0b3d95ab99a0c811fc0164de1e1df604.ppt
- Количество слайдов: 34
 Chapter 14 Forensic Challenges Fundamentals of Forensic DNA Typing Slides prepared by John M. Butler June 2009
Chapter 14 Forensic Challenges Fundamentals of Forensic DNA Typing Slides prepared by John M. Butler June 2009
 Chapter 14 – Forensic Challenges Chapter Summary DNA can be damaged or destroyed as it is exposed to environmental elements present at crime or disaster scenes. Degraded DNA often results in partial STR profiles or none at all. The use of reduced size PCR products, also known as “mini. STRs”, can in some cases enable recovery of information from degraded DNA or samples containing PCR inhibitors. Mixtures of DNA from two or more sources occur in some crime scenes and resolving the components of a mixture can be challenging in many situations. Depending on the type of mixture obtained including the ratio of contributors, the peak heights of STR alleles observed in a mixture can be used to group alleles to decipher the mixture component genotypes. Low-level DNA analysis, sometimes referred to as “low-copy number” or LCN testing, involves attempting to detect a DNA profile from only a few cells often through boosting the number of PCR cycles to enhance sensitivity. LCN testing is prone to problems, such as allelic drop-out (due to stochastic amplification effects) and allelic drop-in (due to sporadic contamination), and rigorous rules are generally applied including generating composite profiles of replicated alleles from multiple amplifications of the same DNA extract.
