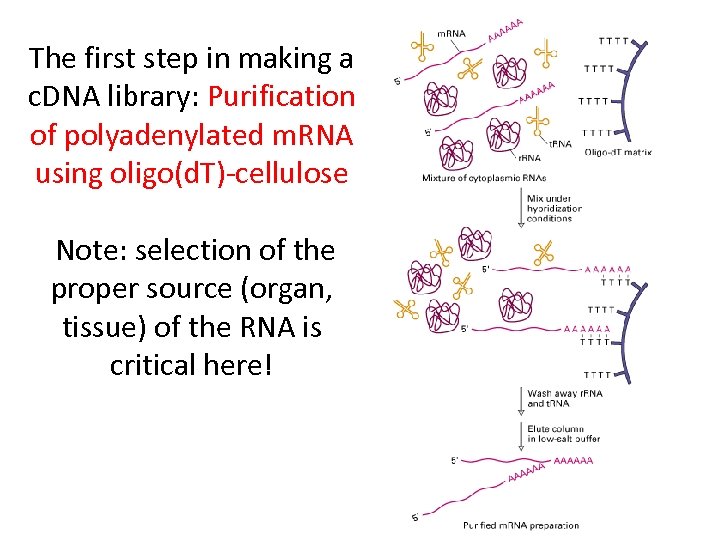 The first step in making a c. DNA library: Purification of polyadenylated m. RNA using oligo(d. T)-cellulose Note: selection of the proper source (organ, tissue) of the RNA is critical here!
The first step in making a c. DNA library: Purification of polyadenylated m. RNA using oligo(d. T)-cellulose Note: selection of the proper source (organ, tissue) of the RNA is critical here!
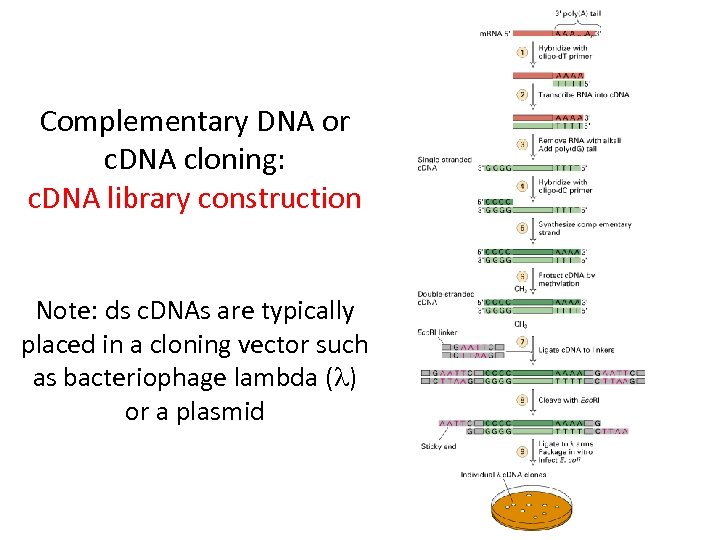 Complementary DNA or c. DNA cloning: c. DNA library construction Note: ds c. DNAs are typically placed in a cloning vector such as bacteriophage lambda (l) or a plasmid
Complementary DNA or c. DNA cloning: c. DNA library construction Note: ds c. DNAs are typically placed in a cloning vector such as bacteriophage lambda (l) or a plasmid
 There are several possible ways to screen a c. DNA library • Using a DNA probe with a homologous sequence (e. g. , a homologous c. DNA or gene clone from a related species) • Using an oligonucleotide probe based on a known amino acid sequence (requires purification of the protein and some peptide sequencing) • Using an antibody against the protein of interest (note: this requires use of an expression vector)
There are several possible ways to screen a c. DNA library • Using a DNA probe with a homologous sequence (e. g. , a homologous c. DNA or gene clone from a related species) • Using an oligonucleotide probe based on a known amino acid sequence (requires purification of the protein and some peptide sequencing) • Using an antibody against the protein of interest (note: this requires use of an expression vector)
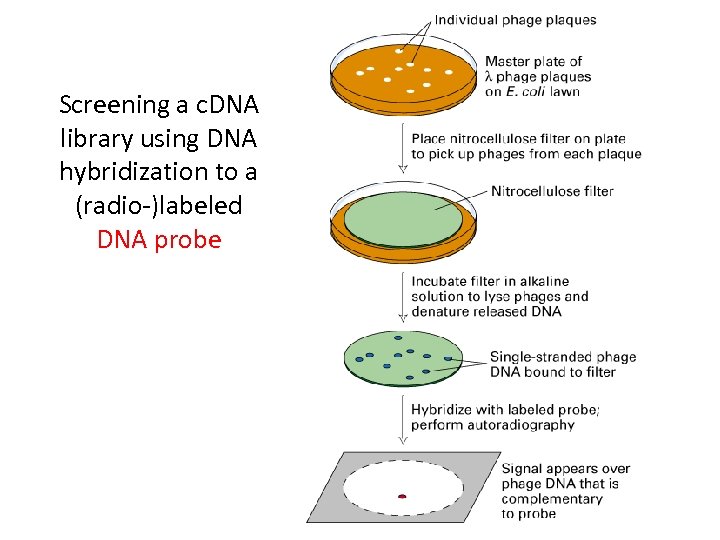 Screening a c. DNA library using DNA hybridization to a (radio-)labeled DNA probe
Screening a c. DNA library using DNA hybridization to a (radio-)labeled DNA probe
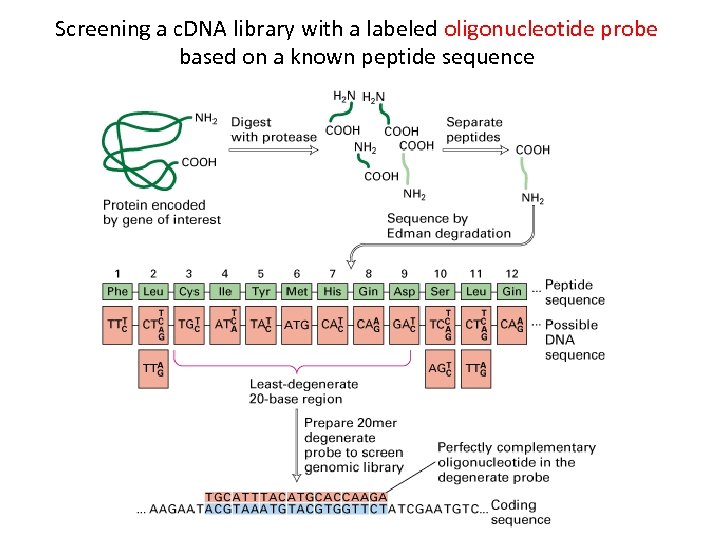 Screening a c. DNA library with a labeled oligonucleotide probe based on a known peptide sequence
Screening a c. DNA library with a labeled oligonucleotide probe based on a known peptide sequence
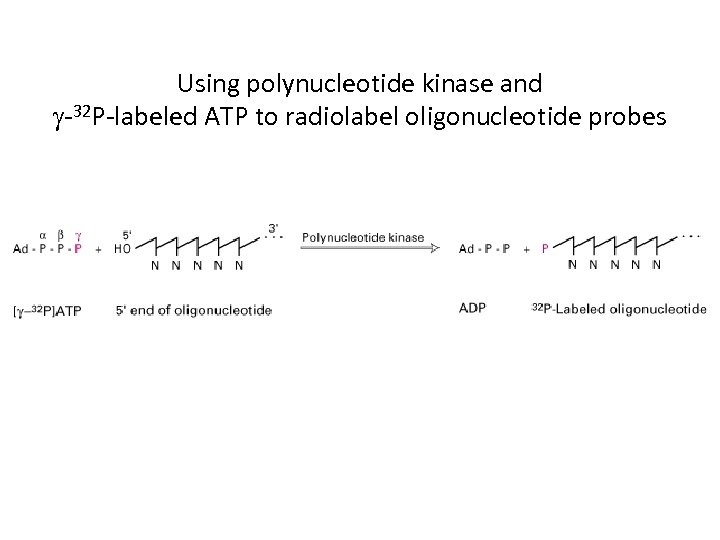 Using polynucleotide kinase and g-32 P-labeled ATP to radiolabel oligonucleotide probes
Using polynucleotide kinase and g-32 P-labeled ATP to radiolabel oligonucleotide probes
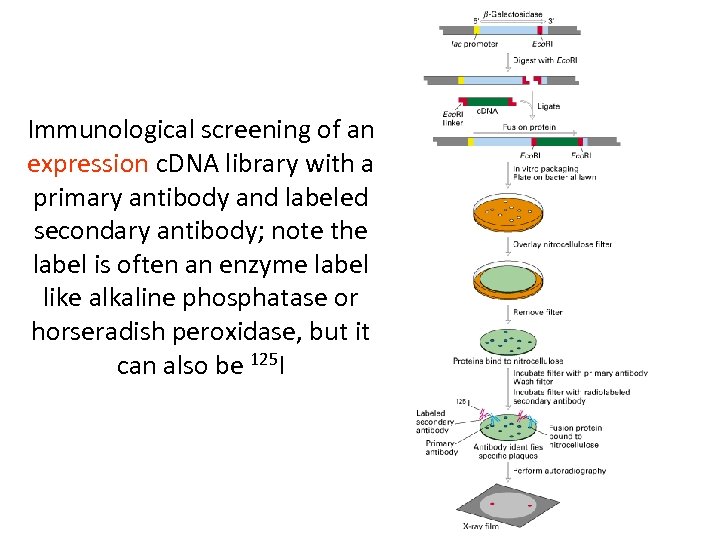 Immunological screening of an expression c. DNA library with a primary antibody and labeled secondary antibody; note the label is often an enzyme label like alkaline phosphatase or horseradish peroxidase, but it can also be 125 I
Immunological screening of an expression c. DNA library with a primary antibody and labeled secondary antibody; note the label is often an enzyme label like alkaline phosphatase or horseradish peroxidase, but it can also be 125 I
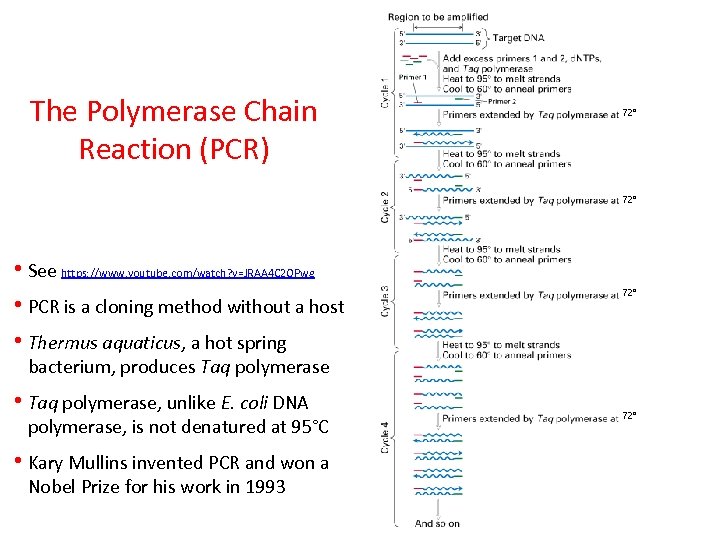 The Polymerase Chain Reaction (PCR) 72° • See https: //www. youtube. com/watch? v=JRAA 4 C 2 OPwg • PCR is a cloning method without a host • Thermus aquaticus, a hot spring 72° bacterium, produces Taq polymerase • Taq polymerase, unlike E. coli DNA polymerase, is not denatured at 95°C • Kary Mullins invented PCR and won a Nobel Prize for his work in 1993 72°
