b62d5752e1009f7b446a1adc67a4de9f.ppt
- Количество слайдов: 45
 Chapter 18~Regulaton of Gene Expression
Chapter 18~Regulaton of Gene Expression
 Control of Prokaryotic (Bacterial) Genes 2007 -2008
Control of Prokaryotic (Bacterial) Genes 2007 -2008
 Bacterial metabolism • Bacteria need to respond quickly to changes in their environment – if they have enough of a product, need to stop production STOP • why? waste of energy to produce more • how? stop production of enzymes for synthesis – if they find new food/energy source, need to utilize it quickly GO • why? metabolism, growth, reproduction • how? start production of enzymes for digestion
Bacterial metabolism • Bacteria need to respond quickly to changes in their environment – if they have enough of a product, need to stop production STOP • why? waste of energy to produce more • how? stop production of enzymes for synthesis – if they find new food/energy source, need to utilize it quickly GO • why? metabolism, growth, reproduction • how? start production of enzymes for digestion
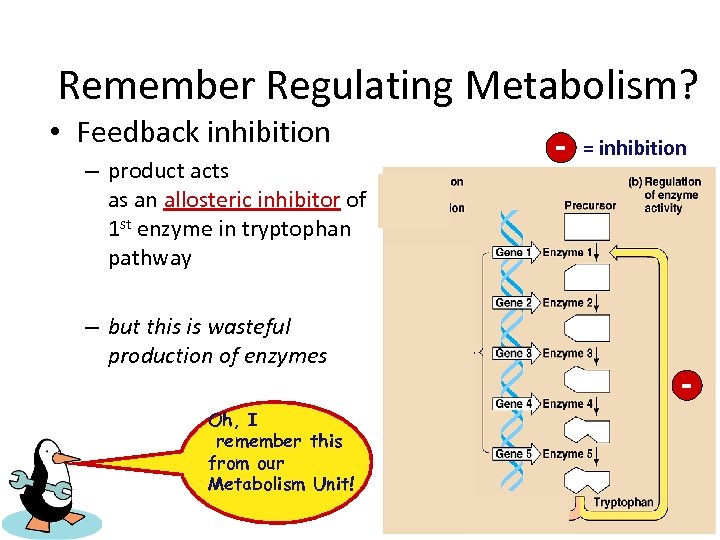 Remember Regulating Metabolism? • Feedback inhibition – product acts as an allosteric inhibitor of 1 st enzyme in tryptophan pathway – but this is wasteful production of enzymes Oh, I remember this from our Metabolism Unit! - = inhibition -
Remember Regulating Metabolism? • Feedback inhibition – product acts as an allosteric inhibitor of 1 st enzyme in tryptophan pathway – but this is wasteful production of enzymes Oh, I remember this from our Metabolism Unit! - = inhibition -
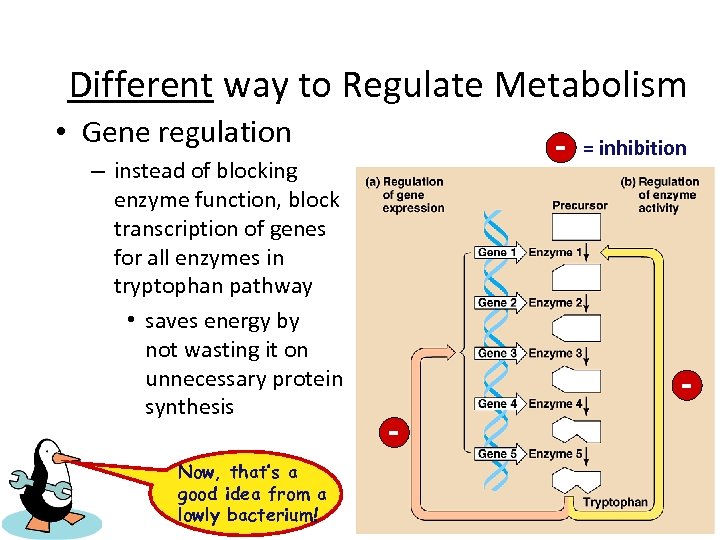 Different way to Regulate Metabolism • Gene regulation – instead of blocking enzyme function, block transcription of genes for all enzymes in tryptophan pathway • saves energy by not wasting it on unnecessary protein synthesis Now, that’s a good idea from a lowly bacterium! - = inhibition -
Different way to Regulate Metabolism • Gene regulation – instead of blocking enzyme function, block transcription of genes for all enzymes in tryptophan pathway • saves energy by not wasting it on unnecessary protein synthesis Now, that’s a good idea from a lowly bacterium! - = inhibition -
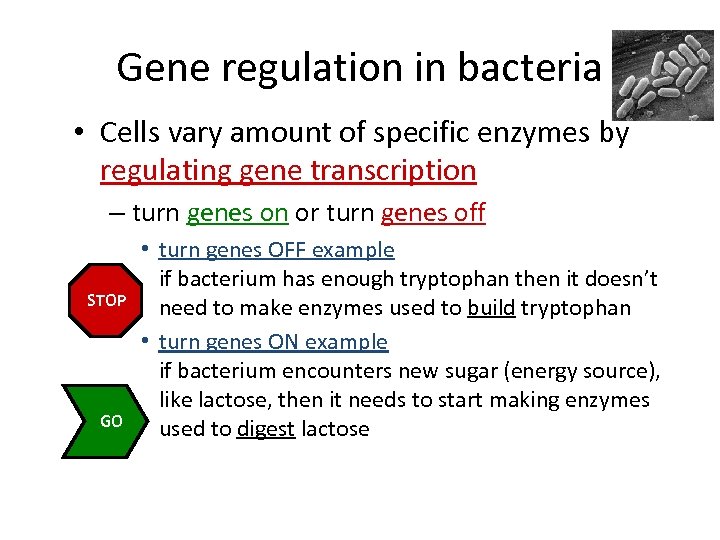 Gene regulation in bacteria • Cells vary amount of specific enzymes by regulating gene transcription – turn genes on or turn genes off STOP GO • turn genes OFF example if bacterium has enough tryptophan then it doesn’t need to make enzymes used to build tryptophan • turn genes ON example if bacterium encounters new sugar (energy source), like lactose, then it needs to start making enzymes used to digest lactose
Gene regulation in bacteria • Cells vary amount of specific enzymes by regulating gene transcription – turn genes on or turn genes off STOP GO • turn genes OFF example if bacterium has enough tryptophan then it doesn’t need to make enzymes used to build tryptophan • turn genes ON example if bacterium encounters new sugar (energy source), like lactose, then it needs to start making enzymes used to digest lactose