Chapter 14 – Forensic Challenges Chapter Summary DNA can be damaged or destroyed as it is exposed to environmental elements present at crime or disaster scenes. Degraded DNA often results in partial STR profiles or none at all. The use of reduced size PCR products, also known as “mini. STRs”, can in some cases enable recovery of information from degraded DNA or samples containing PCR inhibitors. Mixtures of DNA from two or more sources occur in some crime scenes and resolving the components of a mixture can be challenging in many situations. Depending on the type of mixture obtained including the ratio of contributors, the peak heights of STR alleles observed in a mixture can be used to group alleles to decipher the mixture component genotypes. Low-level DNA analysis, sometimes referred to as “low-copy number” or LCN testing, involves attempting to detect a DNA profile from only a few cells often through boosting the number of PCR cycles to enhance sensitivity. LCN testing is prone to problems, such as allelic drop-out (due to stochastic amplification effects) and allelic drop-in (due to sporadic contamination), and rigorous rules are generally applied including generating composite profiles of replicated alleles from multiple amplifications of the same DNA extract.
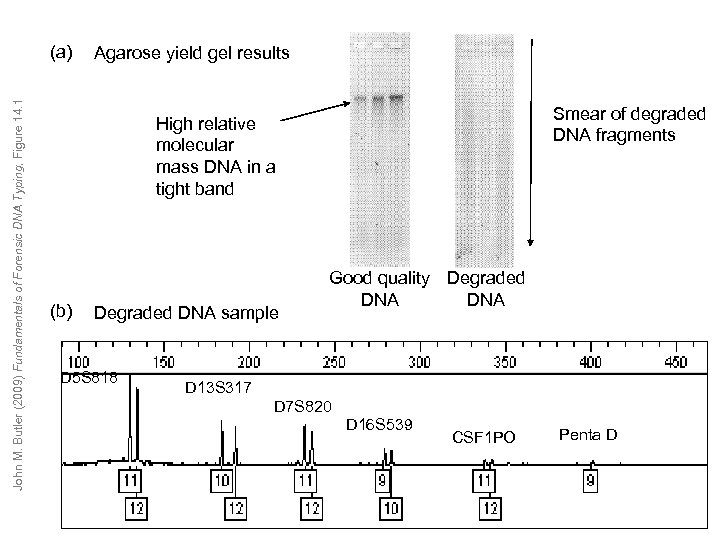 John M. Butler (2009) Fundamentals of Forensic DNA Typing, Figure 14. 1 (a) Agarose yield gel results Smear of degraded DNA fragments High relative molecular mass DNA in a tight band (b) Degraded DNA sample D 5 S 818 Good quality Degraded DNA D 13 S 317 D 7 S 820 D 16 S 539 CSF 1 PO Penta D