The Polymerase Chain Reaction (PCR) 72° • See https: //www. youtube. com/watch? v=JRAA 4 C 2 OPwg • PCR is a cloning method without a host • Thermus aquaticus, a hot spring 72° bacterium, produces Taq polymerase • Taq polymerase, unlike E. coli DNA polymerase, is not denatured at 95°C • Kary Mullins invented PCR and won a Nobel Prize for his work in 1993 72°
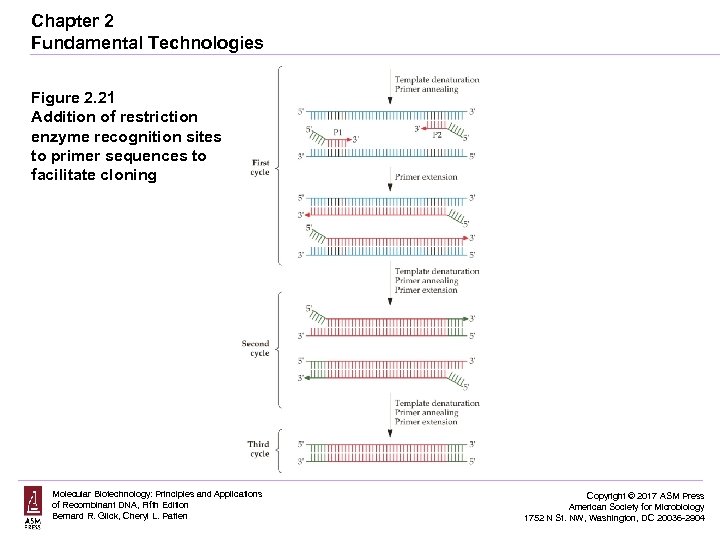 Chapter 2 Fundamental Technologies Figure 2. 21 Addition of restriction enzyme recognition sites to primer sequences to facilitate cloning Molecular Biotechnology: Principles and Applications of Recombinant DNA, Fifth Edition Bernard R. Glick, Cheryl L. Patten Copyright © 2017 ASM Press American Society for Microbiology 1752 N St. NW, Washington, DC 20036 -2904
Chapter 2 Fundamental Technologies Figure 2. 21 Addition of restriction enzyme recognition sites to primer sequences to facilitate cloning Molecular Biotechnology: Principles and Applications of Recombinant DNA, Fifth Edition Bernard R. Glick, Cheryl L. Patten Copyright © 2017 ASM Press American Society for Microbiology 1752 N St. NW, Washington, DC 20036 -2904
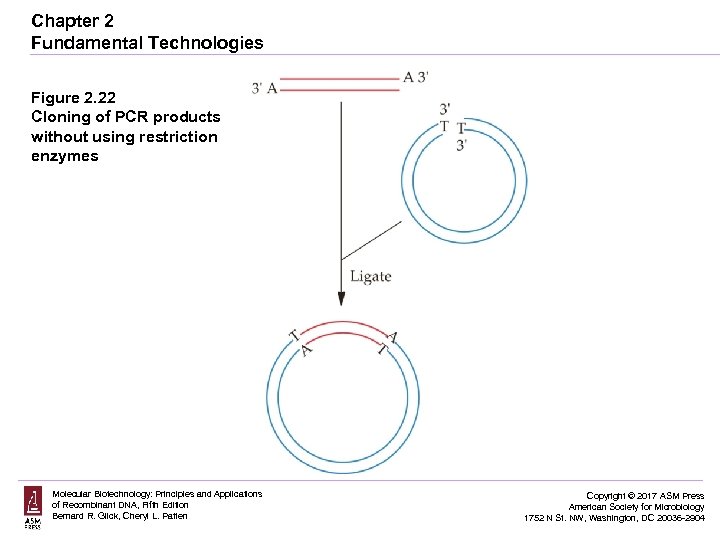 Chapter 2 Fundamental Technologies Figure 2. 22 Cloning of PCR products without using restriction enzymes Molecular Biotechnology: Principles and Applications of Recombinant DNA, Fifth Edition Bernard R. Glick, Cheryl L. Patten Copyright © 2017 ASM Press American Society for Microbiology 1752 N St. NW, Washington, DC 20036 -2904
Chapter 2 Fundamental Technologies Figure 2. 22 Cloning of PCR products without using restriction enzymes Molecular Biotechnology: Principles and Applications of Recombinant DNA, Fifth Edition Bernard R. Glick, Cheryl L. Patten Copyright © 2017 ASM Press American Society for Microbiology 1752 N St. NW, Washington, DC 20036 -2904
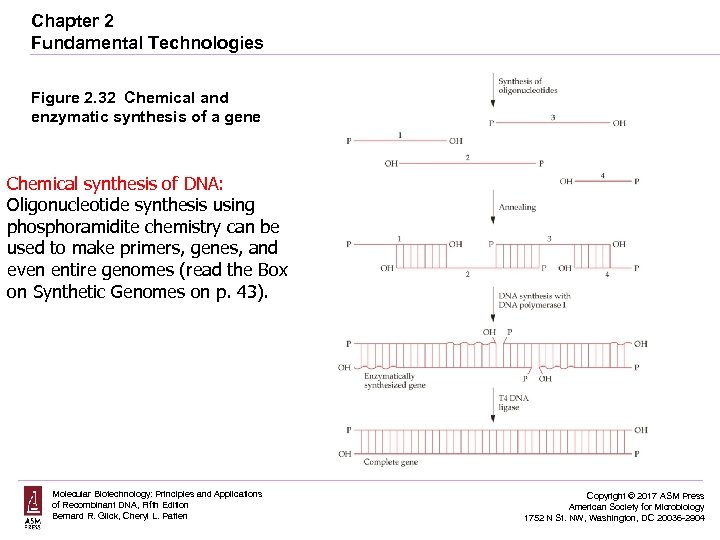 Chapter 2 Fundamental Technologies Figure 2. 32 Chemical and enzymatic synthesis of a gene Chemical synthesis of DNA: Oligonucleotide synthesis using phosphoramidite chemistry can be used to make primers, genes, and even entire genomes (read the Box on Synthetic Genomes on p. 43). Molecular Biotechnology: Principles and Applications of Recombinant DNA, Fifth Edition Bernard R. Glick, Cheryl L. Patten Copyright © 2017 ASM Press American Society for Microbiology 1752 N St. NW, Washington, DC 20036 -2904
Chapter 2 Fundamental Technologies Figure 2. 32 Chemical and enzymatic synthesis of a gene Chemical synthesis of DNA: Oligonucleotide synthesis using phosphoramidite chemistry can be used to make primers, genes, and even entire genomes (read the Box on Synthetic Genomes on p. 43). Molecular Biotechnology: Principles and Applications of Recombinant DNA, Fifth Edition Bernard R. Glick, Cheryl L. Patten Copyright © 2017 ASM Press American Society for Microbiology 1752 N St. NW, Washington, DC 20036 -2904
 DNA Sequencing Technologies • • • Sanger sequencing (Dideoxy Sequencing) Pyrosequencing Sequencing using reversible chain terminators (i. e. , sequencing by synthesis) Sequencing by single molecule synthesis See https: //www. youtube. com/watch? v=j. FCD 8 Q 6 q. STM for explanations of Sanger sequencing, sequencing by synthesis, sequencing by ligation, and ion semiconductor sequencing See also https: //www. youtube. com/watch? v=Wq 35 ZXyayu. U for Nanopore sequencing with the Min. ION device- very cool!