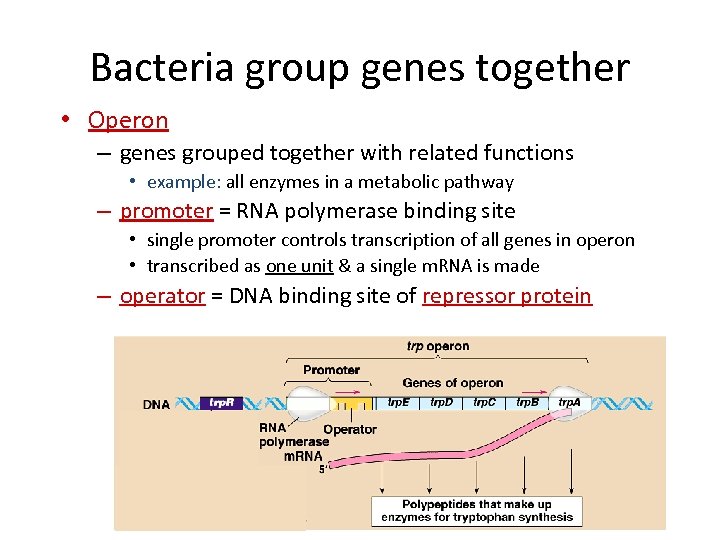 Bacteria group genes together • Operon – genes grouped together with related functions • example: all enzymes in a metabolic pathway – promoter = RNA polymerase binding site • single promoter controls transcription of all genes in operon • transcribed as one unit & a single m. RNA is made – operator = DNA binding site of repressor protein
Bacteria group genes together • Operon – genes grouped together with related functions • example: all enzymes in a metabolic pathway – promoter = RNA polymerase binding site • single promoter controls transcription of all genes in operon • transcribed as one unit & a single m. RNA is made – operator = DNA binding site of repressor protein
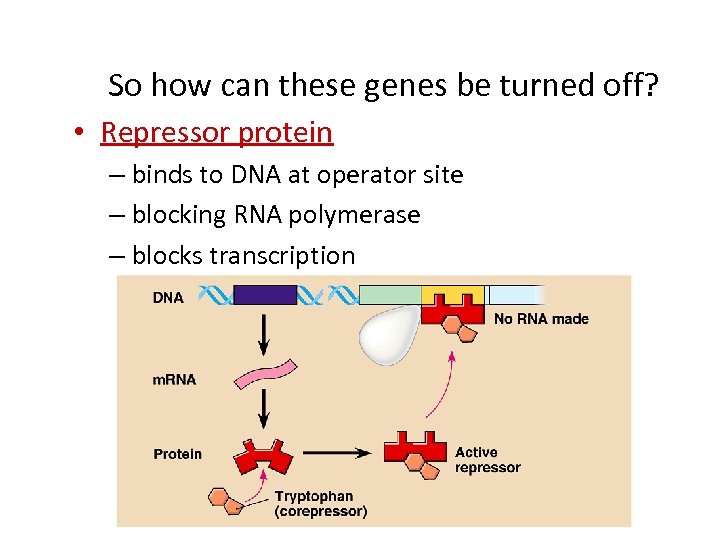 So how can these genes be turned off? • Repressor protein – binds to DNA at operator site – blocking RNA polymerase – blocks transcription
So how can these genes be turned off? • Repressor protein – binds to DNA at operator site – blocking RNA polymerase – blocks transcription
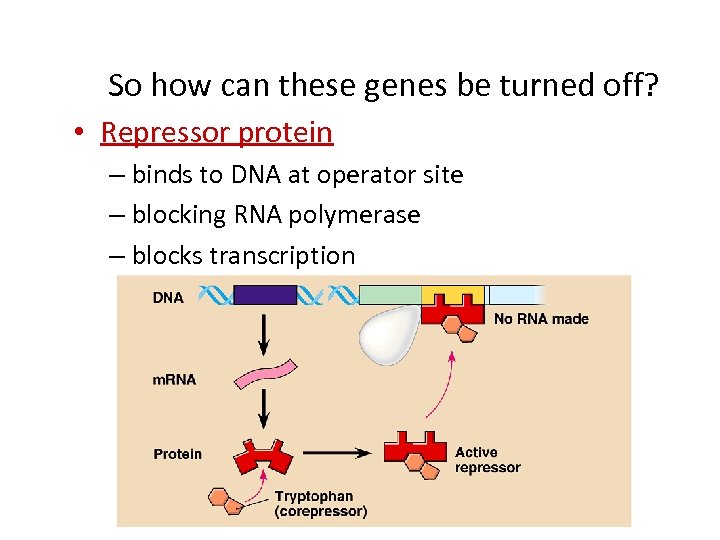 So how can these genes be turned off? • Repressor protein – binds to DNA at operator site – blocking RNA polymerase – blocks transcription
So how can these genes be turned off? • Repressor protein – binds to DNA at operator site – blocking RNA polymerase – blocks transcription
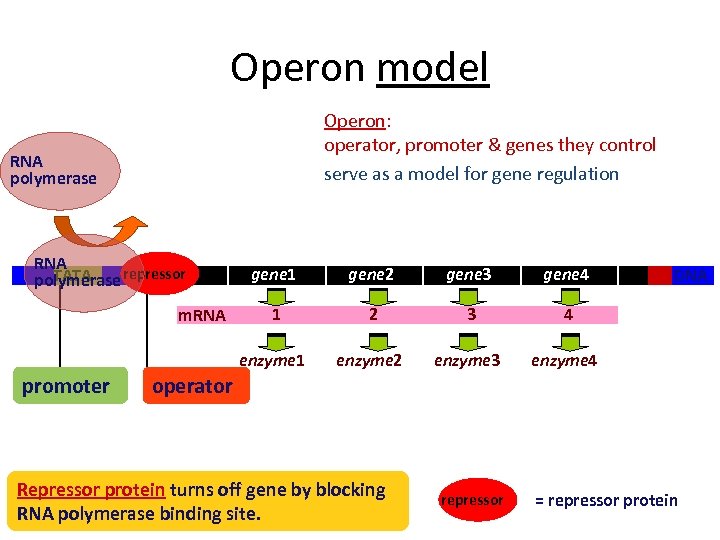 Operon model Operon: operator, promoter & genes they control serve as a model for gene regulation RNA polymerase RNA TATA polymerase repressor promoter gene 2 gene 3 gene 4 1 2 3 4 enzyme 1 m. RNA gene 1 enzyme 2 enzyme 3 enzyme 4 DNA operator Repressor protein turns off gene by blocking RNA polymerase binding site. repressor = repressor protein
Operon model Operon: operator, promoter & genes they control serve as a model for gene regulation RNA polymerase RNA TATA polymerase repressor promoter gene 2 gene 3 gene 4 1 2 3 4 enzyme 1 m. RNA gene 1 enzyme 2 enzyme 3 enzyme 4 DNA operator Repressor protein turns off gene by blocking RNA polymerase binding site. repressor = repressor protein
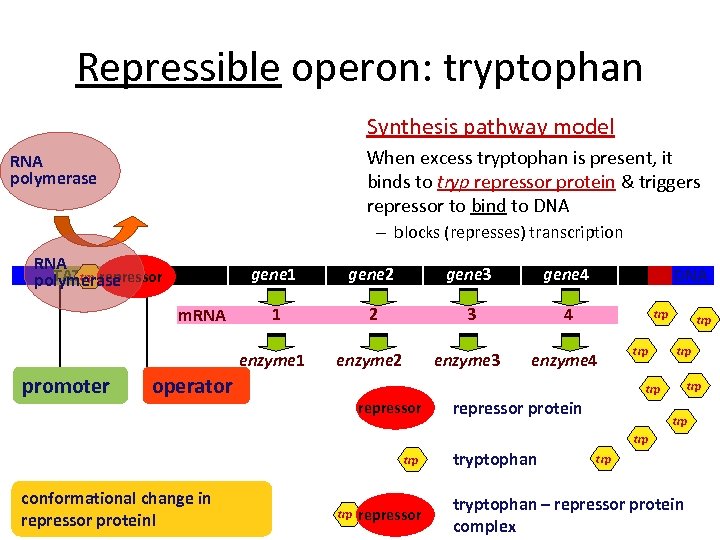 Repressible operon: tryptophan Synthesis pathway model When excess tryptophan is present, it binds to tryp repressor protein & triggers repressor to bind to DNA RNA polymerase – blocks (represses) transcription RNA trp TATA repressor polymerase gene 1 promoter gene 3 gene 4 1 2 3 4 enzyme 1 m. RNA gene 2 enzyme 3 enzyme 4 operator DNA trp trp trp repressor protein trp trp conformational change in repressor protein! trp repressor tryptophan trp tryptophan – repressor protein complex
Repressible operon: tryptophan Synthesis pathway model When excess tryptophan is present, it binds to tryp repressor protein & triggers repressor to bind to DNA RNA polymerase – blocks (represses) transcription RNA trp TATA repressor polymerase gene 1 promoter gene 3 gene 4 1 2 3 4 enzyme 1 m. RNA gene 2 enzyme 3 enzyme 4 operator DNA trp trp trp repressor protein trp trp conformational change in repressor protein! trp repressor tryptophan trp tryptophan – repressor protein complex
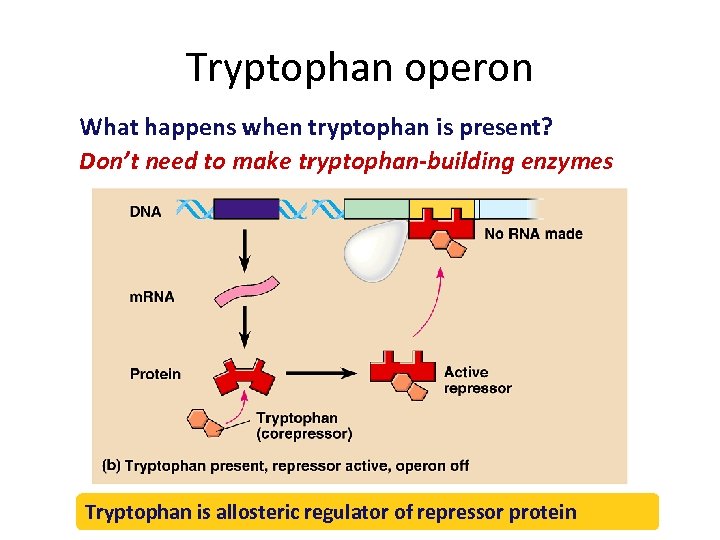 Tryptophan operon What happens when tryptophan is present? Don’t need to make tryptophan-building enzymes Tryptophan is allosteric regulator of repressor protein
Tryptophan operon What happens when tryptophan is present? Don’t need to make tryptophan-building enzymes Tryptophan is allosteric regulator of repressor protein