John M. Butler (2009) Fundamentals of Forensic DNA Typing, Figure 14. 1 (a) Agarose yield gel results Smear of degraded DNA fragments High relative molecular mass DNA in a tight band (b) Degraded DNA sample D 5 S 818 Good quality Degraded DNA D 13 S 317 D 7 S 820 D 16 S 539 CSF 1 PO Penta D
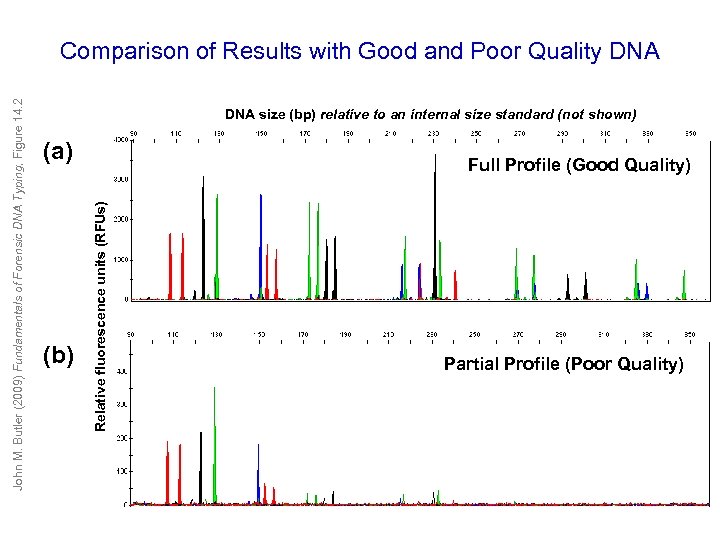 DNA size (bp) relative to an internal size standard (not shown) (a) (b) Full Profile (Good Quality) Relative fluorescence units (RFUs) John M. Butler (2009) Fundamentals of Forensic DNA Typing, Figure 14. 2 Comparison of Results with Good and Poor Quality DNA Partial Profile (Poor Quality)
DNA size (bp) relative to an internal size standard (not shown) (a) (b) Full Profile (Good Quality) Relative fluorescence units (RFUs) John M. Butler (2009) Fundamentals of Forensic DNA Typing, Figure 14. 2 Comparison of Results with Good and Poor Quality DNA Partial Profile (Poor Quality)
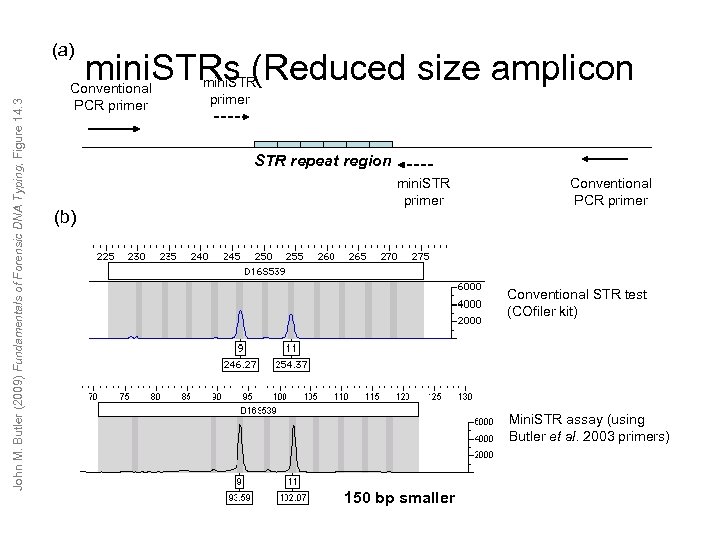 John M. Butler (2009) Fundamentals of Forensic DNA Typing, Figure 14. 3 (a) mini. STRs (Reduced size amplicon Conventional PCR primer mini. STR primer STR repeat region (b) mini. STR primer Conventional PCR primer Conventional STR test (COfiler kit) Mini. STR assay (using Butler et al. 2003 primers) 150 bp smaller
John M. Butler (2009) Fundamentals of Forensic DNA Typing, Figure 14. 3 (a) mini. STRs (Reduced size amplicon Conventional PCR primer mini. STR primer STR repeat region (b) mini. STR primer Conventional PCR primer Conventional STR test (COfiler kit) Mini. STR assay (using Butler et al. 2003 primers) 150 bp smaller