DNA Sequencing Technologies • • • Sanger sequencing (Dideoxy Sequencing) Pyrosequencing Sequencing using reversible chain terminators (i. e. , sequencing by synthesis) Sequencing by single molecule synthesis See https: //www. youtube. com/watch? v=j. FCD 8 Q 6 q. STM for explanations of Sanger sequencing, sequencing by synthesis, sequencing by ligation, and ion semiconductor sequencing See also https: //www. youtube. com/watch? v=Wq 35 ZXyayu. U for Nanopore sequencing with the Min. ION device- very cool!
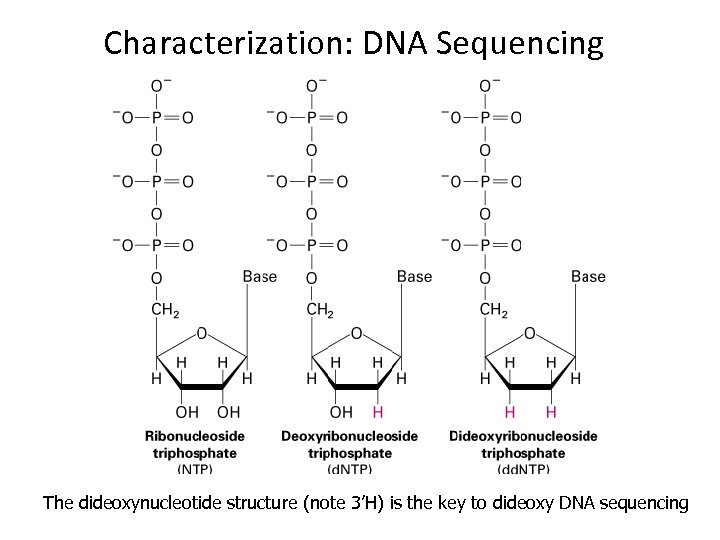 Characterization: DNA Sequencing The dideoxynucleotide structure (note 3’H) is the key to dideoxy DNA sequencing
Characterization: DNA Sequencing The dideoxynucleotide structure (note 3’H) is the key to dideoxy DNA sequencing
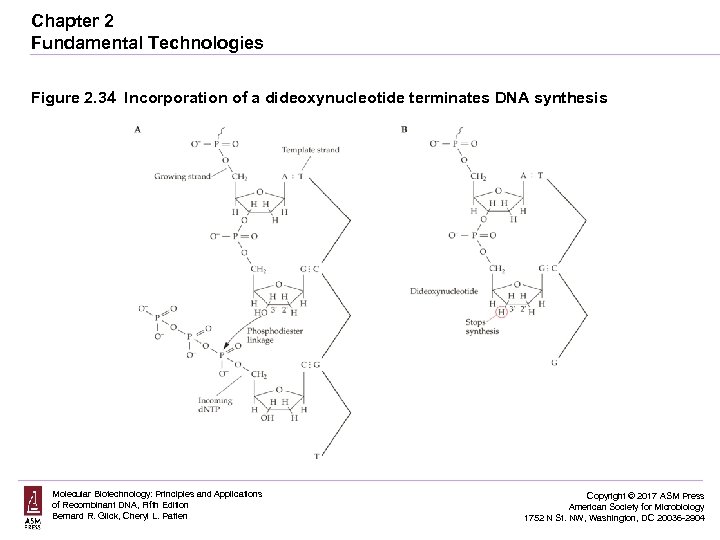 Chapter 2 Fundamental Technologies Figure 2. 34 Incorporation of a dideoxynucleotide terminates DNA synthesis Molecular Biotechnology: Principles and Applications of Recombinant DNA, Fifth Edition Bernard R. Glick, Cheryl L. Patten Copyright © 2017 ASM Press American Society for Microbiology 1752 N St. NW, Washington, DC 20036 -2904
Chapter 2 Fundamental Technologies Figure 2. 34 Incorporation of a dideoxynucleotide terminates DNA synthesis Molecular Biotechnology: Principles and Applications of Recombinant DNA, Fifth Edition Bernard R. Glick, Cheryl L. Patten Copyright © 2017 ASM Press American Society for Microbiology 1752 N St. NW, Washington, DC 20036 -2904
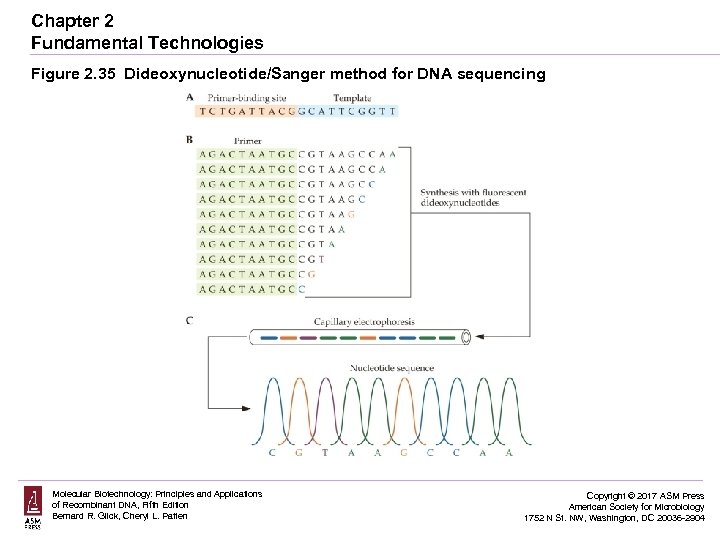 Chapter 2 Fundamental Technologies Figure 2. 35 Dideoxynucleotide/Sanger method for DNA sequencing Molecular Biotechnology: Principles and Applications of Recombinant DNA, Fifth Edition Bernard R. Glick, Cheryl L. Patten Copyright © 2017 ASM Press American Society for Microbiology 1752 N St. NW, Washington, DC 20036 -2904
Chapter 2 Fundamental Technologies Figure 2. 35 Dideoxynucleotide/Sanger method for DNA sequencing Molecular Biotechnology: Principles and Applications of Recombinant DNA, Fifth Edition Bernard R. Glick, Cheryl L. Patten Copyright © 2017 ASM Press American Society for Microbiology 1752 N St. NW, Washington, DC 20036 -2904
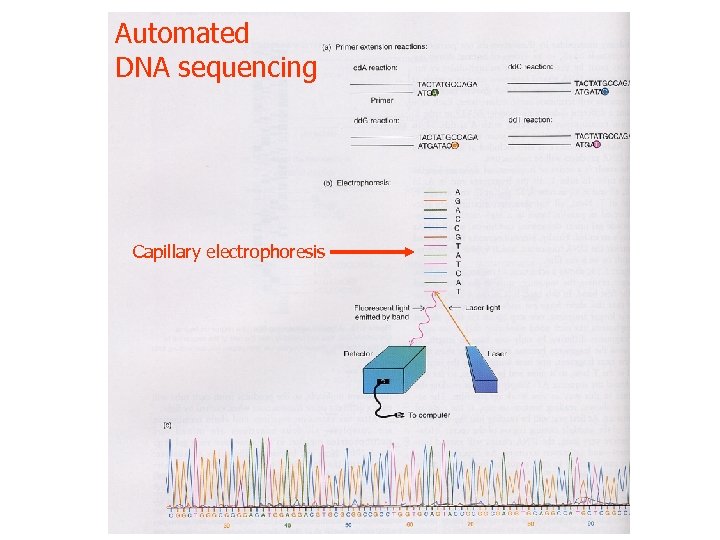 Automated DNA sequencing Capillary electrophoresis
Automated DNA sequencing Capillary electrophoresis
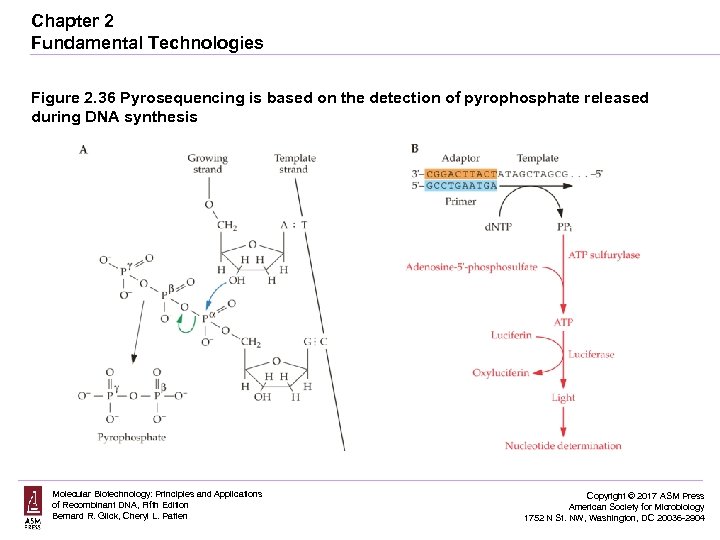 Chapter 2 Fundamental Technologies Figure 2. 36 Pyrosequencing is based on the detection of pyrophosphate released during DNA synthesis Molecular Biotechnology: Principles and Applications of Recombinant DNA, Fifth Edition Bernard R. Glick, Cheryl L. Patten Copyright © 2017 ASM Press American Society for Microbiology 1752 N St. NW, Washington, DC 20036 -2904
