727d862d85ab2a6c514d768d627d094f.ppt
- Количество слайдов: 37
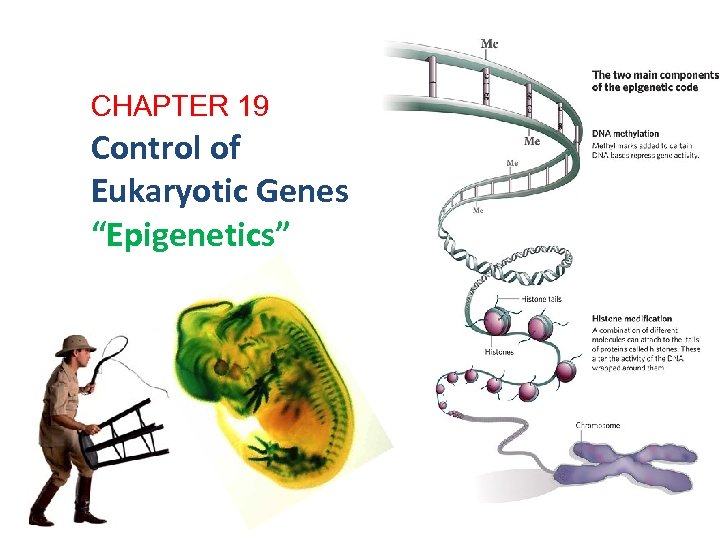 CHAPTER 19 Control of Eukaryotic Genes “Epigenetics” 2007 -2008
CHAPTER 19 Control of Eukaryotic Genes “Epigenetics” 2007 -2008
 The BIG Questions… • How are genes turned on & off in eukaryotes? • How do cells with the same genes differentiate to perform completely different, specialized functions?
The BIG Questions… • How are genes turned on & off in eukaryotes? • How do cells with the same genes differentiate to perform completely different, specialized functions?
 REVIEW Evolution of gene regulation • Prokaryotes – single-celled – evolved to grow & divide rapidly – must respond quickly to changes in external environment • exploit transient resources • Gene regulation = (? ) Operons – turn genes on & off rapidly • flexibility & reversibility – adjust levels of enzymes for synthesis & digestion
REVIEW Evolution of gene regulation • Prokaryotes – single-celled – evolved to grow & divide rapidly – must respond quickly to changes in external environment • exploit transient resources • Gene regulation = (? ) Operons – turn genes on & off rapidly • flexibility & reversibility – adjust levels of enzymes for synthesis & digestion
 Evolution of gene regulation • Eukaryotes – Multicellular = only expresses a fraction of its genes – evolved to maintain constant internal conditions even with changing conditions • (? ) Homeostasis • must REGULATE the body as a whole rather than serve the needs of individual cells – Also need to regulate: • (? ) growth & development – long term processes • (? ) specialization – turn on & off large number of genes
Evolution of gene regulation • Eukaryotes – Multicellular = only expresses a fraction of its genes – evolved to maintain constant internal conditions even with changing conditions • (? ) Homeostasis • must REGULATE the body as a whole rather than serve the needs of individual cells – Also need to regulate: • (? ) growth & development – long term processes • (? ) specialization – turn on & off large number of genes
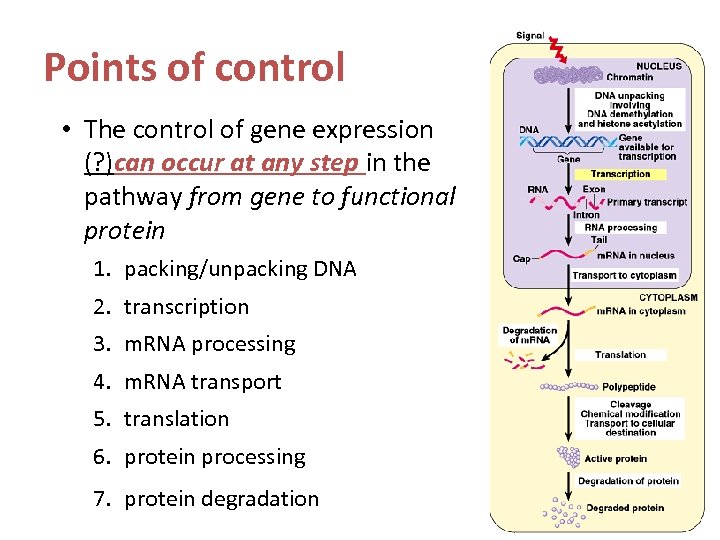 Points of control • The control of gene expression (? )can occur at any step in the pathway from gene to functional protein 1. packing/unpacking DNA 2. transcription 3. m. RNA processing 4. m. RNA transport 5. translation 6. protein processing 7. protein degradation
Points of control • The control of gene expression (? )can occur at any step in the pathway from gene to functional protein 1. packing/unpacking DNA 2. transcription 3. m. RNA processing 4. m. RNA transport 5. translation 6. protein processing 7. protein degradation
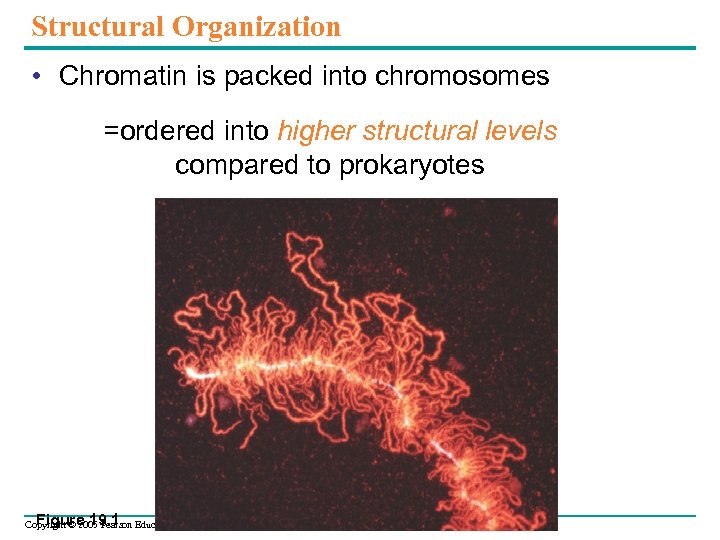 Structural Organization • Chromatin is packed into chromosomes =ordered into higher structural levels compared to prokaryotes Figure 19. 1 Copyright © 2005 Pearson Education, Inc. publishing as Benjamin Cummings