 Compromised Sample Improvements • Better DNA extraction/recovery • Continued use of mini. STRs – to improve success rates for recovery of information from compromised DNA evidence • Replicate results for reproducibility – to improve reliability with low-template DNA testing
Compromised Sample Improvements • Better DNA extraction/recovery • Continued use of mini. STRs – to improve success rates for recovery of information from compromised DNA evidence • Replicate results for reproducibility – to improve reliability with low-template DNA testing
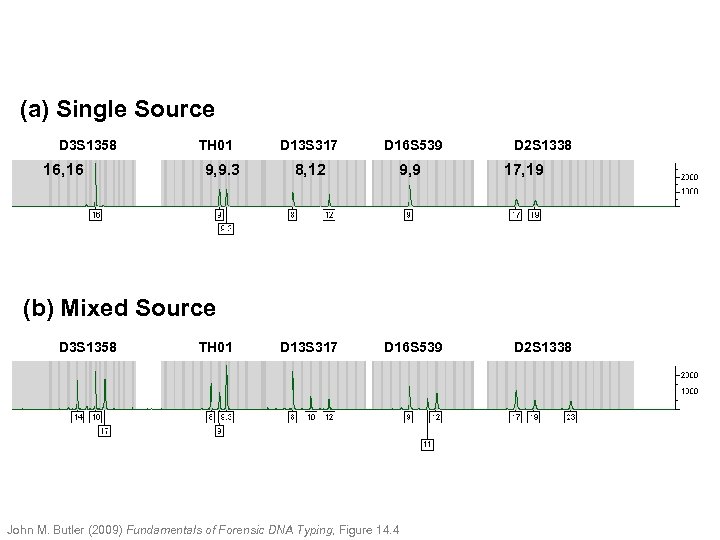 (a) Single Source D 3 S 1358 16, 16 TH 01 9, 9. 3 D 13 S 317 D 16 S 539 8, 12 9, 9 D 13 S 317 D 16 S 539 D 2 S 1338 17, 19 (b) Mixed Source D 3 S 1358 TH 01 John M. Butler (2009) Fundamentals of Forensic DNA Typing, Figure 14. 4 D 2 S 1338
(a) Single Source D 3 S 1358 16, 16 TH 01 9, 9. 3 D 13 S 317 D 16 S 539 8, 12 9, 9 D 13 S 317 D 16 S 539 D 2 S 1338 17, 19 (b) Mixed Source D 3 S 1358 TH 01 John M. Butler (2009) Fundamentals of Forensic DNA Typing, Figure 14. 4 D 2 S 1338
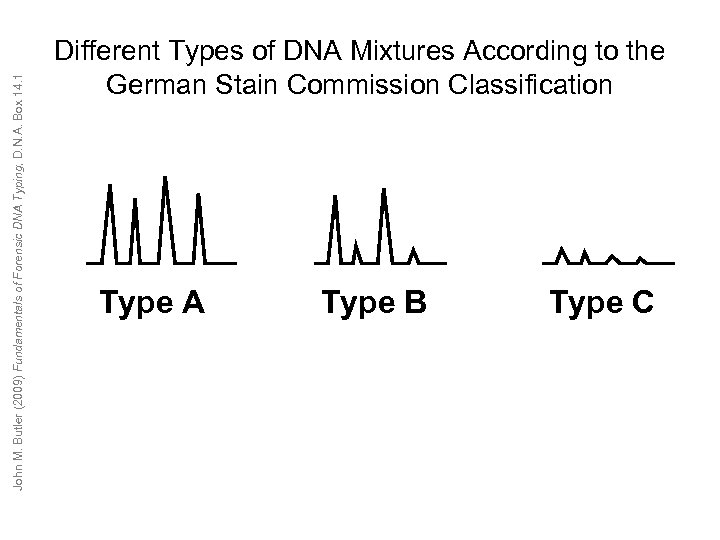 John M. Butler (2009) Fundamentals of Forensic DNA Typing, D. N. A. Box 14. 1 Different Types of DNA Mixtures According to the German Stain Commission Classification Type A Type B Type C
John M. Butler (2009) Fundamentals of Forensic DNA Typing, D. N. A. Box 14. 1 Different Types of DNA Mixtures According to the German Stain Commission Classification Type A Type B Type C
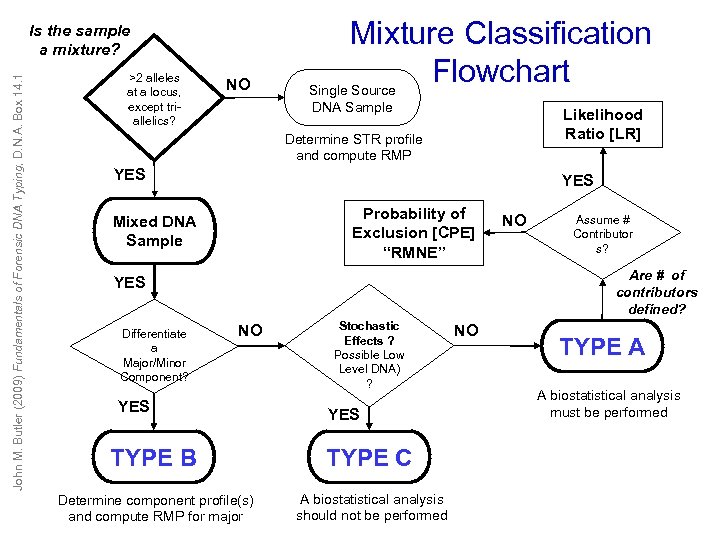 John M. Butler (2009) Fundamentals of Forensic DNA Typing, D. N. A. Box 14. 