Chapter 2 Fundamental Technologies Figure 2. 36 Pyrosequencing is based on the detection of pyrophosphate released during DNA synthesis Molecular Biotechnology: Principles and Applications of Recombinant DNA, Fifth Edition Bernard R. Glick, Cheryl L. Patten Copyright © 2017 ASM Press American Society for Microbiology 1752 N St. NW, Washington, DC 20036 -2904
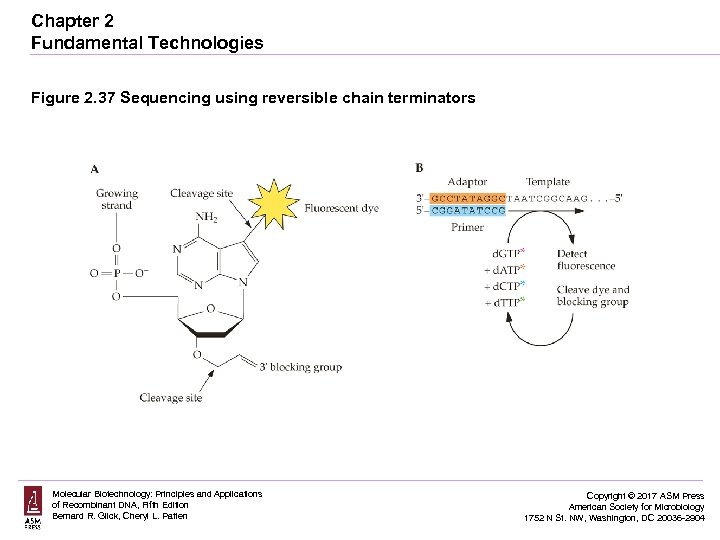 Chapter 2 Fundamental Technologies Figure 2. 37 Sequencing using reversible chain terminators Molecular Biotechnology: Principles and Applications of Recombinant DNA, Fifth Edition Bernard R. Glick, Cheryl L. Patten Copyright © 2017 ASM Press American Society for Microbiology 1752 N St. NW, Washington, DC 20036 -2904
Chapter 2 Fundamental Technologies Figure 2. 37 Sequencing using reversible chain terminators Molecular Biotechnology: Principles and Applications of Recombinant DNA, Fifth Edition Bernard R. Glick, Cheryl L. Patten Copyright © 2017 ASM Press American Society for Microbiology 1752 N St. NW, Washington, DC 20036 -2904
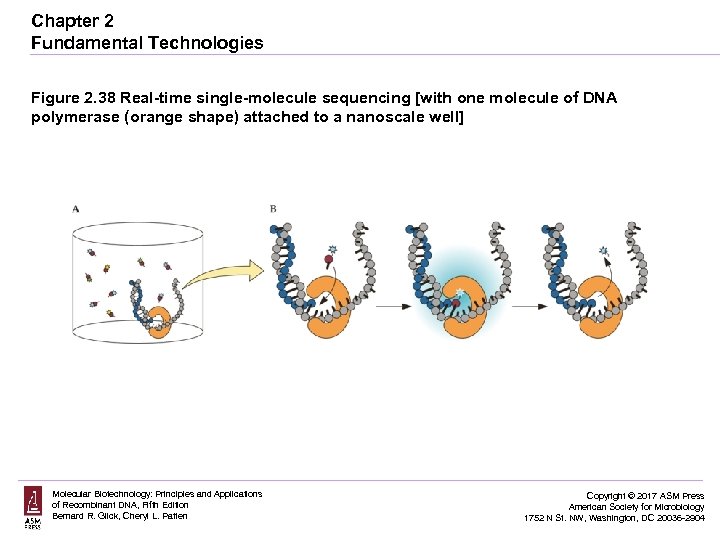 Chapter 2 Fundamental Technologies Figure 2. 38 Real-time single-molecule sequencing [with one molecule of DNA polymerase (orange shape) attached to a nanoscale well] Molecular Biotechnology: Principles and Applications of Recombinant DNA, Fifth Edition Bernard R. Glick, Cheryl L. Patten Copyright © 2017 ASM Press American Society for Microbiology 1752 N St. NW, Washington, DC 20036 -2904
Chapter 2 Fundamental Technologies Figure 2. 38 Real-time single-molecule sequencing [with one molecule of DNA polymerase (orange shape) attached to a nanoscale well] Molecular Biotechnology: Principles and Applications of Recombinant DNA, Fifth Edition Bernard R. Glick, Cheryl L. Patten Copyright © 2017 ASM Press American Society for Microbiology 1752 N St. NW, Washington, DC 20036 -2904
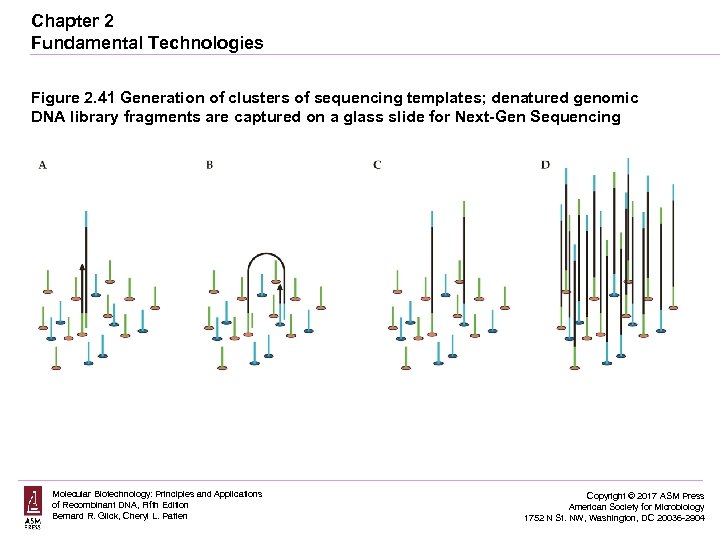 Chapter 2 Fundamental Technologies Figure 2. 41 Generation of clusters of sequencing templates; denatured genomic DNA library fragments are captured on a glass slide for Next-Gen Sequencing Molecular Biotechnology: Principles and Applications of Recombinant DNA, Fifth Edition Bernard R. Glick, Cheryl L. Patten Copyright © 2017 ASM Press American Society for Microbiology 1752 N St. NW, Washington, DC 20036 -2904
Chapter 2 Fundamental Technologies Figure 2. 41 Generation of clusters of sequencing templates; denatured genomic DNA library fragments are captured on a glass slide for Next-Gen Sequencing Molecular Biotechnology: Principles and Applications of Recombinant DNA, Fifth Edition Bernard R. Glick, Cheryl L. Patten Copyright © 2017 ASM Press American Society for Microbiology 1752 N St. NW, Washington, DC 20036 -2904