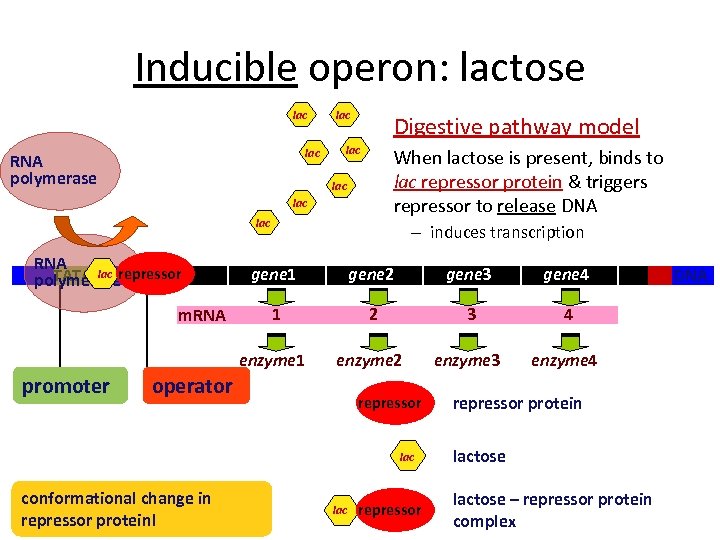 Inducible operon: lactose lac RNA polymerase lac Digestive pathway model lac When lactose is present, binds to lac repressor protein & triggers repressor to release DNA lac lac RNA TATA lac polymeraserepressor – induces transcription promoter gene 2 gene 3 gene 4 1 2 3 4 enzyme 1 m. RNA gene 1 enzyme 2 enzyme 3 enzyme 4 operator repressor lac conformational change in repressor protein! lac repressor protein lactose – repressor protein complex DNA
Inducible operon: lactose lac RNA polymerase lac Digestive pathway model lac When lactose is present, binds to lac repressor protein & triggers repressor to release DNA lac lac RNA TATA lac polymeraserepressor – induces transcription promoter gene 2 gene 3 gene 4 1 2 3 4 enzyme 1 m. RNA gene 1 enzyme 2 enzyme 3 enzyme 4 operator repressor lac conformational change in repressor protein! lac repressor protein lactose – repressor protein complex DNA
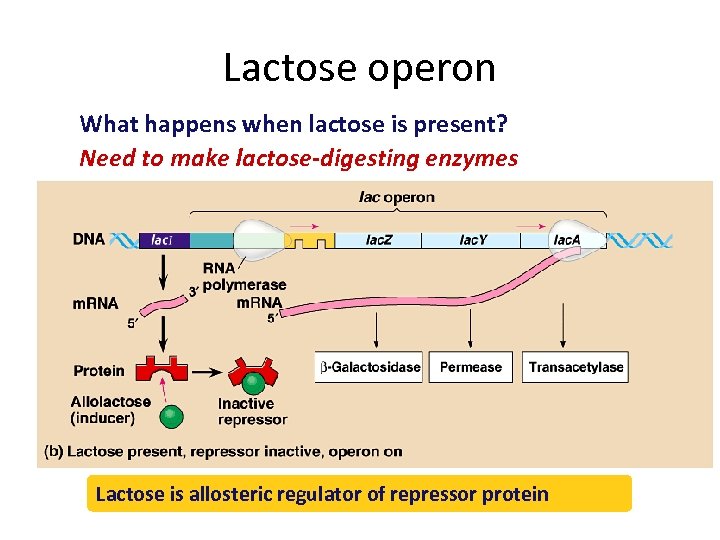 Lactose operon What happens when lactose is present? Need to make lactose-digesting enzymes Lactose is allosteric regulator of repressor protein
Lactose operon What happens when lactose is present? Need to make lactose-digesting enzymes Lactose is allosteric regulator of repressor protein
 1961 | 1965 Jacob & Monod: lac Operon • Francois Jacob & Jacques Monod – first to describe operon system – coined the phrase “operon” Jacques Monod Francois Jacob
1961 | 1965 Jacob & Monod: lac Operon • Francois Jacob & Jacques Monod – first to describe operon system – coined the phrase “operon” Jacques Monod Francois Jacob
 Operon summary • Repressible operon – usually functions in anabolic pathways • synthesizing end products – when end product is present in excess, cell allocates resources to other uses • Inducible operon – usually functions in catabolic pathways, • digesting nutrients to simpler molecules – produce enzymes only when nutrient is available • cell avoids making proteins that have nothing to do, cell allocates resources to other uses
Operon summary • Repressible operon – usually functions in anabolic pathways • synthesizing end products – when end product is present in excess, cell allocates resources to other uses • Inducible operon – usually functions in catabolic pathways, • digesting nutrients to simpler molecules – produce enzymes only when nutrient is available • cell avoids making proteins that have nothing to do, cell allocates resources to other uses
 Positive gene control • occurs when an activator molecule interacts directly with the genome to switch transcription on. • Even if the lac operon is turned on by the presence of allolactose, the degree of transcription depends on the concentrations of other substrates. • The cellular metabolism is biased toward the utilization of glucose.
Positive gene control • occurs when an activator molecule interacts directly with the genome to switch transcription on. • Even if the lac operon is turned on by the presence of allolactose, the degree of transcription depends on the concentrations of other substrates. • The cellular metabolism is biased toward the utilization of glucose.