Structural Organization • Chromatin is packed into chromosomes =ordered into higher structural levels compared to prokaryotes Figure 19. 1 Copyright © 2005 Pearson Education, Inc. publishing as Benjamin Cummings
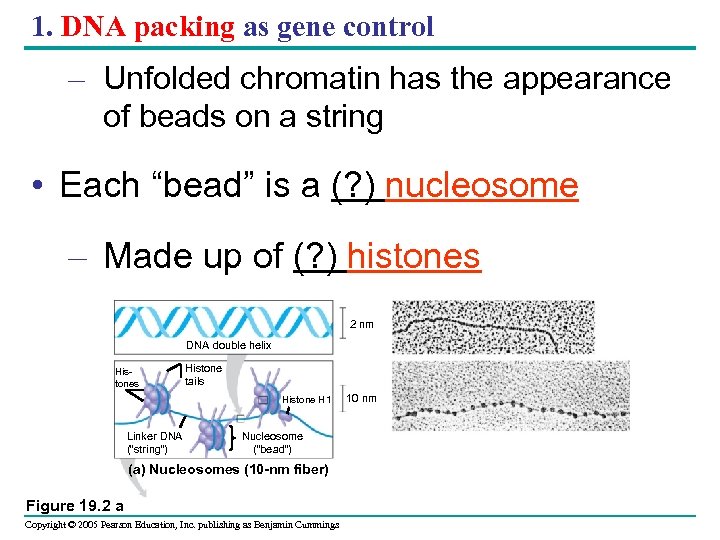 1. DNA packing as gene control – Unfolded chromatin has the appearance of beads on a string • Each “bead” is a (? ) nucleosome – Made up of (? ) histones 2 nm DNA double helix Histones Histone tails Histone H 1 Linker DNA (“string”) Nucleosome (“bead”) (a) Nucleosomes (10 -nm fiber) Figure 19. 2 a Copyright © 2005 Pearson Education, Inc. publishing as Benjamin Cummings 10 nm
1. DNA packing as gene control – Unfolded chromatin has the appearance of beads on a string • Each “bead” is a (? ) nucleosome – Made up of (? ) histones 2 nm DNA double helix Histones Histone tails Histone H 1 Linker DNA (“string”) Nucleosome (“bead”) (a) Nucleosomes (10 -nm fiber) Figure 19. 2 a Copyright © 2005 Pearson Education, Inc. publishing as Benjamin Cummings 10 nm
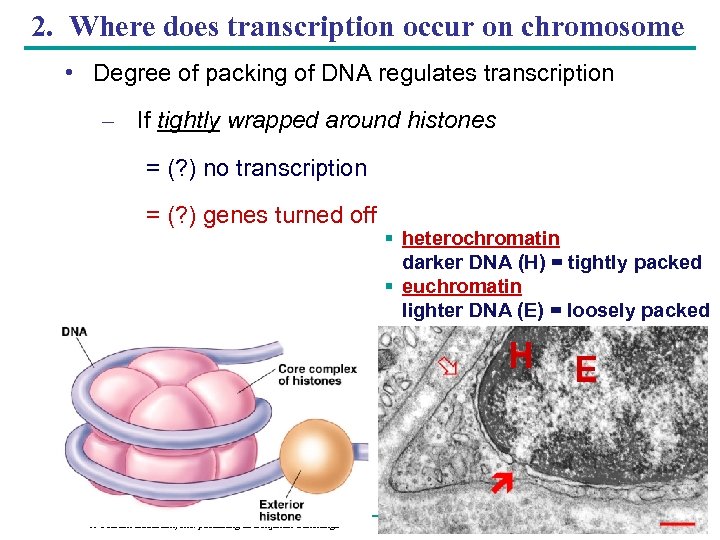 2. Where does transcription occur on chromosome • Degree of packing of DNA regulates transcription – If tightly wrapped around histones = (? ) no transcription = (? ) genes turned off § heterochromatin darker DNA (H) = tightly packed § euchromatin lighter DNA (E) = loosely packed H Copyright © 2005 Pearson Education, Inc. publishing as Benjamin Cummings E
2. Where does transcription occur on chromosome • Degree of packing of DNA regulates transcription – If tightly wrapped around histones = (? ) no transcription = (? ) genes turned off § heterochromatin darker DNA (H) = tightly packed § euchromatin lighter DNA (E) = loosely packed H Copyright © 2005 Pearson Education, Inc. publishing as Benjamin Cummings E
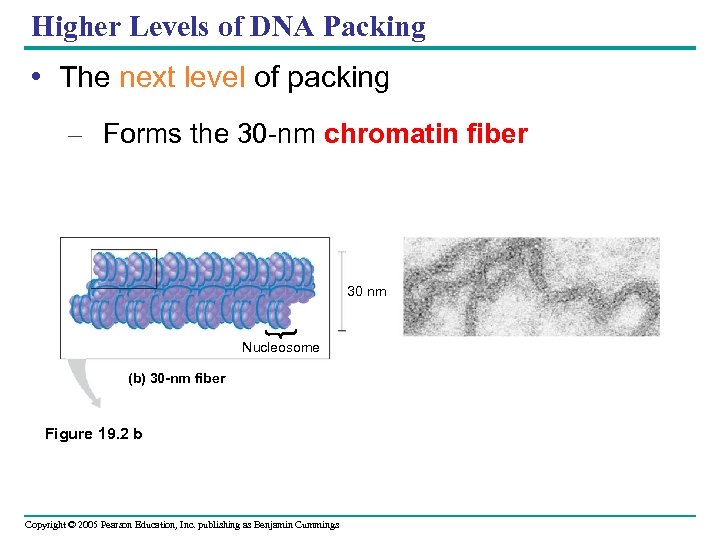 Higher Levels of DNA Packing • The next level of packing – Forms the 30 -nm chromatin fiber 30 nm Nucleosome (b) 30 -nm fiber Figure 19. 2 b Copyright © 2005 Pearson Education, Inc. publishing as Benjamin Cummings
Higher Levels of DNA Packing • The next level of packing – Forms the 30 -nm chromatin fiber 30 nm Nucleosome (b) 30 -nm fiber Figure 19. 2 b Copyright © 2005 Pearson Education, Inc. publishing as Benjamin Cummings
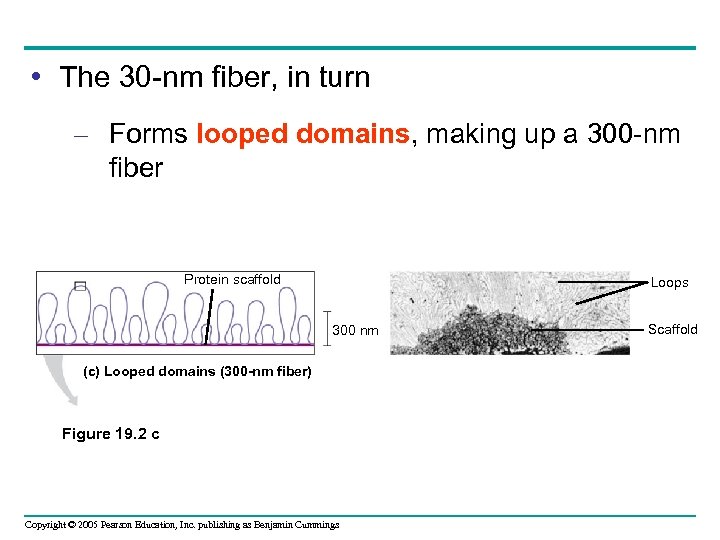 • The 30 -nm fiber, in turn – Forms looped domains, making up a 300 -nm fiber Protein scaffold Loops 300 nm (c) Looped domains (300 -nm fiber) Figure 19. 2 c Copyright © 2005 Pearson Education, Inc. publishing as Benjamin Cummings Scaffold
• The 30 -nm fiber, in turn – Forms looped domains, making up a 300 -nm fiber Protein scaffold Loops 300 nm (c) Looped domains (300 -nm fiber) Figure 19. 2 c Copyright © 2005 Pearson Education, Inc. publishing as Benjamin Cummings Scaffold