1 Is the sample a mixture? >2 alleles at a locus, except triallelics? NO Mixture Classification Flowchart Single Source DNA Sample Likelihood Ratio [LR] Determine STR profile and compute RMP YES Probability of Exclusion [CPE] “RMNE” Mixed DNA Sample Assume # Contributor s? Are # of contributors defined? YES Differentiate a Major/Minor Component? NO NO YES Stochastic Effects ? Possible Low Level DNA) ? YES TYPE B TYPE C Determine component profile(s) and compute RMP for major A biostatistical analysis should not be performed NO TYPE A A biostatistical analysis must be performed
John M. Butler (2009) Fundamentals of Forensic DNA Typing, D. N. A. Box 14. 1 Is the sample a mixture? >2 alleles at a locus, except triallelics? NO Mixture Classification Flowchart Single Source DNA Sample Likelihood Ratio [LR] Determine STR profile and compute RMP YES Probability of Exclusion [CPE] “RMNE” Mixed DNA Sample Assume # Contributor s? Are # of contributors defined? YES Differentiate a Major/Minor Component? NO NO YES Stochastic Effects ? Possible Low Level DNA) ? YES TYPE B TYPE C Determine component profile(s) and compute RMP for major A biostatistical analysis should not be performed NO TYPE A A biostatistical analysis must be performed
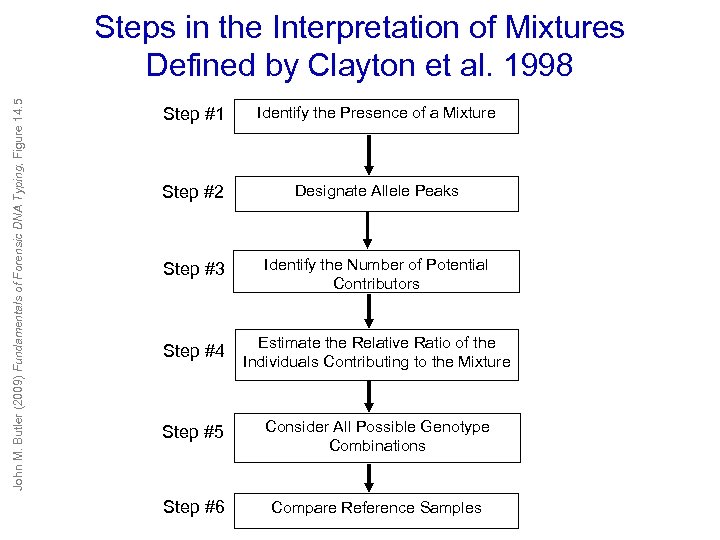 John M. Butler (2009) Fundamentals of Forensic DNA Typing, Figure 14. 5 Steps in the Interpretation of Mixtures Defined by Clayton et al. 1998 Step #1 Identify the Presence of a Mixture Step #2 Designate Allele Peaks Step #3 Identify the Number of Potential Contributors Step #4 Estimate the Relative Ratio of the Individuals Contributing to the Mixture Step #5 Consider All Possible Genotype Combinations Step #6 Compare Reference Samples