 Positive Gene Regulation • Some operons are also subject to positive control through a stimulatory protein, such as catabolite activator protein (CAP), an activator of transcription • When glucose (a preferred food source of E. coli) is scarce, CAP is activated by binding with cyclic AMP • Activated CAP attaches to the promoter of the lac operon and increases the affinity of RNA polymerase, thus accelerating transcription
Positive Gene Regulation • Some operons are also subject to positive control through a stimulatory protein, such as catabolite activator protein (CAP), an activator of transcription • When glucose (a preferred food source of E. coli) is scarce, CAP is activated by binding with cyclic AMP • Activated CAP attaches to the promoter of the lac operon and increases the affinity of RNA polymerase, thus accelerating transcription
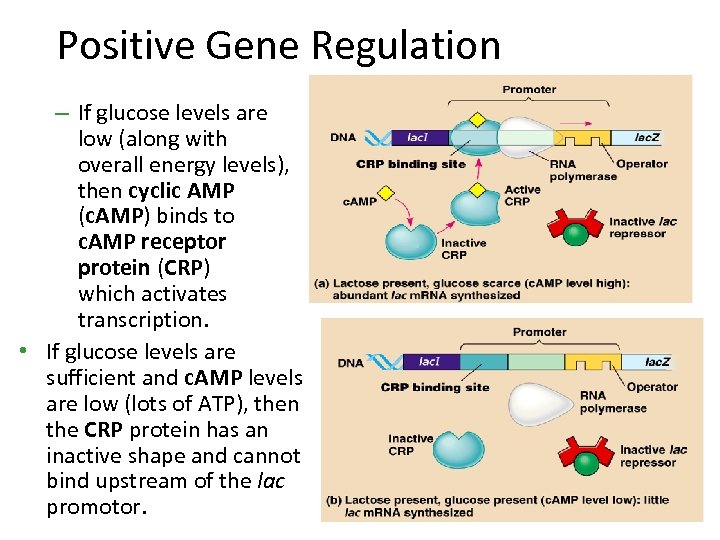 Positive Gene Regulation – If glucose levels are low (along with overall energy levels), then cyclic AMP (c. AMP) binds to c. AMP receptor protein (CRP) which activates transcription. • If glucose levels are sufficient and c. AMP levels are low (lots of ATP), then the CRP protein has an inactive shape and cannot bind upstream of the lac promotor.
Positive Gene Regulation – If glucose levels are low (along with overall energy levels), then cyclic AMP (c. AMP) binds to c. AMP receptor protein (CRP) which activates transcription. • If glucose levels are sufficient and c. AMP levels are low (lots of ATP), then the CRP protein has an inactive shape and cannot bind upstream of the lac promotor.
 Control of Eukaryotic Genes 2007 -2008
Control of Eukaryotic Genes 2007 -2008
 The BIG Questions… • How are genes turned on & off in eukaryotes? • How do cells with the same genes differentiate to perform completely different, specialized functions?
The BIG Questions… • How are genes turned on & off in eukaryotes? • How do cells with the same genes differentiate to perform completely different, specialized functions?
 Evolution of gene regulation • Prokaryotes – single-celled – evolved to grow & divide rapidly – must respond quickly to changes in external environment • exploit transient resources • Gene regulation – turn genes on & off rapidly • flexibility & reversibility – adjust levels of enzymes for synthesis & digestion
Evolution of gene regulation • Prokaryotes – single-celled – evolved to grow & divide rapidly – must respond quickly to changes in external environment • exploit transient resources • Gene regulation – turn genes on & off rapidly • flexibility & reversibility – adjust levels of enzymes for synthesis & digestion
 Evolution of gene regulation • Eukaryotes – multicellular – evolved to maintain constant internal conditions while facing changing external conditions • homeostasis – regulate body as a whole • growth & development – long term processes • specialization – turn on & off large number of genes • must coordinate the body as a whole rather than serve the needs of individual cells