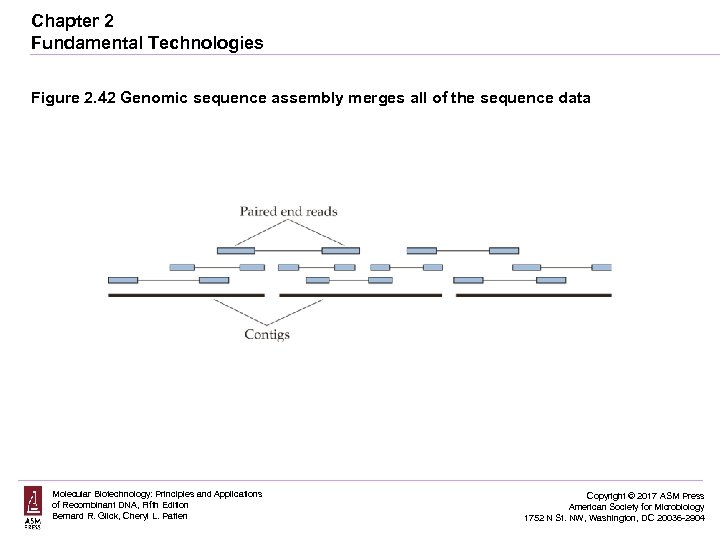 Chapter 2 Fundamental Technologies Figure 2. 42 Genomic sequence assembly merges all of the sequence data Molecular Biotechnology: Principles and Applications of Recombinant DNA, Fifth Edition Bernard R. Glick, Cheryl L. Patten Copyright © 2017 ASM Press American Society for Microbiology 1752 N St. NW, Washington, DC 20036 -2904
Chapter 2 Fundamental Technologies Figure 2. 42 Genomic sequence assembly merges all of the sequence data Molecular Biotechnology: Principles and Applications of Recombinant DNA, Fifth Edition Bernard R. Glick, Cheryl L. Patten Copyright © 2017 ASM Press American Society for Microbiology 1752 N St. NW, Washington, DC 20036 -2904
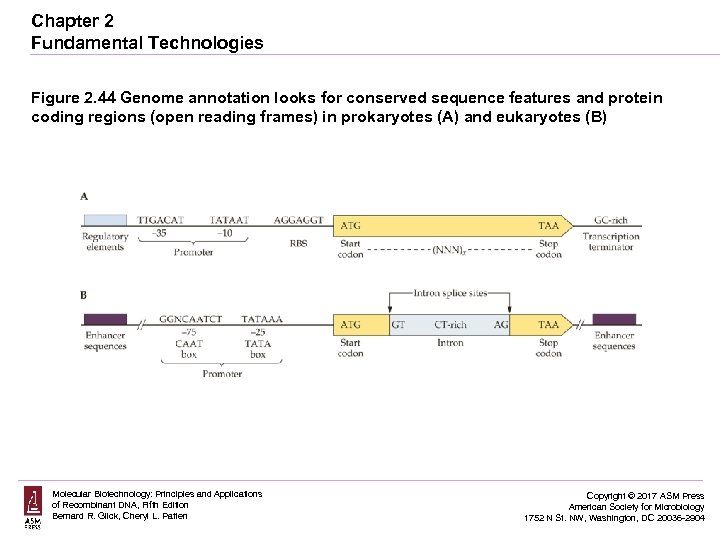 Chapter 2 Fundamental Technologies Figure 2. 44 Genome annotation looks for conserved sequence features and protein coding regions (open reading frames) in prokaryotes (A) and eukaryotes (B) Molecular Biotechnology: Principles and Applications of Recombinant DNA, Fifth Edition Bernard R. Glick, Cheryl L. Patten Copyright © 2017 ASM Press American Society for Microbiology 1752 N St. NW, Washington, DC 20036 -2904
Chapter 2 Fundamental Technologies Figure 2. 44 Genome annotation looks for conserved sequence features and protein coding regions (open reading frames) in prokaryotes (A) and eukaryotes (B) Molecular Biotechnology: Principles and Applications of Recombinant DNA, Fifth Edition Bernard R. Glick, Cheryl L. Patten Copyright © 2017 ASM Press American Society for Microbiology 1752 N St. NW, Washington, DC 20036 -2904
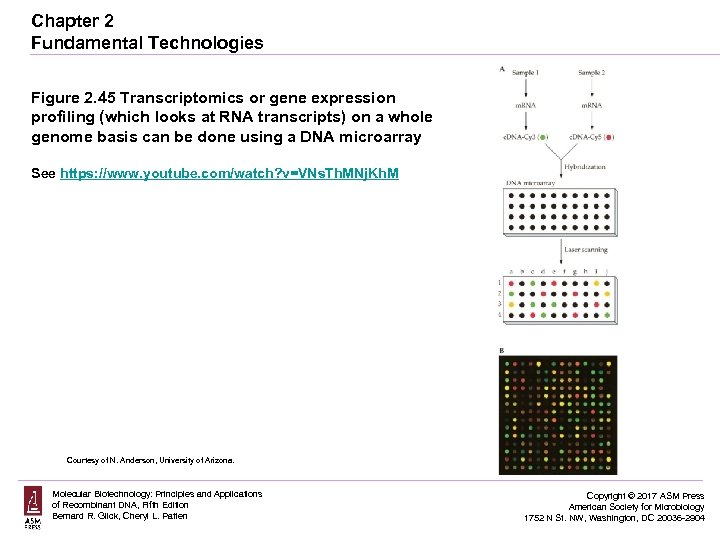 Chapter 2 Fundamental Technologies Figure 2. 45 Transcriptomics or gene expression profiling (which looks at RNA transcripts) on a whole genome basis can be done using a DNA microarray See https: //www. youtube. com/watch? v=VNs. Th. MNj. Kh. M Courtesy of N. Anderson, University of Arizona. Molecular Biotechnology: Principles and Applications of Recombinant DNA, Fifth Edition Bernard R. Glick, Cheryl L. Patten Copyright © 2017 ASM Press American Society for Microbiology 1752 N St. NW, Washington, DC 20036 -2904
Chapter 2 Fundamental Technologies Figure 2. 45 Transcriptomics or gene expression profiling (which looks at RNA transcripts) on a whole genome basis can be done using a DNA microarray See https: //www. youtube. com/watch? v=VNs. Th. MNj. Kh. M Courtesy of N. Anderson, University of Arizona. Molecular Biotechnology: Principles and Applications of Recombinant DNA, Fifth Edition Bernard R. Glick, Cheryl L. Patten Copyright © 2017 ASM Press American Society for Microbiology 1752 N St. NW, Washington, DC 20036 -2904
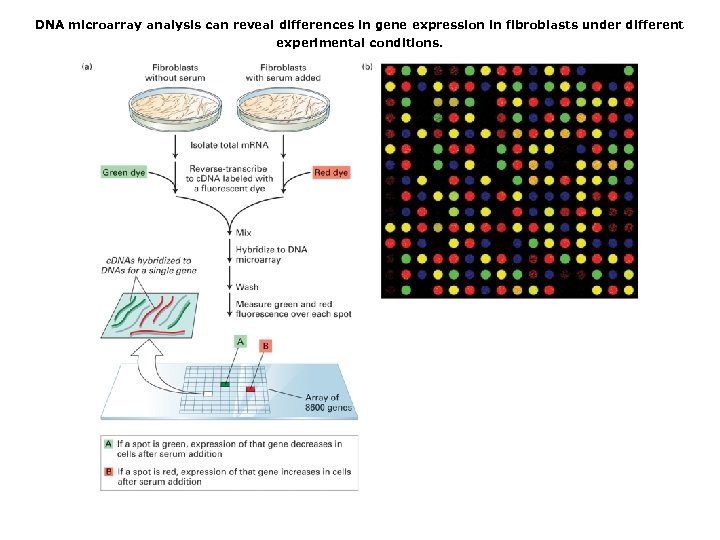 DNA microarray analysis can reveal differences in gene expression in fibroblasts under different experimental conditions.