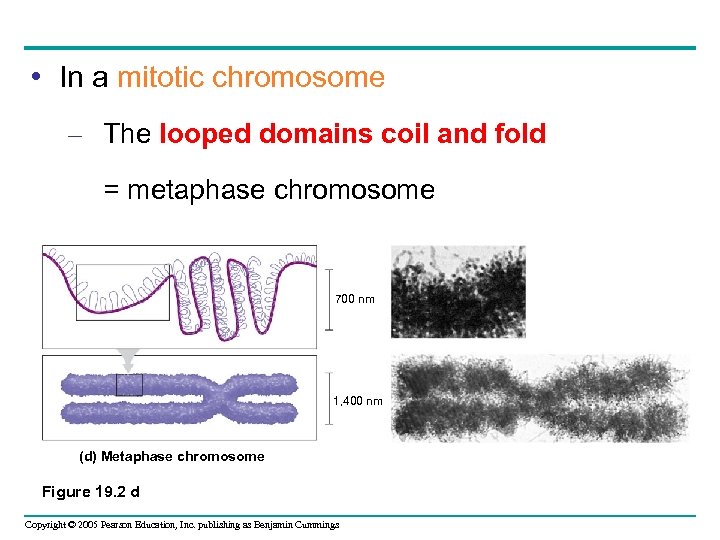 • In a mitotic chromosome – The looped domains coil and fold = metaphase chromosome 700 nm 1, 400 nm (d) Metaphase chromosome Figure 19. 2 d Copyright © 2005 Pearson Education, Inc. publishing as Benjamin Cummings
• In a mitotic chromosome – The looped domains coil and fold = metaphase chromosome 700 nm 1, 400 nm (d) Metaphase chromosome Figure 19. 2 d Copyright © 2005 Pearson Education, Inc. publishing as Benjamin Cummings
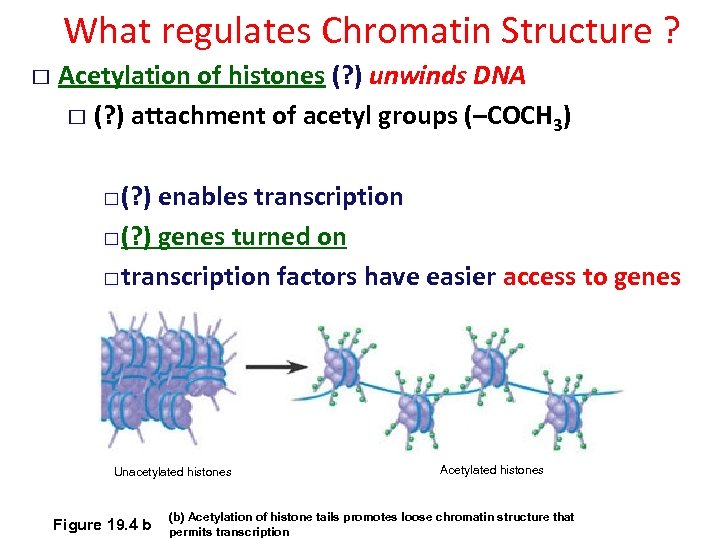 What regulates Chromatin Structure ? Acetylation of histones (? ) unwinds DNA (? ) attachment of acetyl groups (–COCH 3) (? ) enables transcription (? ) genes turned on transcription factors have easier access to genes Unacetylated histones Figure 19. 4 b Acetylated histones (b) Acetylation of histone tails promotes loose chromatin structure that permits transcription
What regulates Chromatin Structure ? Acetylation of histones (? ) unwinds DNA (? ) attachment of acetyl groups (–COCH 3) (? ) enables transcription (? ) genes turned on transcription factors have easier access to genes Unacetylated histones Figure 19. 4 b Acetylated histones (b) Acetylation of histone tails promotes loose chromatin structure that permits transcription
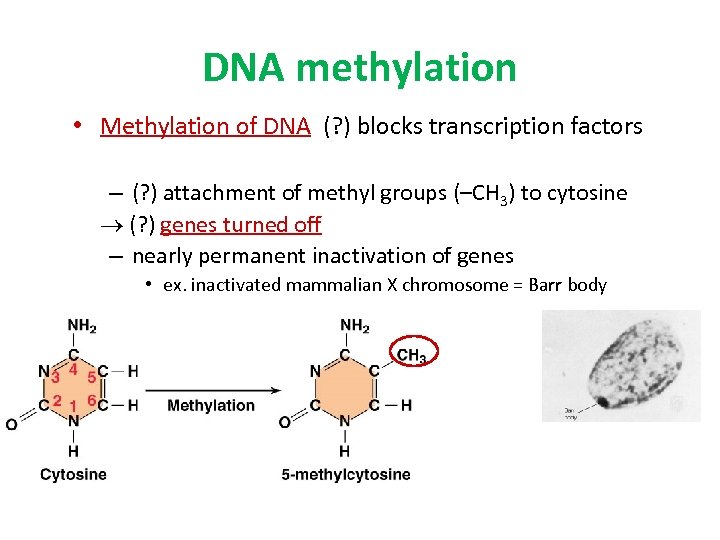 DNA methylation • Methylation of DNA (? ) blocks transcription factors – (? ) attachment of methyl groups (–CH 3) to cytosine (? ) genes turned off – nearly permanent inactivation of genes • ex. inactivated mammalian X chromosome = Barr body
DNA methylation • Methylation of DNA (? ) blocks transcription factors – (? ) attachment of methyl groups (–CH 3) to cytosine (? ) genes turned off – nearly permanent inactivation of genes • ex. inactivated mammalian X chromosome = Barr body
 Epigenetic Inheritence =Terminology for gene expression - ** DNA sequence NOT changed, just the expression of the gene (on or off) - Can Chromatin modifications be passed offspring? (sometimes – poorly understood) **** In a new embryo, all tags are removed except for “imprinted” tags for getting development started
Epigenetic Inheritence =Terminology for gene expression - ** DNA sequence NOT changed, just the expression of the gene (on or off) - Can Chromatin modifications be passed offspring? (sometimes – poorly understood) **** In a new embryo, all tags are removed except for “imprinted” tags for getting development started
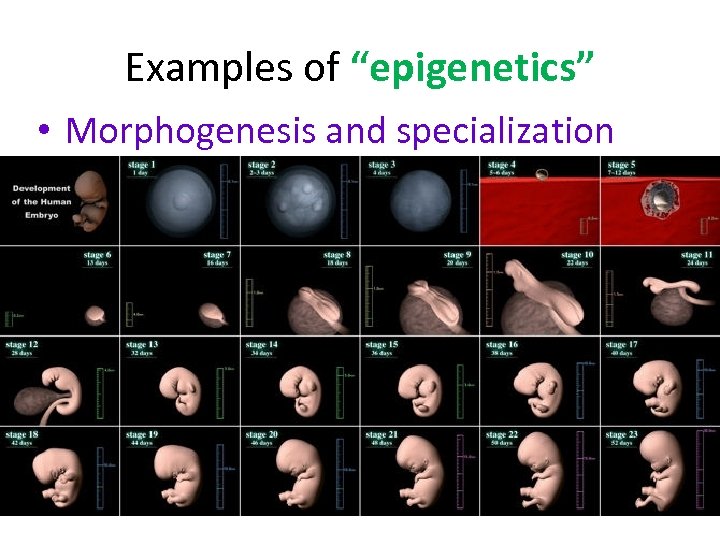 Examples of “epigenetics” • Morphogenesis and specialization
Examples of “epigenetics” • Morphogenesis and specialization
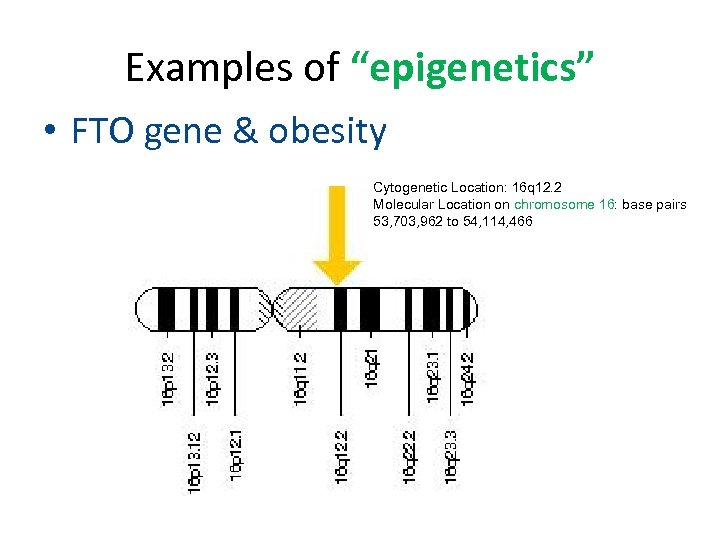 Examples of “epigenetics” • FTO gene & obesity Cytogenetic Location: 16 q 12. 2 Molecular Location on chromosome 16: base pairs 53, 703, 962 to 54, 114, 466