Evolution of gene regulation • Eukaryotes – multicellular – evolved to maintain constant internal conditions while facing changing external conditions • homeostasis – regulate body as a whole • growth & development – long term processes • specialization – turn on & off large number of genes • must coordinate the body as a whole rather than serve the needs of individual cells
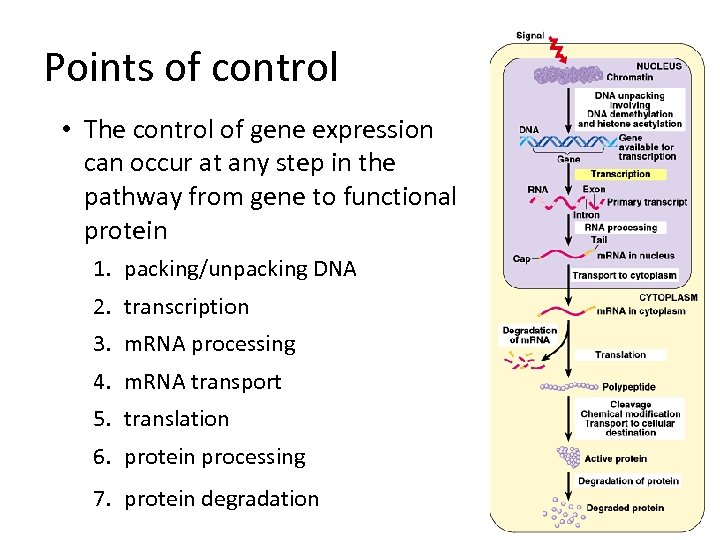 Points of control • The control of gene expression can occur at any step in the pathway from gene to functional protein 1. packing/unpacking DNA 2. transcription 3. m. RNA processing 4. m. RNA transport 5. translation 6. protein processing 7. protein degradation
Points of control • The control of gene expression can occur at any step in the pathway from gene to functional protein 1. packing/unpacking DNA 2. transcription 3. m. RNA processing 4. m. RNA transport 5. translation 6. protein processing 7. protein degradation
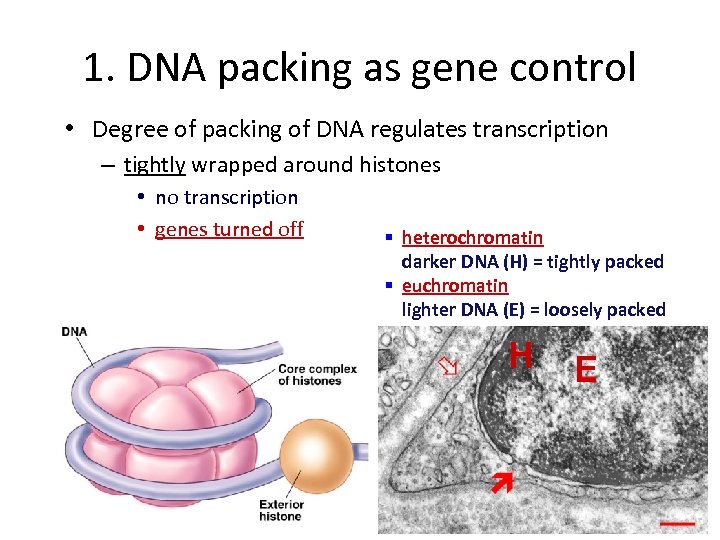 1. DNA packing as gene control • Degree of packing of DNA regulates transcription – tightly wrapped around histones • no transcription • genes turned off § heterochromatin darker DNA (H) = tightly packed § euchromatin lighter DNA (E) = loosely packed H E
1. DNA packing as gene control • Degree of packing of DNA regulates transcription – tightly wrapped around histones • no transcription • genes turned off § heterochromatin darker DNA (H) = tightly packed § euchromatin lighter DNA (E) = loosely packed H E
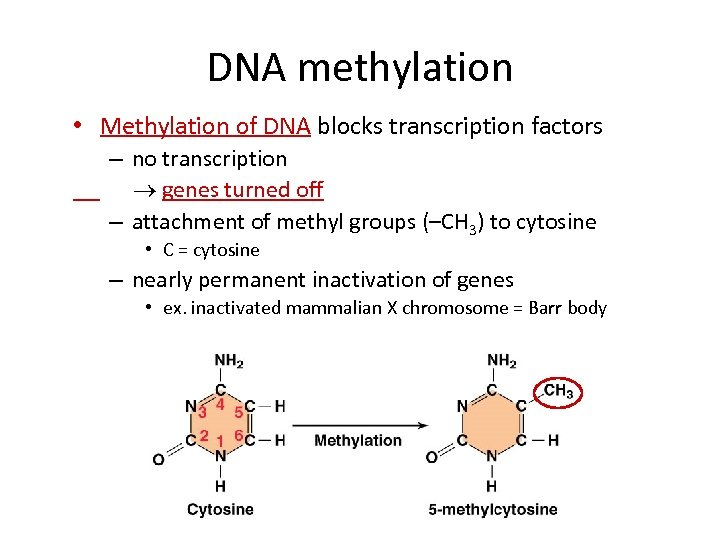 DNA methylation • Methylation of DNA blocks transcription factors – no transcription genes turned off – attachment of methyl groups (–CH 3) to cytosine • C = cytosine – nearly permanent inactivation of genes • ex. inactivated mammalian X chromosome = Barr body
DNA methylation • Methylation of DNA blocks transcription factors – no transcription genes turned off – attachment of methyl groups (–CH 3) to cytosine • C = cytosine – nearly permanent inactivation of genes • ex. inactivated mammalian X chromosome = Barr body
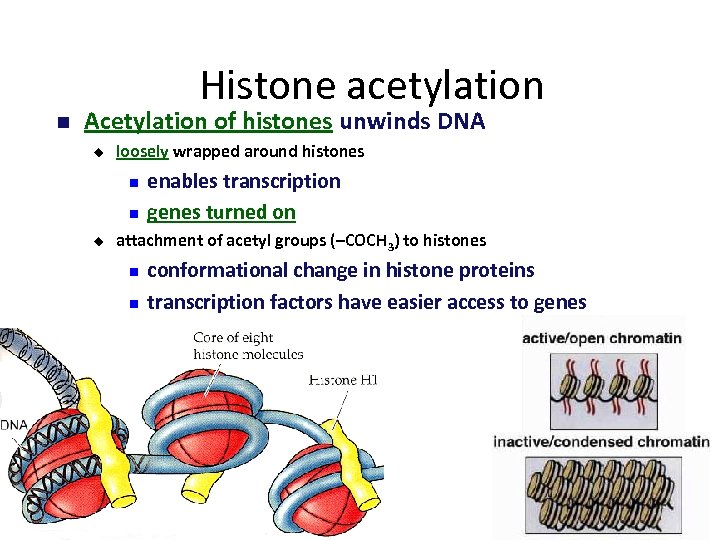 Histone acetylation Acetylation of histones unwinds DNA u loosely wrapped around histones u enables transcription genes turned on attachment of acetyl groups (–COCH 3) to histones conformational change in histone proteins transcription factors have easier access to genes
Histone acetylation Acetylation of histones unwinds DNA u loosely wrapped around histones u enables transcription genes turned on attachment of acetyl groups (–COCH 3) to histones conformational change in histone proteins transcription factors have easier access to genes
 Epigenetic Inheritance • Although the chromatin modifications just discussed do not alter DNA sequence, they may be passed to future generations of cells • The inheritance of traits transmitted by mechanisms not directly involving the nucleotide sequence is called epigenetic inheritance
Epigenetic Inheritance • Although the chromatin modifications just discussed do not alter DNA sequence, they may be passed to future generations of cells • The inheritance of traits transmitted by mechanisms not directly involving the nucleotide sequence is called epigenetic inheritance