DNA microarray analysis can reveal differences in gene expression in fibroblasts under different experimental conditions.
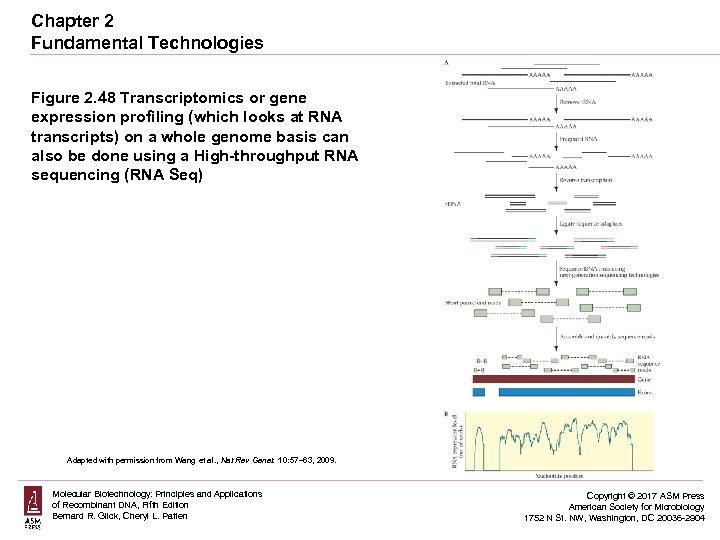 Chapter 2 Fundamental Technologies Figure 2. 48 Transcriptomics or gene expression profiling (which looks at RNA transcripts) on a whole genome basis can also be done using a High-throughput RNA sequencing (RNA Seq) Adapted with permission from Wang et al. , Nat Rev Genet. 10: 57– 63, 2009. Molecular Biotechnology: Principles and Applications of Recombinant DNA, Fifth Edition Bernard R. Glick, Cheryl L. Patten Copyright © 2017 ASM Press American Society for Microbiology 1752 N St. NW, Washington, DC 20036 -2904
Chapter 2 Fundamental Technologies Figure 2. 48 Transcriptomics or gene expression profiling (which looks at RNA transcripts) on a whole genome basis can also be done using a High-throughput RNA sequencing (RNA Seq) Adapted with permission from Wang et al. , Nat Rev Genet. 10: 57– 63, 2009. Molecular Biotechnology: Principles and Applications of Recombinant DNA, Fifth Edition Bernard R. Glick, Cheryl L. Patten Copyright © 2017 ASM Press American Society for Microbiology 1752 N St. NW, Washington, DC 20036 -2904
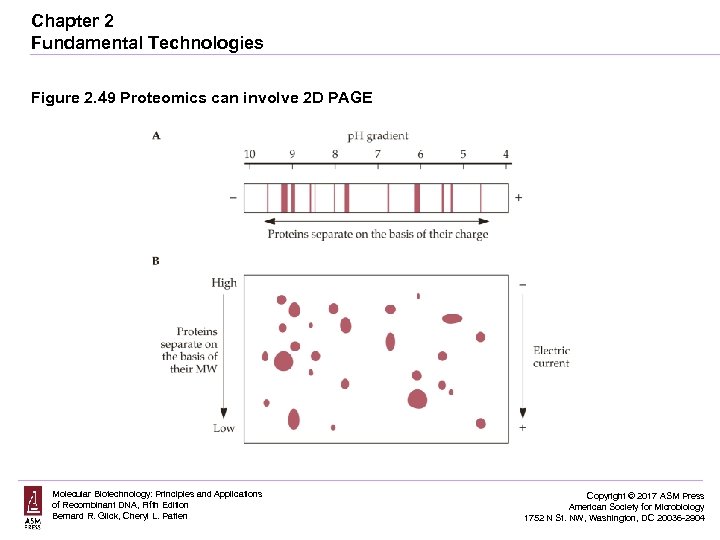 Chapter 2 Fundamental Technologies Figure 2. 49 Proteomics can involve 2 D PAGE Molecular Biotechnology: Principles and Applications of Recombinant DNA, Fifth Edition Bernard R. Glick, Cheryl L. Patten Copyright © 2017 ASM Press American Society for Microbiology 1752 N St. NW, Washington, DC 20036 -2904
Chapter 2 Fundamental Technologies Figure 2. 49 Proteomics can involve 2 D PAGE Molecular Biotechnology: Principles and Applications of Recombinant DNA, Fifth Edition Bernard R. Glick, Cheryl L. Patten Copyright © 2017 ASM Press American Society for Microbiology 1752 N St. NW, Washington, DC 20036 -2904
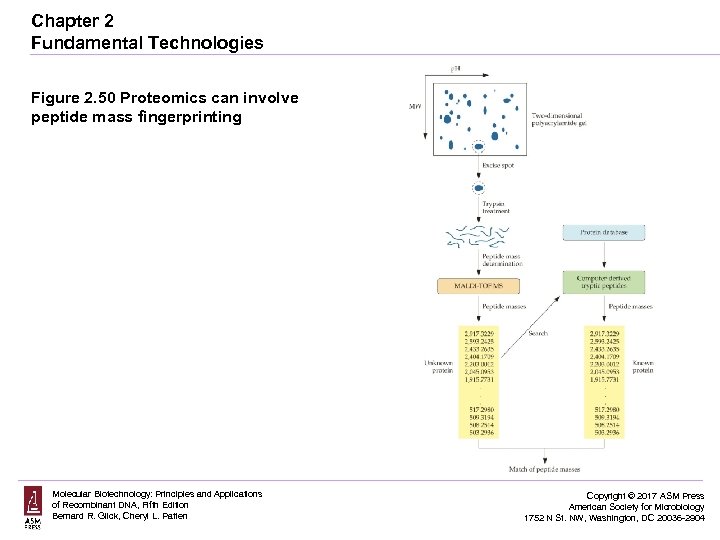 Chapter 2 Fundamental Technologies Figure 2. 50 Proteomics can involve peptide mass fingerprinting Molecular Biotechnology: Principles and Applications of Recombinant DNA, Fifth Edition Bernard R. Glick, Cheryl L. Patten Copyright © 2017 ASM Press American Society for Microbiology 1752 N St. NW, Washington, DC 20036 -2904
Chapter 2 Fundamental Technologies Figure 2. 50 Proteomics can involve peptide mass fingerprinting Molecular Biotechnology: Principles and Applications of Recombinant DNA, Fifth Edition Bernard R. Glick, Cheryl L. Patten Copyright © 2017 ASM Press American Society for Microbiology 1752 N St. NW, Washington, DC 20036 -2904
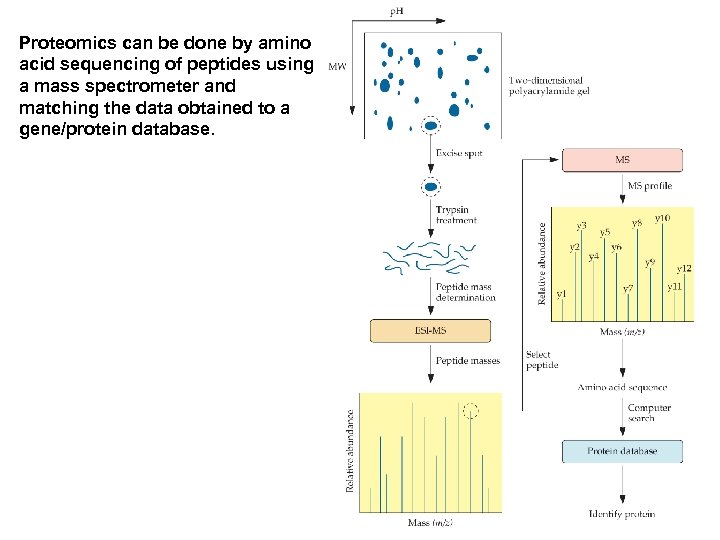 Proteomics can be done by amino acid sequencing of peptides using a mass spectrometer and matching the data obtained to a gene/protein database.
Proteomics can be done by amino acid sequencing of peptides using a mass spectrometer and matching the data obtained to a gene/protein database.