Examples of “epigenetics” • FTO gene & obesity Cytogenetic Location: 16 q 12. 2 Molecular Location on chromosome 16: base pairs 53, 703, 962 to 54, 114, 466
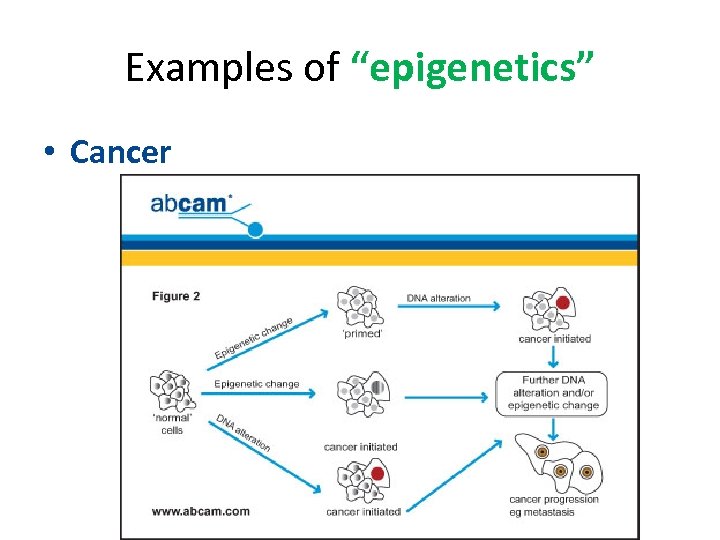 Examples of “epigenetics” • Cancer
Examples of “epigenetics” • Cancer
 Examples of “epigenetics” • Twin Studies…”epigenetic drift” • http: //learn. genetics. utah. edu/content/epigenetics/twins/
Examples of “epigenetics” • Twin Studies…”epigenetic drift” • http: //learn. genetics. utah. edu/content/epigenetics/twins/
 III. PROCESS CONTROLS – (? ) transcription controls seem to be the most “important” factor in gene expression Copyright © 2005 Pearson Education, Inc. publishing as Benjamin Cummings
III. PROCESS CONTROLS – (? ) transcription controls seem to be the most “important” factor in gene expression Copyright © 2005 Pearson Education, Inc. publishing as Benjamin Cummings
 A. The Roles of Transcription Factors • To initiate transcription – (? ) RNA polymerase requires transcription factors (proteins) to bind to the (? ) promotor region Copyright © 2005 Pearson Education, Inc. publishing as Benjamin Cummings
A. The Roles of Transcription Factors • To initiate transcription – (? ) RNA polymerase requires transcription factors (proteins) to bind to the (? ) promotor region Copyright © 2005 Pearson Education, Inc. publishing as Benjamin Cummings
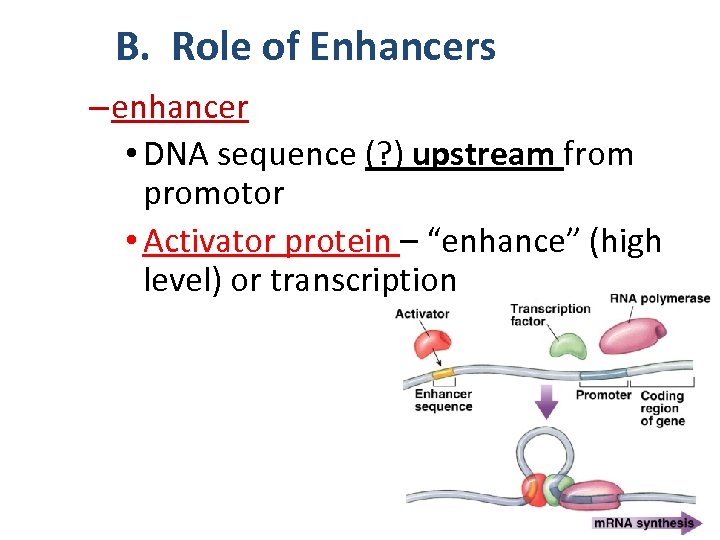 B. Role of Enhancers – enhancer • DNA sequence (? ) upstream from promotor • Activator protein – “enhance” (high level) or transcription
B. Role of Enhancers – enhancer • DNA sequence (? ) upstream from promotor • Activator protein – “enhance” (high level) or transcription
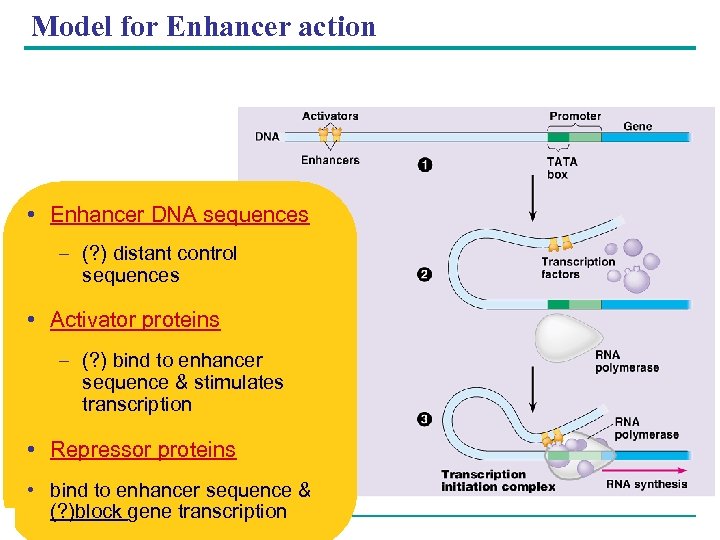 Model for Enhancer action • Enhancer DNA sequences – (? ) distant control sequences • Activator proteins – (? ) bind to enhancer sequence & stimulates transcription • Repressor proteins • bind to enhancer sequence & (? )block gene transcription Copyright © 2005 Pearson Education, Inc. publishing as Benjamin Cummings
Model for Enhancer action • Enhancer DNA sequences – (? ) distant control sequences • Activator proteins – (? ) bind to enhancer sequence & stimulates transcription • Repressor proteins • bind to enhancer sequence & (? )block gene transcription Copyright © 2005 Pearson Education, Inc. publishing as Benjamin Cummings
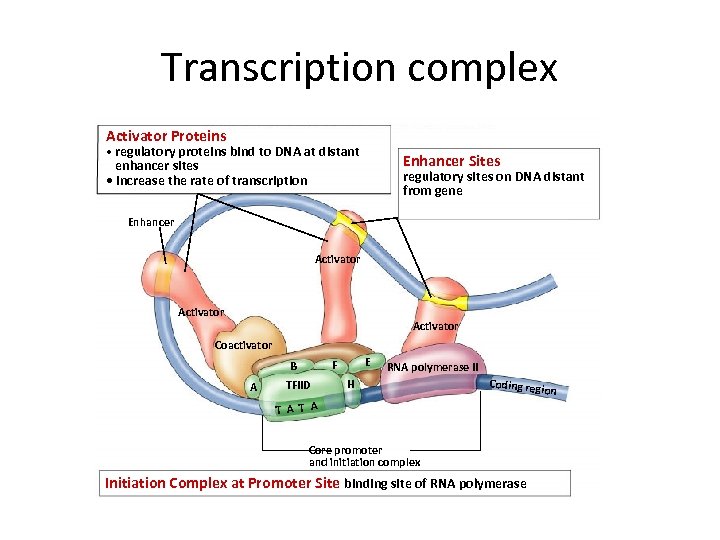 Transcription complex Activator Proteins • regulatory proteins bind to DNA at distant Enhancer Sites enhancer sites • increase the rate of transcription regulatory sites on DNA distant from gene Enhancer Activator Coactivator A E F B TFIID RNA polymerase II H Coding reg T A Core promoter and initiation complex Initiation Complex at Promoter Site binding site of RNA polymerase ion