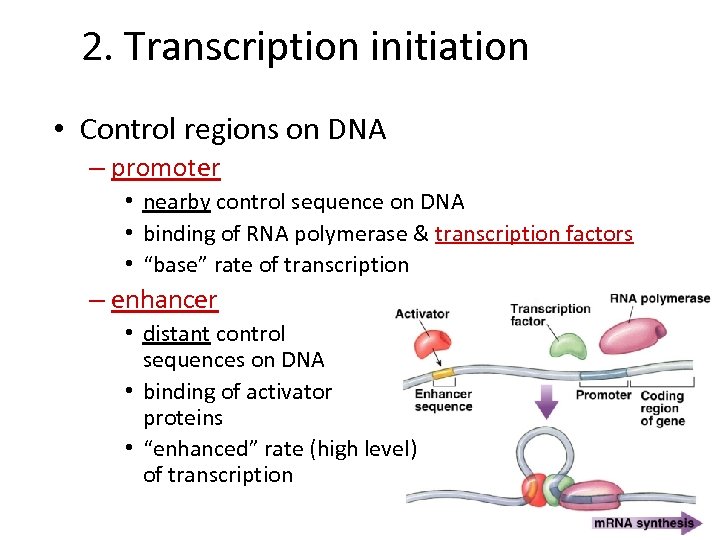 2. Transcription initiation • Control regions on DNA – promoter • nearby control sequence on DNA • binding of RNA polymerase & transcription factors • “base” rate of transcription – enhancer • distant control sequences on DNA • binding of activator proteins • “enhanced” rate (high level) of transcription
2. Transcription initiation • Control regions on DNA – promoter • nearby control sequence on DNA • binding of RNA polymerase & transcription factors • “base” rate of transcription – enhancer • distant control sequences on DNA • binding of activator proteins • “enhanced” rate (high level) of transcription
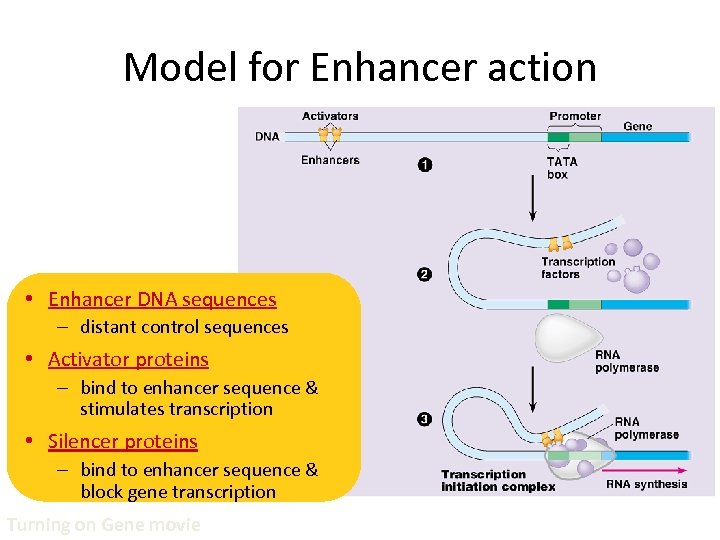 Model for Enhancer action • Enhancer DNA sequences – distant control sequences • Activator proteins – bind to enhancer sequence & stimulates transcription • Silencer proteins – bind to enhancer sequence & block gene transcription Turning on Gene movie
Model for Enhancer action • Enhancer DNA sequences – distant control sequences • Activator proteins – bind to enhancer sequence & stimulates transcription • Silencer proteins – bind to enhancer sequence & block gene transcription Turning on Gene movie
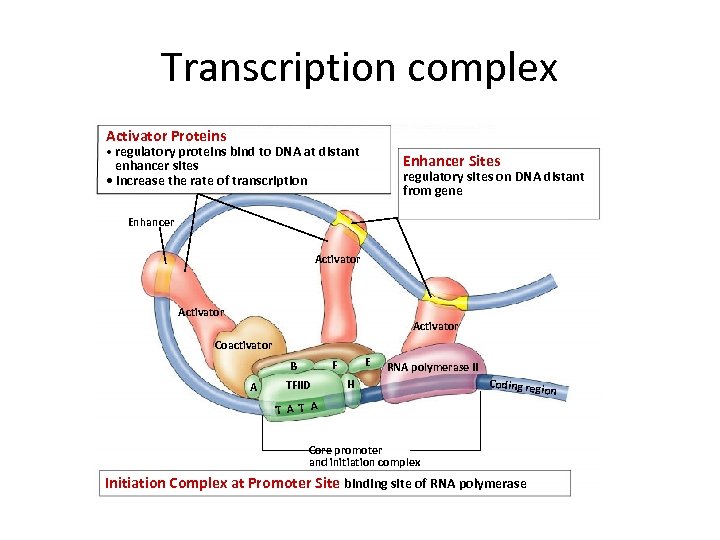 Transcription complex Activator Proteins • regulatory proteins bind to DNA at distant Enhancer Sites enhancer sites • increase the rate of transcription regulatory sites on DNA distant from gene Enhancer Activator Coactivator A E F B TFIID RNA polymerase II H Coding reg T A Core promoter and initiation complex Initiation Complex at Promoter Site binding site of RNA polymerase ion
Transcription complex Activator Proteins • regulatory proteins bind to DNA at distant Enhancer Sites enhancer sites • increase the rate of transcription regulatory sites on DNA distant from gene Enhancer Activator Coactivator A E F B TFIID RNA polymerase II H Coding reg T A Core promoter and initiation complex Initiation Complex at Promoter Site binding site of RNA polymerase ion
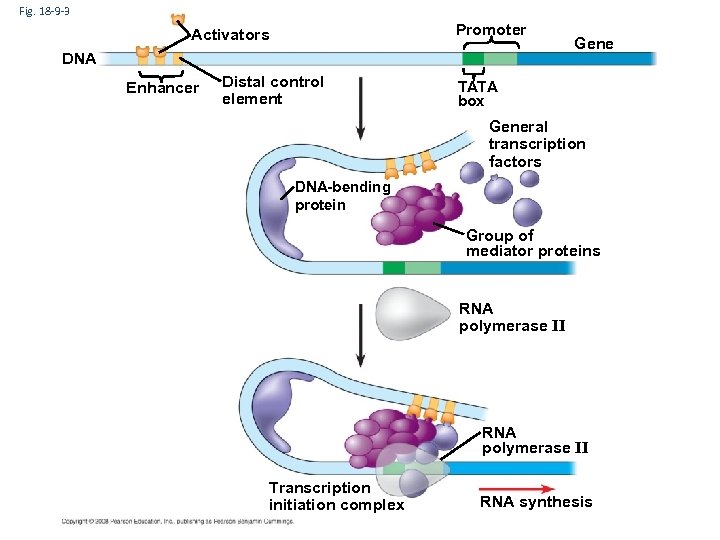 Fig. 18 -9 -3 Promoter Activators DNA Enhancer Distal control element Gene TATA box General transcription factors DNA-bending protein Group of mediator proteins RNA polymerase II Transcription initiation complex RNA synthesis
Fig. 18 -9 -3 Promoter Activators DNA Enhancer Distal control element Gene TATA box General transcription factors DNA-bending protein Group of mediator proteins RNA polymerase II Transcription initiation complex RNA synthesis
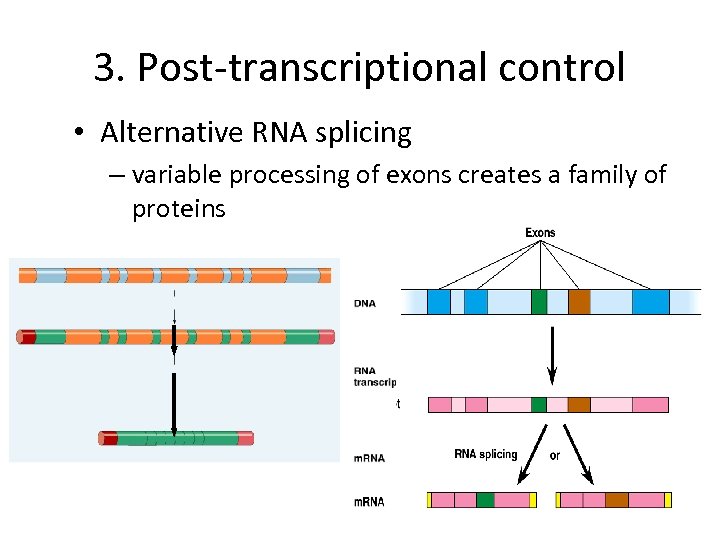 3. Post-transcriptional control • Alternative RNA splicing – variable processing of exons creates a family of proteins
3. Post-transcriptional control • Alternative RNA splicing – variable processing of exons creates a family of proteins
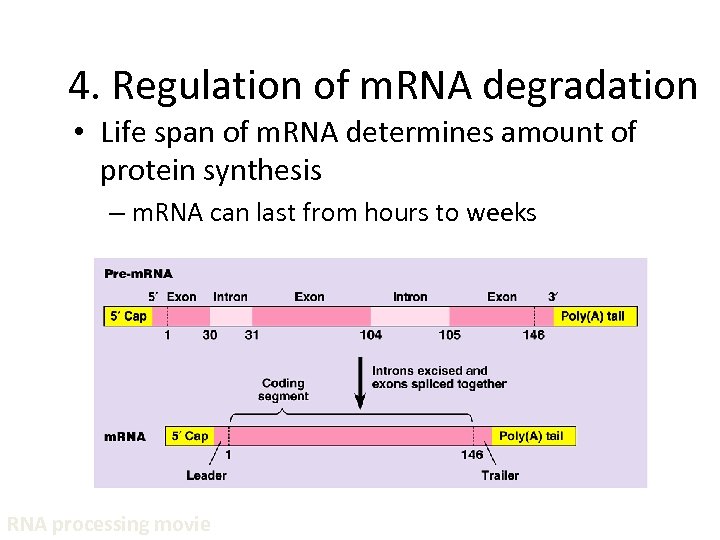 4. Regulation of m. RNA degradation • Life span of m. RNA determines amount of protein synthesis – m. RNA can last from hours to weeks RNA processing movie
4. Regulation of m. RNA degradation • Life span of m. RNA determines amount of protein synthesis – m. RNA can last from hours to weeks RNA processing movie