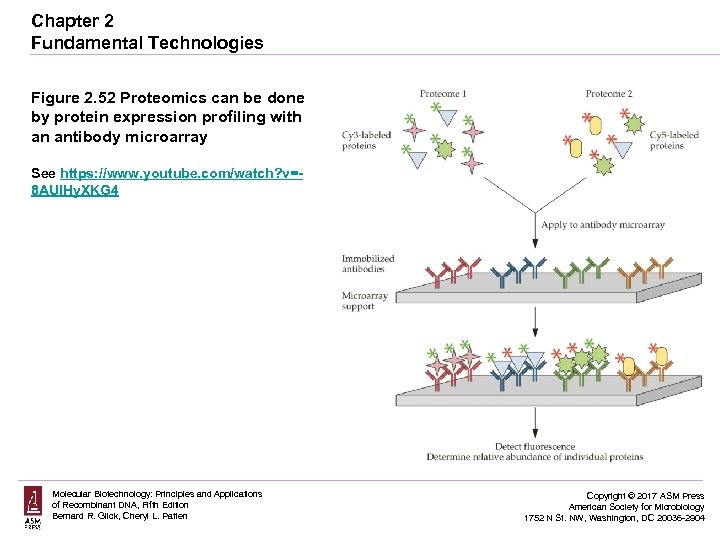 Chapter 2 Fundamental Technologies Figure 2. 52 Proteomics can be done by protein expression profiling with an antibody microarray See https: //www. youtube. com/watch? v=8 AUIHy. XKG 4 Molecular Biotechnology: Principles and Applications of Recombinant DNA, Fifth Edition Bernard R. Glick, Cheryl L. Patten Copyright © 2017 ASM Press American Society for Microbiology 1752 N St. NW, Washington, DC 20036 -2904
Chapter 2 Fundamental Technologies Figure 2. 52 Proteomics can be done by protein expression profiling with an antibody microarray See https: //www. youtube. com/watch? v=8 AUIHy. XKG 4 Molecular Biotechnology: Principles and Applications of Recombinant DNA, Fifth Edition Bernard R. Glick, Cheryl L. Patten Copyright © 2017 ASM Press American Society for Microbiology 1752 N St. NW, Washington, DC 20036 -2904
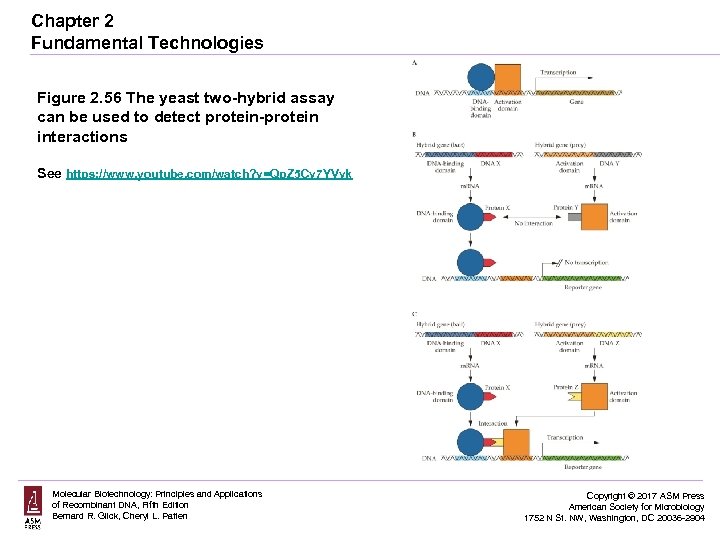 Chapter 2 Fundamental Technologies Figure 2. 56 The yeast two-hybrid assay can be used to detect protein-protein interactions See https: //www. youtube. com/watch? v=Qp. Z 5 Cv 7 YVvk Molecular Biotechnology: Principles and Applications of Recombinant DNA, Fifth Edition Bernard R. Glick, Cheryl L. Patten Copyright © 2017 ASM Press American Society for Microbiology 1752 N St. NW, Washington, DC 20036 -2904
Chapter 2 Fundamental Technologies Figure 2. 56 The yeast two-hybrid assay can be used to detect protein-protein interactions See https: //www. youtube. com/watch? v=Qp. Z 5 Cv 7 YVvk Molecular Biotechnology: Principles and Applications of Recombinant DNA, Fifth Edition Bernard R. Glick, Cheryl L. Patten Copyright © 2017 ASM Press American Society for Microbiology 1752 N St. NW, Washington, DC 20036 -2904
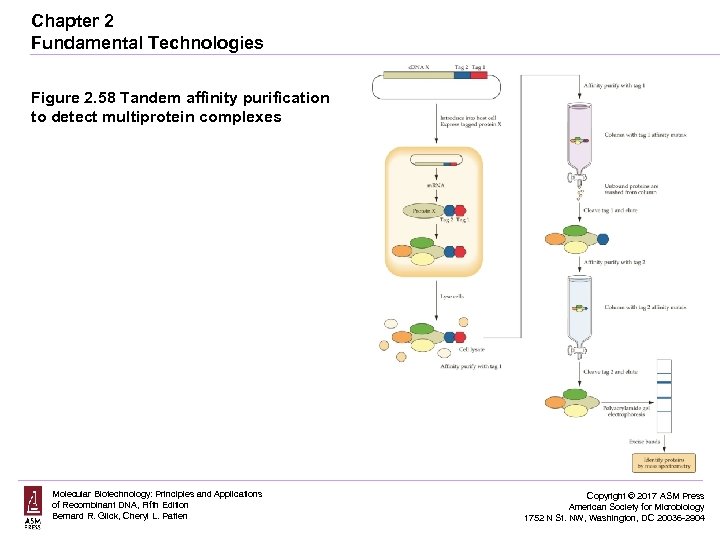 Chapter 2 Fundamental Technologies Figure 2. 58 Tandem affinity purification to detect multiprotein complexes Molecular Biotechnology: Principles and Applications of Recombinant DNA, Fifth Edition Bernard R. Glick, Cheryl L. Patten Copyright © 2017 ASM Press American Society for Microbiology 1752 N St. NW, Washington, DC 20036 -2904
Chapter 2 Fundamental Technologies Figure 2. 58 Tandem affinity purification to detect multiprotein complexes Molecular Biotechnology: Principles and Applications of Recombinant DNA, Fifth Edition Bernard R. Glick, Cheryl L. Patten Copyright © 2017 ASM Press American Society for Microbiology 1752 N St. NW, Washington, DC 20036 -2904
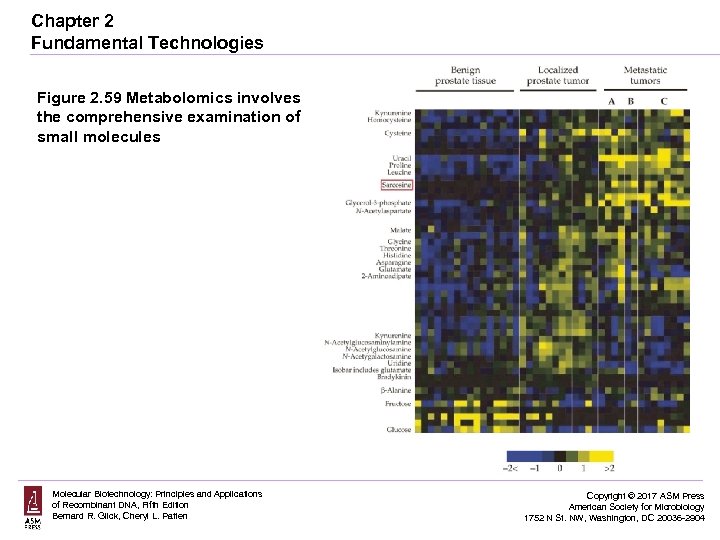 Chapter 2 Fundamental Technologies Figure 2. 59 Metabolomics involves the comprehensive examination of small molecules Molecular Biotechnology: Principles and Applications of Recombinant DNA, Fifth Edition Bernard R. Glick, Cheryl L. Patten Copyright © 2017 ASM Press American Society for Microbiology 1752 N St. NW, Washington, DC 20036 -2904
Chapter 2 Fundamental Technologies Figure 2. 59 Metabolomics involves the comprehensive examination of small molecules Molecular Biotechnology: Principles and Applications of Recombinant DNA, Fifth Edition Bernard R. Glick, Cheryl L. Patten Copyright © 2017 ASM Press American Society for Microbiology 1752 N St. NW, Washington, DC 20036 -2904