Transcription complex Activator Proteins • regulatory proteins bind to DNA at distant Enhancer Sites enhancer sites • increase the rate of transcription regulatory sites on DNA distant from gene Enhancer Activator Coactivator A E F B TFIID RNA polymerase II H Coding reg T A Core promoter and initiation complex Initiation Complex at Promoter Site binding site of RNA polymerase ion
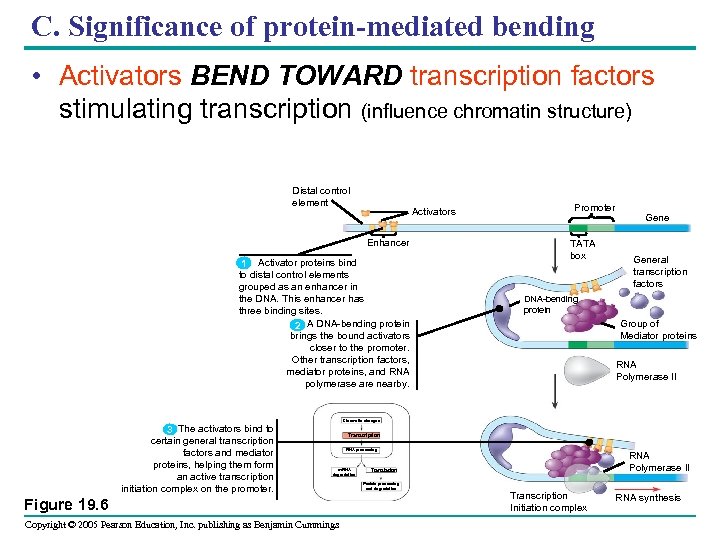 C. Significance of protein-mediated bending • Activators BEND TOWARD transcription factors stimulating transcription (influence chromatin structure) Distal control element Activators Enhancer 1 Activator proteins bind to distal control elements grouped as an enhancer in the DNA. This enhancer has three binding sites. 2 A DNA-bending protein brings the bound activators closer to the promoter. Other transcription factors, mediator proteins, and RNA polymerase are nearby. Promoter TATA box General transcription factors DNA-bending protein Group of Mediator proteins RNA Polymerase II Chromatin changes 3 The activators bind to certain general transcription factors and mediator proteins, helping them form an active transcription initiation complex on the promoter. Transcription RNA processing m. RNA degradation Figure 19. 6 Copyright © 2005 Pearson Education, Inc. publishing as Benjamin Cummings RNA Polymerase II Translation Protein processing and degradation Transcription Initiation complex RNA synthesis
C. Significance of protein-mediated bending • Activators BEND TOWARD transcription factors stimulating transcription (influence chromatin structure) Distal control element Activators Enhancer 1 Activator proteins bind to distal control elements grouped as an enhancer in the DNA. This enhancer has three binding sites. 2 A DNA-bending protein brings the bound activators closer to the promoter. Other transcription factors, mediator proteins, and RNA polymerase are nearby. Promoter TATA box General transcription factors DNA-bending protein Group of Mediator proteins RNA Polymerase II Chromatin changes 3 The activators bind to certain general transcription factors and mediator proteins, helping them form an active transcription initiation complex on the promoter. Transcription RNA processing m. RNA degradation Figure 19. 6 Copyright © 2005 Pearson Education, Inc. publishing as Benjamin Cummings RNA Polymerase II Translation Protein processing and degradation Transcription Initiation complex RNA synthesis
 2. Post-transcriptional controls = RNA modification * Gene expression can be blocked or stimulated during RNA modification Copyright © 2005 Pearson Education, Inc. publishing as Benjamin Cummings
2. Post-transcriptional controls = RNA modification * Gene expression can be blocked or stimulated during RNA modification Copyright © 2005 Pearson Education, Inc. publishing as Benjamin Cummings