John M. Butler (2009) Fundamentals of Forensic DNA Typing, Figure 14. 5 Steps in the Interpretation of Mixtures Defined by Clayton et al. 1998 Step #1 Identify the Presence of a Mixture Step #2 Designate Allele Peaks Step #3 Identify the Number of Potential Contributors Step #4 Estimate the Relative Ratio of the Individuals Contributing to the Mixture Step #5 Consider All Possible Genotype Combinations Step #6 Compare Reference Samples
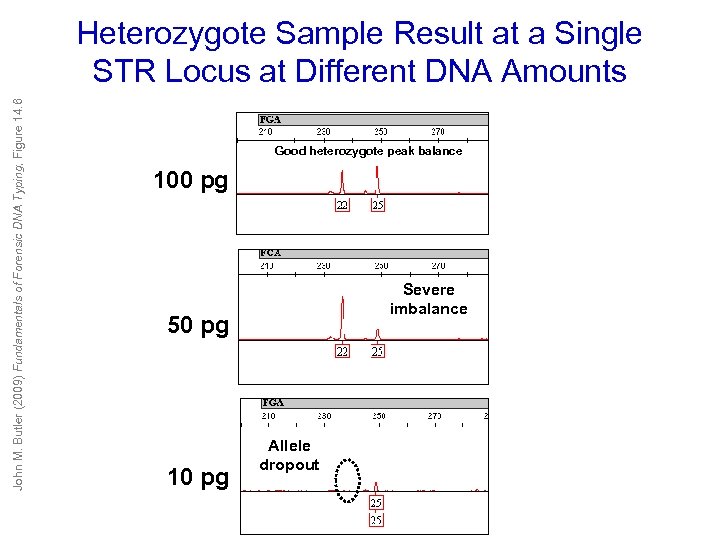 John M. Butler (2009) Fundamentals of Forensic DNA Typing, Figure 14. 6 Heterozygote Sample Result at a Single STR Locus at Different DNA Amounts Good heterozygote peak balance 100 pg Severe imbalance 50 pg 10 pg Allele dropout
John M. Butler (2009) Fundamentals of Forensic DNA Typing, Figure 14. 6 Heterozygote Sample Result at a Single STR Locus at Different DNA Amounts Good heterozygote peak balance 100 pg Severe imbalance 50 pg 10 pg Allele dropout
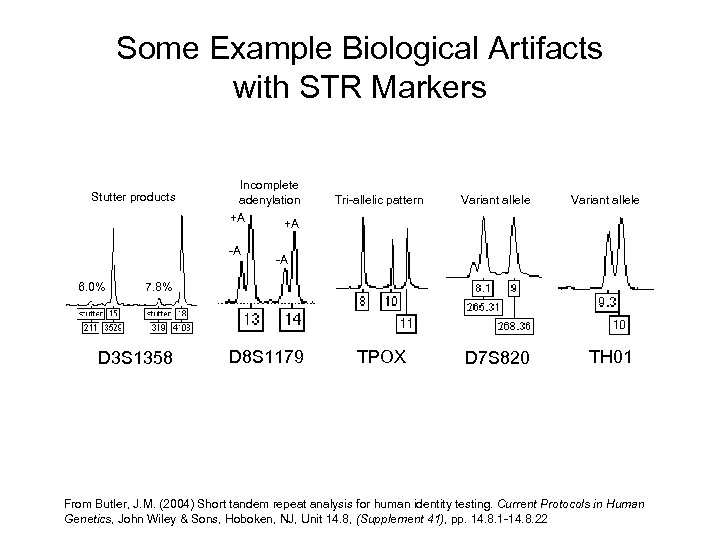 Some Example Biological Artifacts with STR Markers Stutter products Incomplete adenylation +A +A -A 6. 0% Tri-allelic pattern Variant allele TPOX D 7 S 820 Variant allele -A 7. 8% D 3 S 1358 D 8 S 1179 TH 01 From Butler, J. M. (2004) Short tandem repeat analysis for human identity testing. Current Protocols in Human Genetics, John Wiley & Sons, Hoboken, NJ, Unit 14. 8, (Supplement 41), pp. 14. 8. 1 -14. 8. 22
Some Example Biological Artifacts with STR Markers Stutter products Incomplete adenylation +A +A -A 6. 0% Tri-allelic pattern Variant allele TPOX D 7 S 820 Variant allele -A 7. 8% D 3 S 1358 D 8 S 1179 TH 01 From Butler, J. M. (2004) Short tandem repeat analysis for human identity testing. Current Protocols in Human Genetics, John Wiley & Sons, Hoboken, NJ, Unit 14. 8, (Supplement 41), pp. 14. 8. 1 -14. 8. 22