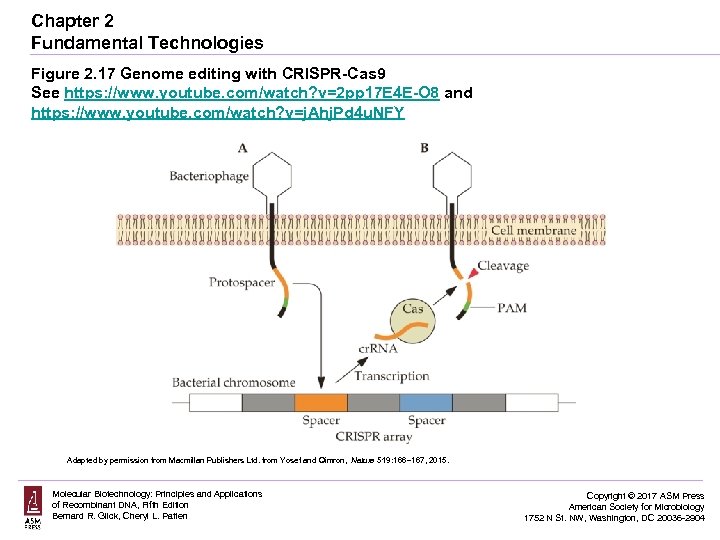 Chapter 2 Fundamental Technologies Figure 2. 17 Genome editing with CRISPR-Cas 9 See https: //www. youtube. com/watch? v=2 pp 17 E 4 E-O 8 and https: //www. youtube. com/watch? v=j. Ahj. Pd 4 u. NFY Adapted by permission from Macmillan Publishers Ltd. from Yosef and Qimron, Nature 519: 166– 167, 2015. Molecular Biotechnology: Principles and Applications of Recombinant DNA, Fifth Edition Bernard R. Glick, Cheryl L. Patten Copyright © 2017 ASM Press American Society for Microbiology 1752 N St. NW, Washington, DC 20036 -2904
Chapter 2 Fundamental Technologies Figure 2. 17 Genome editing with CRISPR-Cas 9 See https: //www. youtube. com/watch? v=2 pp 17 E 4 E-O 8 and https: //www. youtube. com/watch? v=j. Ahj. Pd 4 u. NFY Adapted by permission from Macmillan Publishers Ltd. from Yosef and Qimron, Nature 519: 166– 167, 2015. Molecular Biotechnology: Principles and Applications of Recombinant DNA, Fifth Edition Bernard R. Glick, Cheryl L. Patten Copyright © 2017 ASM Press American Society for Microbiology 1752 N St. NW, Washington, DC 20036 -2904
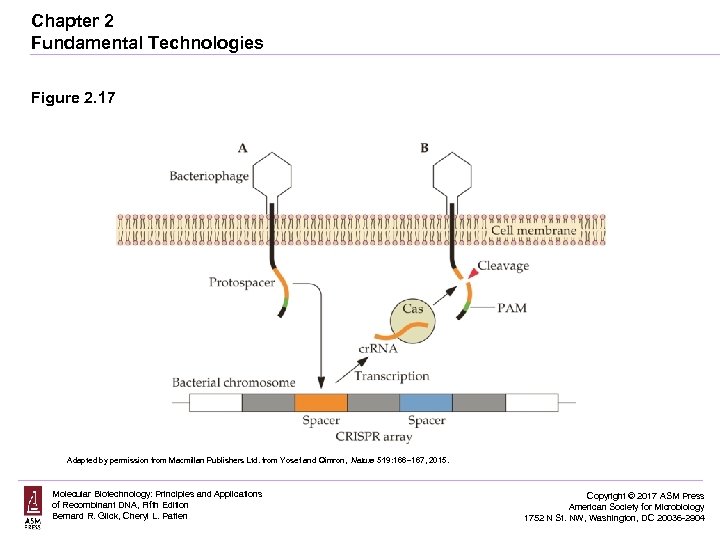 Chapter 2 Fundamental Technologies Figure 2. 17 Adapted by permission from Macmillan Publishers Ltd. from Yosef and Qimron, Nature 519: 166– 167, 2015. Molecular Biotechnology: Principles and Applications of Recombinant DNA, Fifth Edition Bernard R. Glick, Cheryl L. Patten Copyright © 2017 ASM Press American Society for Microbiology 1752 N St. NW, Washington, DC 20036 -2904
Chapter 2 Fundamental Technologies Figure 2. 17 Adapted by permission from Macmillan Publishers Ltd. from Yosef and Qimron, Nature 519: 166– 167, 2015. Molecular Biotechnology: Principles and Applications of Recombinant DNA, Fifth Edition Bernard R. Glick, Cheryl L. Patten Copyright © 2017 ASM Press American Society for Microbiology 1752 N St. NW, Washington, DC 20036 -2904
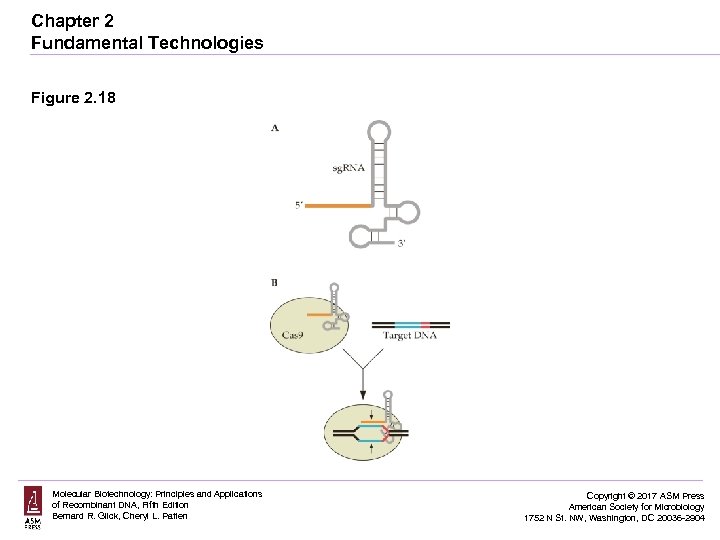 Chapter 2 Fundamental Technologies Figure 2. 18 Molecular Biotechnology: Principles and Applications of Recombinant DNA, Fifth Edition Bernard R. Glick, Cheryl L. Patten Copyright © 2017 ASM Press American Society for Microbiology 1752 N St. NW, Washington, DC 20036 -2904
Chapter 2 Fundamental Technologies Figure 2. 18 Molecular Biotechnology: Principles and Applications of Recombinant DNA, Fifth Edition Bernard R. Glick, Cheryl L. Patten Copyright © 2017 ASM Press American Society for Microbiology 1752 N St. NW, Washington, DC 20036 -2904
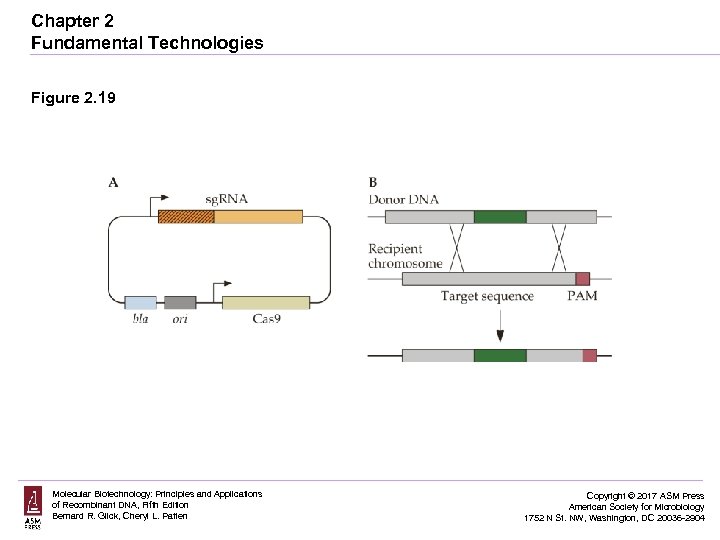 Chapter 2 Fundamental Technologies Figure 2. 19 Molecular Biotechnology: Principles and Applications of Recombinant DNA, Fifth Edition Bernard R. Glick, Cheryl L. Patten Copyright © 2017 ASM Press American Society for Microbiology 1752 N St. NW, Washington, DC 20036 -2904
Chapter 2 Fundamental Technologies Figure 2. 19 Molecular Biotechnology: Principles and Applications of Recombinant DNA, Fifth Edition Bernard R. Glick, Cheryl L. Patten Copyright © 2017 ASM Press American Society for Microbiology 1752 N St. NW, Washington, DC 20036 -2904