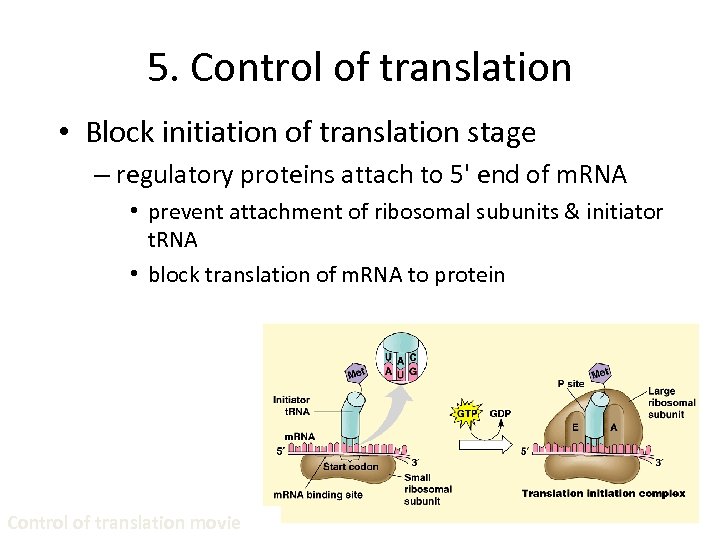 5. Control of translation • Block initiation of translation stage – regulatory proteins attach to 5' end of m. RNA • prevent attachment of ribosomal subunits & initiator t. RNA • block translation of m. RNA to protein Control of translation movie
5. Control of translation • Block initiation of translation stage – regulatory proteins attach to 5' end of m. RNA • prevent attachment of ribosomal subunits & initiator t. RNA • block translation of m. RNA to protein Control of translation movie
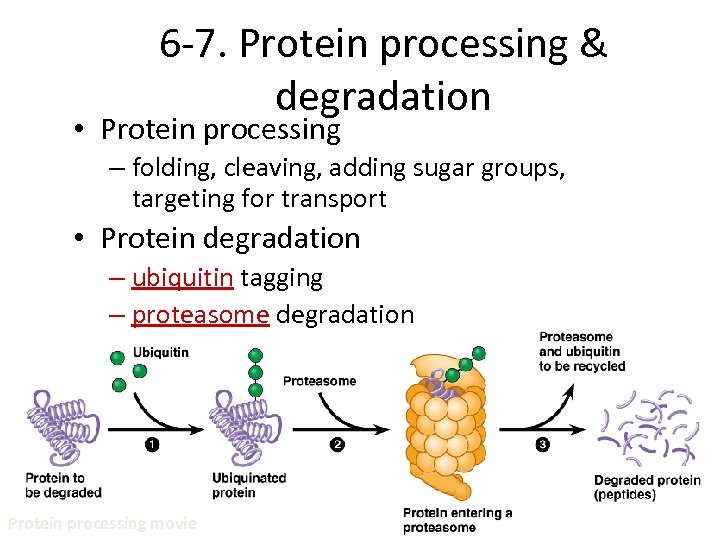 6 -7. Protein processing & degradation • Protein processing – folding, cleaving, adding sugar groups, targeting for transport • Protein degradation – ubiquitin tagging – proteasome degradation Protein processing movie
6 -7. Protein processing & degradation • Protein processing – folding, cleaving, adding sugar groups, targeting for transport • Protein degradation – ubiquitin tagging – proteasome degradation Protein processing movie
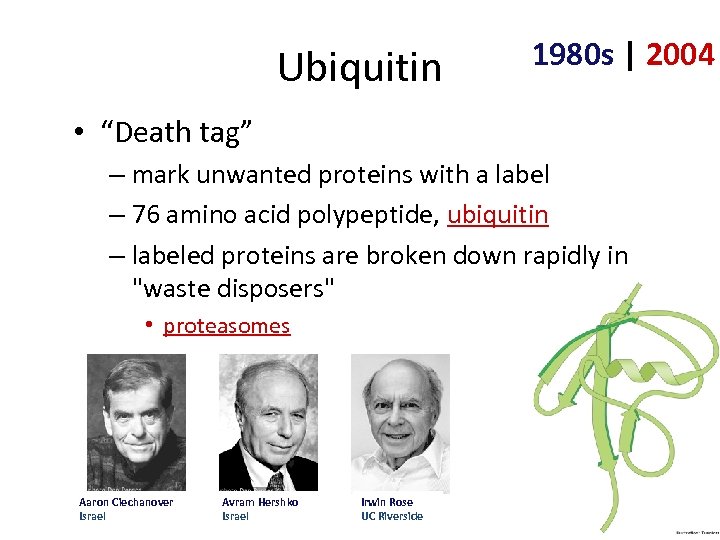 Ubiquitin 1980 s | 2004 • “Death tag” – mark unwanted proteins with a label – 76 amino acid polypeptide, ubiquitin – labeled proteins are broken down rapidly in "waste disposers" • proteasomes Aaron Ciechanover Israel Avram Hershko Israel Irwin Rose UC Riverside
Ubiquitin 1980 s | 2004 • “Death tag” – mark unwanted proteins with a label – 76 amino acid polypeptide, ubiquitin – labeled proteins are broken down rapidly in "waste disposers" • proteasomes Aaron Ciechanover Israel Avram Hershko Israel Irwin Rose UC Riverside
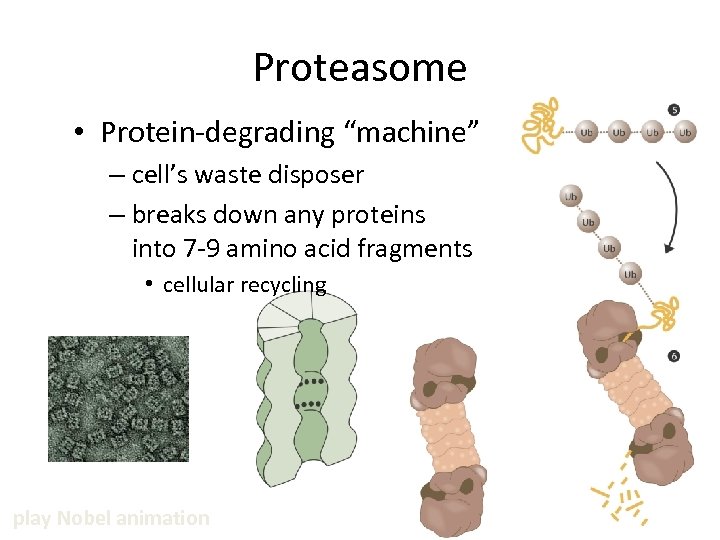 Proteasome • Protein-degrading “machine” – cell’s waste disposer – breaks down any proteins into 7 -9 amino acid fragments • cellular recycling play Nobel animation
Proteasome • Protein-degrading “machine” – cell’s waste disposer – breaks down any proteins into 7 -9 amino acid fragments • cellular recycling play Nobel animation
 Concept 18. 3: Noncoding RNAs play multiple roles in controlling gene expression • Only a small fraction of DNA codes for proteins, r. RNA, and t. RNA • A significant amount of the genome may be transcribed into noncoding RNAs • Noncoding RNAs regulate gene expression at two points: m. RNA translation and chromatin configuration
Concept 18. 3: Noncoding RNAs play multiple roles in controlling gene expression • Only a small fraction of DNA codes for proteins, r. RNA, and t. RNA • A significant amount of the genome may be transcribed into noncoding RNAs • Noncoding RNAs regulate gene expression at two points: m. RNA translation and chromatin configuration
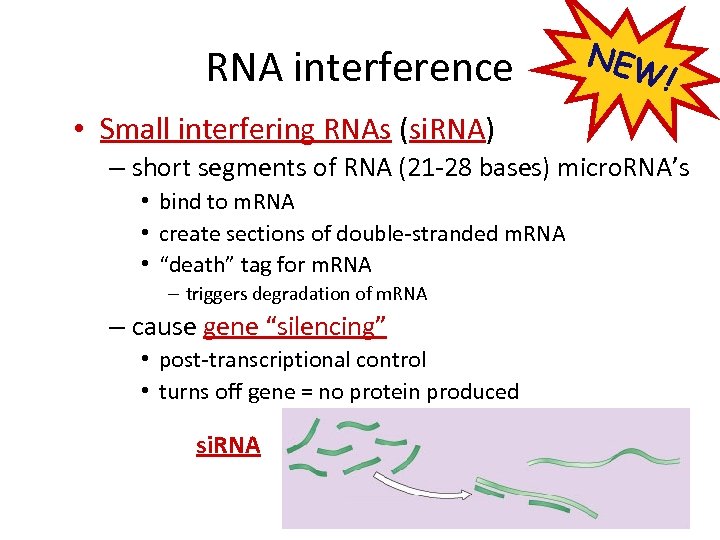 RNA interference NEW ! • Small interfering RNAs (si. RNA) – short segments of RNA (21 -28 bases) micro. RNA’s • bind to m. RNA • create sections of double-stranded m. RNA • “death” tag for m. RNA – triggers degradation of m. RNA – cause gene “silencing” • post-transcriptional control • turns off gene = no protein produced si. RNA
RNA interference NEW ! • Small interfering RNAs (si. RNA) – short segments of RNA (21 -28 bases) micro. RNA’s • bind to m. RNA • create sections of double-stranded m. RNA • “death” tag for m. RNA – triggers degradation of m. RNA – cause gene “silencing” • post-transcriptional control • turns off gene = no protein produced si. RNA