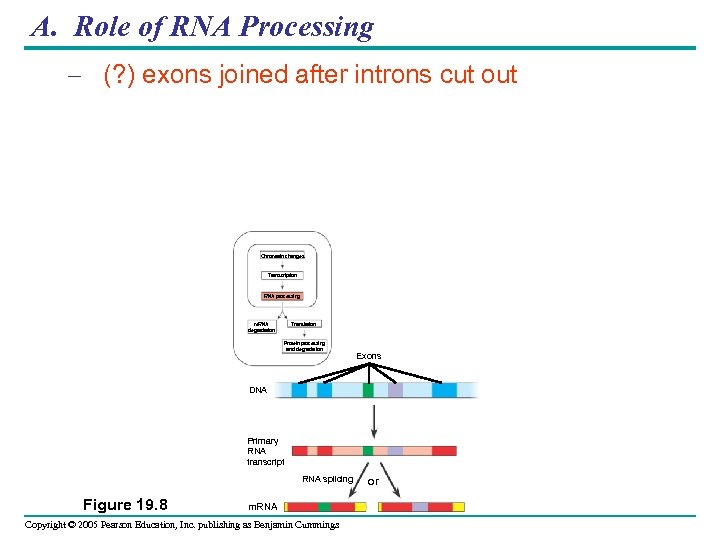 A. Role of RNA Processing – (? ) exons joined after introns cut out Chromatin changes Transcription RNA processing m. RNA degradation Translation Protein processing and degradation Exons DNA Primary RNA transcript RNA splicing Figure 19. 8 m. RNA Copyright © 2005 Pearson Education, Inc. publishing as Benjamin Cummings or
A. Role of RNA Processing – (? ) exons joined after introns cut out Chromatin changes Transcription RNA processing m. RNA degradation Translation Protein processing and degradation Exons DNA Primary RNA transcript RNA splicing Figure 19. 8 m. RNA Copyright © 2005 Pearson Education, Inc. publishing as Benjamin Cummings or
 B. Role of m. RNA Degradation • The life span of m. RNA molecules in the cytoplasm (hours to weeks) – Degradation of the leader (5’cap) and trailer regions (poly-A tail) by enzymes – Prokaryotes vs. Eukaryotes lifespan Copyright © 2005 Pearson Education, Inc. publishing as Benjamin Cummings
B. Role of m. RNA Degradation • The life span of m. RNA molecules in the cytoplasm (hours to weeks) – Degradation of the leader (5’cap) and trailer regions (poly-A tail) by enzymes – Prokaryotes vs. Eukaryotes lifespan Copyright © 2005 Pearson Education, Inc. publishing as Benjamin Cummings
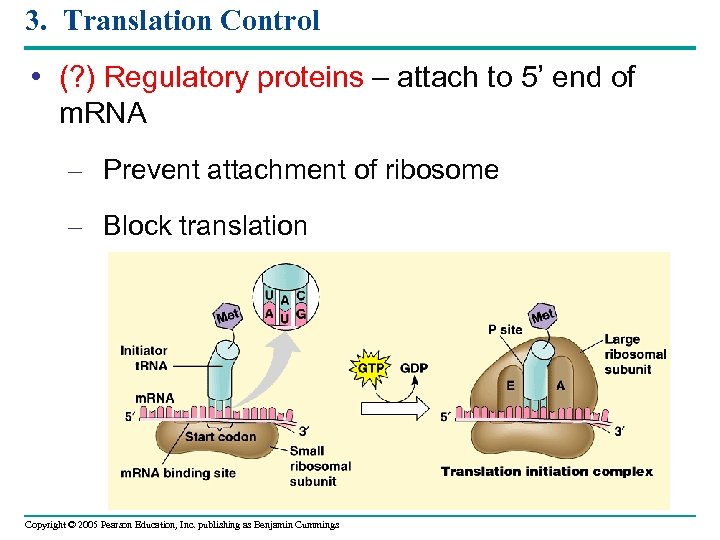 3. Translation Control • (? ) Regulatory proteins – attach to 5’ end of m. RNA – Prevent attachment of ribosome – Block translation Copyright © 2005 Pearson Education, Inc. publishing as Benjamin Cummings
3. Translation Control • (? ) Regulatory proteins – attach to 5’ end of m. RNA – Prevent attachment of ribosome – Block translation Copyright © 2005 Pearson Education, Inc. publishing as Benjamin Cummings
 B. Protein Processing • After translation – (? )protein processing/modification are controlled by cellular events (Endo. System) = folding, cleaving, adding sugar groups, targeting for transport Copyright © 2005 Pearson Education, Inc. publishing as Benjamin Cummings
B. Protein Processing • After translation – (? )protein processing/modification are controlled by cellular events (Endo. System) = folding, cleaving, adding sugar groups, targeting for transport Copyright © 2005 Pearson Education, Inc. publishing as Benjamin Cummings
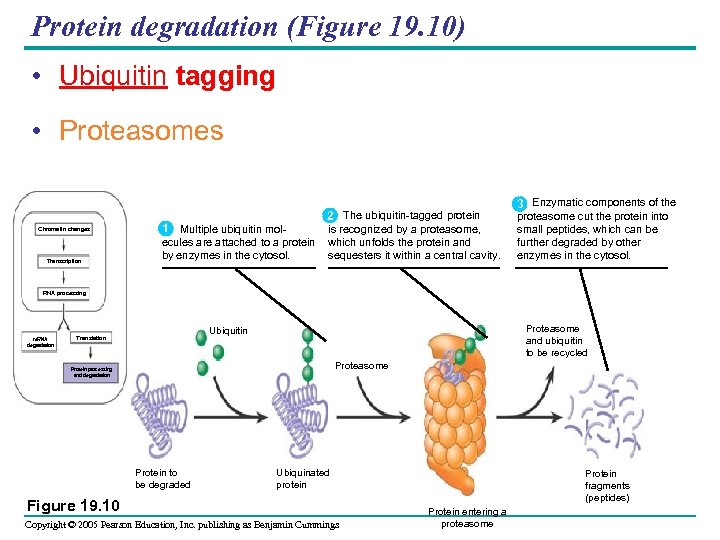 Protein degradation (Figure 19. 10) • Ubiquitin tagging • Proteasomes Chromatin changes Transcription 1 Multiple ubiquitin molecules are attached to a protein by enzymes in the cytosol. 2 The ubiquitin-tagged protein is recognized by a proteasome, which unfolds the protein and sequesters it within a central cavity. 3 Enzymatic components of the proteasome cut the protein into small peptides, which can be further degraded by other enzymes in the cytosol. RNA processing m. RNA degradation Proteasome and ubiquitin to be recycled Ubiquitin Translation Proteasome Protein processing and degradation Protein to be degraded Ubiquinated protein Figure 19. 10 Copyright © 2005 Pearson Education, Inc. publishing as Benjamin Cummings Protein fragments (peptides) Protein entering a proteasome
Protein degradation (Figure 19. 10) • Ubiquitin tagging • Proteasomes Chromatin changes Transcription 1 Multiple ubiquitin molecules are attached to a protein by enzymes in the cytosol. 2 The ubiquitin-tagged protein is recognized by a proteasome, which unfolds the protein and sequesters it within a central cavity. 3 Enzymatic components of the proteasome cut the protein into small peptides, which can be further degraded by other enzymes in the cytosol. RNA processing m. RNA degradation Proteasome and ubiquitin to be recycled Ubiquitin Translation Proteasome Protein processing and degradation Protein to be degraded Ubiquinated protein Figure 19. 10 Copyright © 2005 Pearson Education, Inc. publishing as Benjamin Cummings Protein fragments (peptides) Protein entering a proteasome
 Ubiquitin 1980 s | 2004 • “Death tag” – mark unwanted proteins with a label – 76 amino acid polypeptide = ubiquitin – labeled proteins are broken down rapidly in "waste disposers“ = proteasomes Nobel Prize 2004 Aaron Ciechanover Israel Avram Hershko Israel Irwin Rose UC Riverside
Ubiquitin 1980 s | 2004 • “Death tag” – mark unwanted proteins with a label – 76 amino acid polypeptide = ubiquitin – labeled proteins are broken down rapidly in "waste disposers“ = proteasomes Nobel Prize 2004 Aaron Ciechanover Israel Avram Hershko Israel Irwin Rose UC Riverside
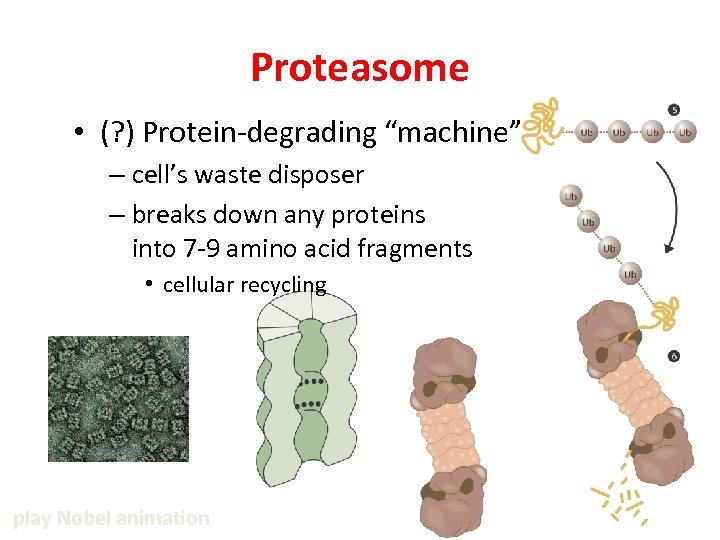 Proteasome • (? ) Protein-degrading “machine” – cell’s waste disposer – breaks down any proteins into 7 -9 amino acid fragments • cellular recycling play Nobel animation
Proteasome • (? ) Protein-degrading “machine” – cell’s waste disposer – breaks down any proteins into 7 -9 amino acid fragments • cellular recycling play Nobel animation
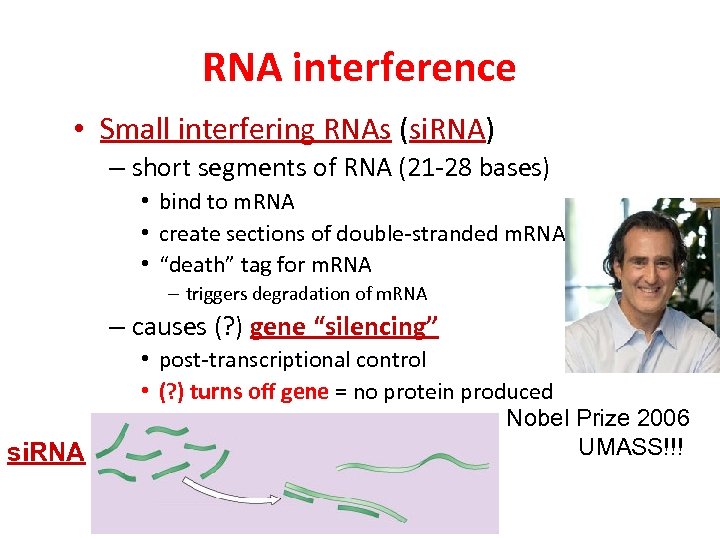 RNA interference • Small interfering RNAs (si. RNA) – short segments of RNA (21 -28 bases) • bind to m. RNA • create sections of double-stranded m. RNA • “death” tag for m. RNA – triggers degradation of m. RNA – causes (? ) gene “silencing” si. RNA • post-transcriptional control • (? ) turns off gene = no protein produced Nobel Prize 2006 UMASS!!!
RNA interference • Small interfering RNAs (si. RNA) – short segments of RNA (21 -28 bases) • bind to m. RNA • create sections of double-stranded m. RNA • “death” tag for m. RNA – triggers degradation of m. RNA – causes (? ) gene “silencing” si. RNA • post-transcriptional control • (? ) turns off gene = no protein produced Nobel Prize 2006 UMASS!!!