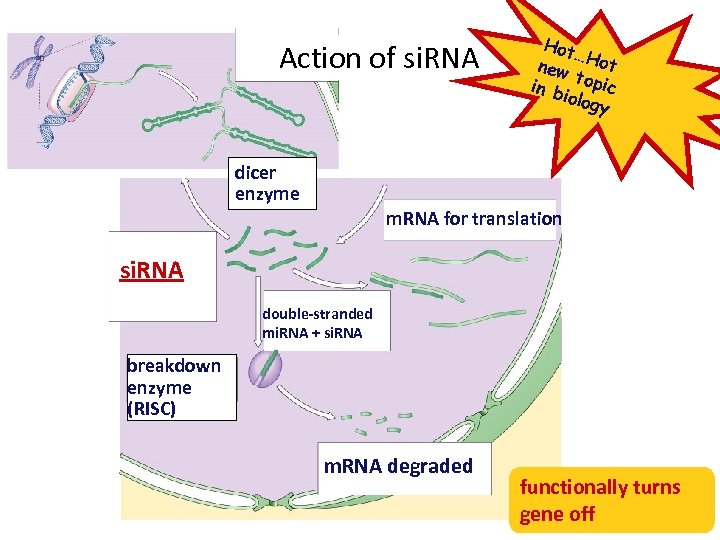 Action of si. RNA dicer enzyme Hot … new Hot t in b opic iolog y m. RNA for translation si. RNA double-stranded mi. RNA + si. RNA breakdown enzyme (RISC) m. RNA degraded functionally turns gene off
Action of si. RNA dicer enzyme Hot … new Hot t in b opic iolog y m. RNA for translation si. RNA double-stranded mi. RNA + si. RNA breakdown enzyme (RISC) m. RNA degraded functionally turns gene off
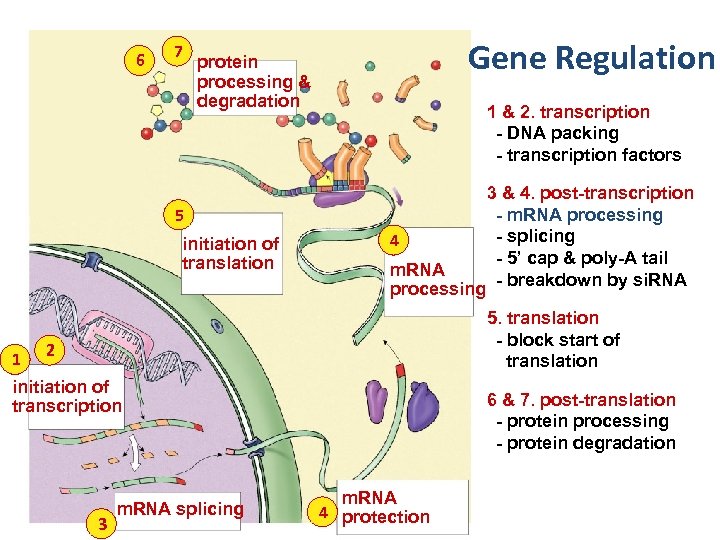 6 7 Gene Regulation protein processing & degradation 1 & 2. transcription - DNA packing - transcription factors 5 initiation of translation 1 4 m. RNA processing 3 & 4. post-transcription - m. RNA processing - splicing - 5’ cap & poly-A tail - breakdown by si. RNA 5. translation - block start of translation 2 initiation of transcription 3 m. RNA splicing 6 & 7. post-translation - protein processing - protein degradation m. RNA 4 protection
6 7 Gene Regulation protein processing & degradation 1 & 2. transcription - DNA packing - transcription factors 5 initiation of translation 1 4 m. RNA processing 3 & 4. post-transcription - m. RNA processing - splicing - 5’ cap & poly-A tail - breakdown by si. RNA 5. translation - block start of translation 2 initiation of transcription 3 m. RNA splicing 6 & 7. post-translation - protein processing - protein degradation m. RNA 4 protection
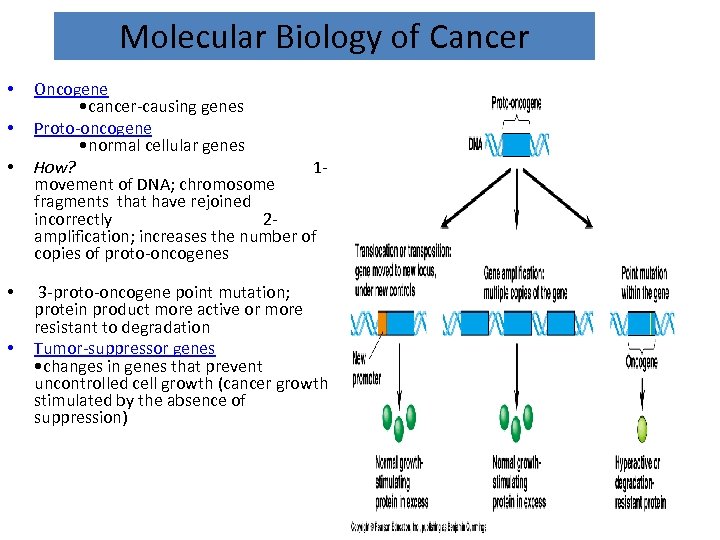 Molecular Biology of Cancer • • • Oncogene • cancer-causing genes Proto-oncogene • normal cellular genes How? 1 movement of DNA; chromosome fragments that have rejoined incorrectly 2 amplification; increases the number of copies of proto-oncogenes 3 -proto-oncogene point mutation; protein product more active or more resistant to degradation Tumor-suppressor genes • changes in genes that prevent uncontrolled cell growth (cancer growth stimulated by the absence of suppression)
Molecular Biology of Cancer • • • Oncogene • cancer-causing genes Proto-oncogene • normal cellular genes How? 1 movement of DNA; chromosome fragments that have rejoined incorrectly 2 amplification; increases the number of copies of proto-oncogenes 3 -proto-oncogene point mutation; protein product more active or more resistant to degradation Tumor-suppressor genes • changes in genes that prevent uncontrolled cell growth (cancer growth stimulated by the absence of suppression)
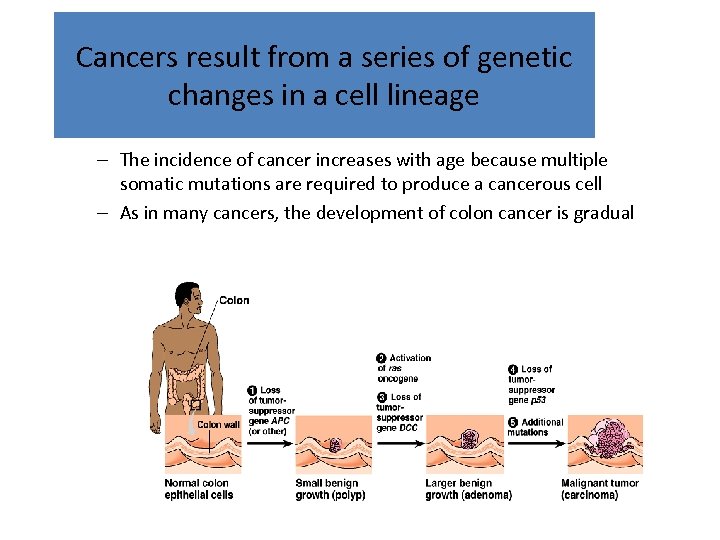 Cancers result from a series of genetic changes in a cell lineage – The incidence of cancer increases with age because multiple somatic mutations are required to produce a cancerous cell – As in many cancers, the development of colon cancer is gradual
Cancers result from a series of genetic changes in a cell lineage – The incidence of cancer increases with age because multiple somatic mutations are required to produce a cancerous cell – As in many cancers, the development of colon cancer is gradual
 Turn your Question Genes on! 2007 -2008
Turn your Question Genes on! 2007 -2008