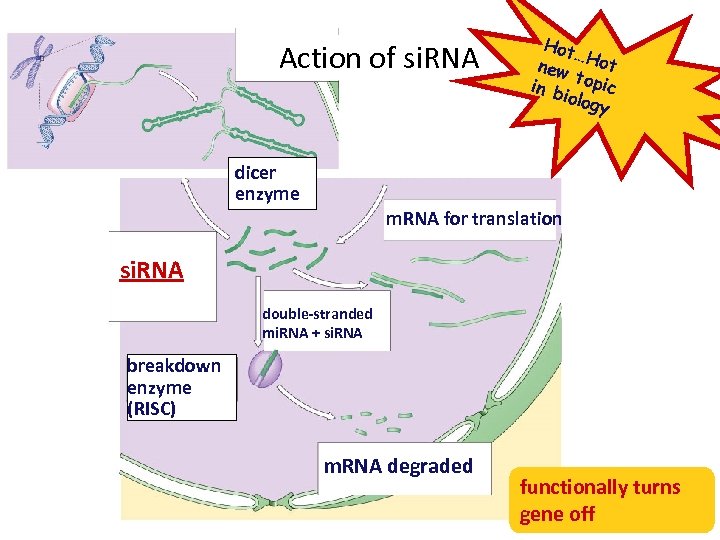 Action of si. RNA dicer enzyme Hot … new Hot t in b opic iolog y m. RNA for translation si. RNA double-stranded mi. RNA + si. RNA breakdown enzyme (RISC) m. RNA degraded functionally turns gene off
Action of si. RNA dicer enzyme Hot … new Hot t in b opic iolog y m. RNA for translation si. RNA double-stranded mi. RNA + si. RNA breakdown enzyme (RISC) m. RNA degraded functionally turns gene off
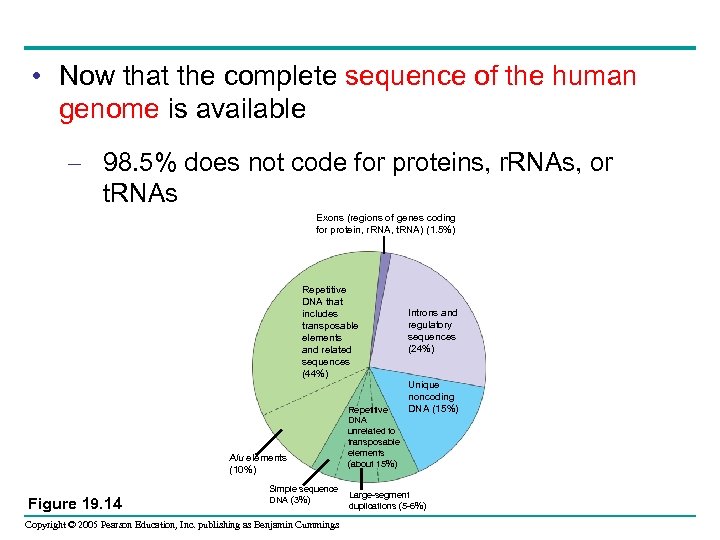 • Now that the complete sequence of the human genome is available – 98. 5% does not code for proteins, r. RNAs, or t. RNAs Exons (regions of genes coding for protein, r. RNA, t. RNA) (1. 5%) Repetitive DNA that includes transposable elements and related sequences (44%) Alu elements (10%) Figure 19. 14 Simple sequence DNA (3%) Copyright © 2005 Pearson Education, Inc. publishing as Benjamin Cummings Repetitive DNA unrelated to transposable elements (about 15%) Introns and regulatory sequences (24%) Unique noncoding DNA (15%) Large-segment duplications (5 -6%)
• Now that the complete sequence of the human genome is available – 98. 5% does not code for proteins, r. RNAs, or t. RNAs Exons (regions of genes coding for protein, r. RNA, t. RNA) (1. 5%) Repetitive DNA that includes transposable elements and related sequences (44%) Alu elements (10%) Figure 19. 14 Simple sequence DNA (3%) Copyright © 2005 Pearson Education, Inc. publishing as Benjamin Cummings Repetitive DNA unrelated to transposable elements (about 15%) Introns and regulatory sequences (24%) Unique noncoding DNA (15%) Large-segment duplications (5 -6%)
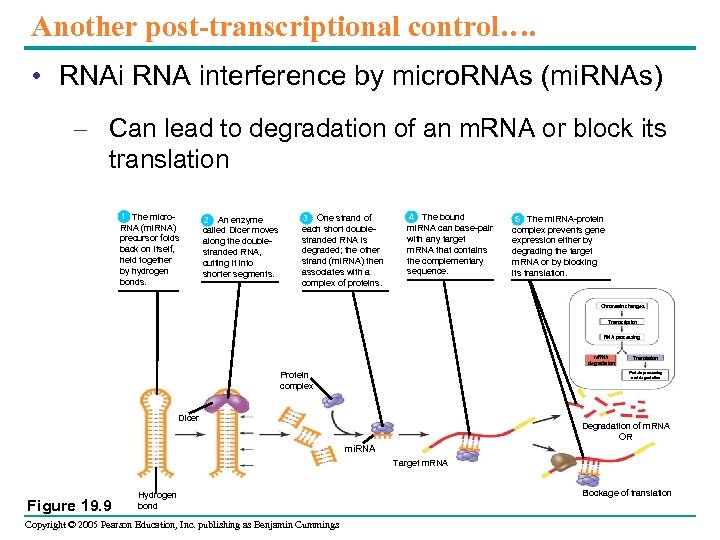 Another post-transcriptional control…. • RNAi RNA interference by micro. RNAs (mi. RNAs) – Can lead to degradation of an m. RNA or block its translation 1 The micro. RNA (mi. RNA) precursor folds back on itself, held together by hydrogen bonds. 2 2 An enzyme called Dicer moves along the doublestranded RNA, cutting it into shorter segments. 3 One strand of each short doublestranded RNA is degraded; the other strand (mi. RNA) then associates with a complex of proteins. 4 The bound mi. RNA can base-pair with any target m. RNA that contains the complementary sequence. 5 The mi. RNA-protein 5 complex prevents gene expression either by degrading the target m. RNA or by blocking its translation. Chromatin changes Transcription RNA processing m. RNA degradation Protein complex Translation Protein processing and degradation Dicer Degradation of m. RNA OR mi. RNA Target m. RNA Figure 19. 9 Hydrogen bond Copyright © 2005 Pearson Education, Inc. publishing as Benjamin Cummings Blockage of translation
Another post-transcriptional control…. • RNAi RNA interference by micro. RNAs (mi. RNAs) – Can lead to degradation of an m. RNA or block its translation 1 The micro. RNA (mi. RNA) precursor folds back on itself, held together by hydrogen bonds. 2 2 An enzyme called Dicer moves along the doublestranded RNA, cutting it into shorter segments. 3 One strand of each short doublestranded RNA is degraded; the other strand (mi. RNA) then associates with a complex of proteins. 4 The bound mi. RNA can base-pair with any target m. RNA that contains the complementary sequence. 5 The mi. RNA-protein 5 complex prevents gene expression either by degrading the target m. RNA or by blocking its translation. Chromatin changes Transcription RNA processing m. RNA degradation Protein complex Translation Protein processing and degradation Dicer Degradation of m. RNA OR mi. RNA Target m. RNA Figure 19. 9 Hydrogen bond Copyright © 2005 Pearson Education, Inc. publishing as Benjamin Cummings Blockage of translation