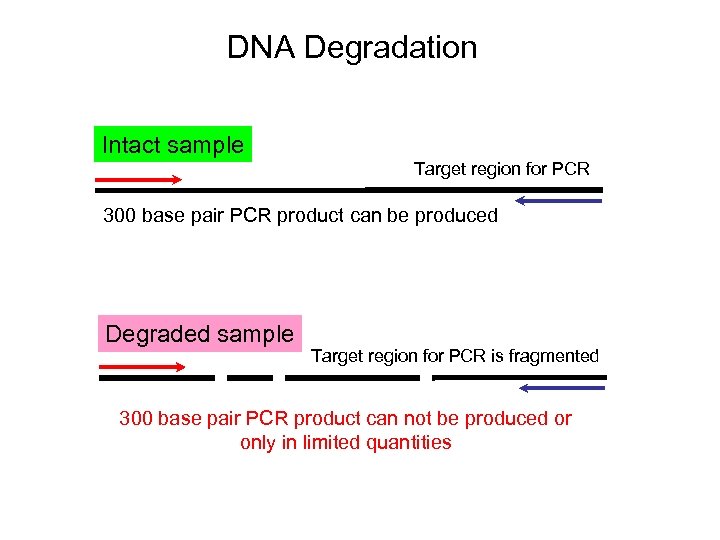 DNA Degradation Intact sample Target region for PCR 300 base pair PCR product can be produced Degraded sample Target region for PCR is fragmented 300 base pair PCR product can not be produced or only in limited quantities
DNA Degradation Intact sample Target region for PCR 300 base pair PCR product can be produced Degraded sample Target region for PCR is fragmented 300 base pair PCR product can not be produced or only in limited quantities
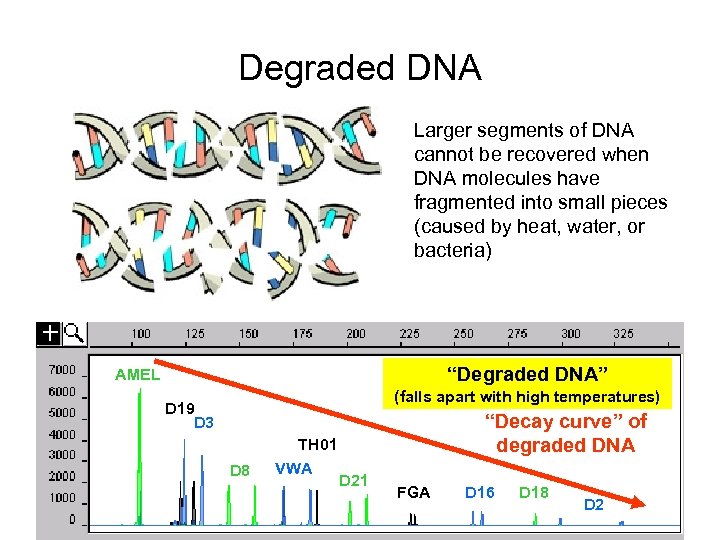 Degraded DNA Larger segments of DNA cannot be recovered when DNA molecules have fragmented into small pieces (caused by heat, water, or bacteria) “Degraded DNA” AMEL (falls apart with high temperatures) D 19 D 3 D 8 TH 01 VWA “Decay curve” of degraded DNA D 21 FGA D 16 D 18 D 2
Degraded DNA Larger segments of DNA cannot be recovered when DNA molecules have fragmented into small pieces (caused by heat, water, or bacteria) “Degraded DNA” AMEL (falls apart with high temperatures) D 19 D 3 D 8 TH 01 VWA “Decay curve” of degraded DNA D 21 FGA D 16 D 18 D 2
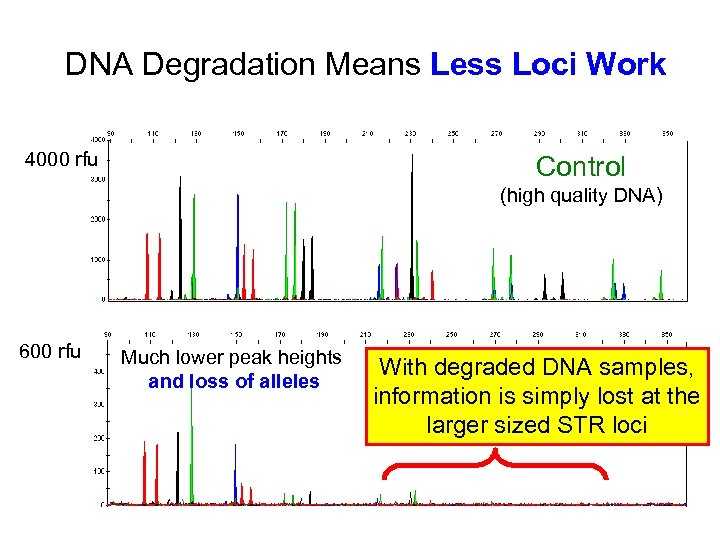 DNA Degradation Means Less Loci Work 4000 rfu Control (high quality DNA) 600 rfu Much lower peak heights and loss of alleles With degraded DNA samples, Degraded information is simply lost at the larger sized STR loci
DNA Degradation Means Less Loci Work 4000 rfu Control (high quality DNA) 600 rfu Much lower peak heights and loss of alleles With degraded DNA samples, Degraded information is simply lost at the larger sized STR loci
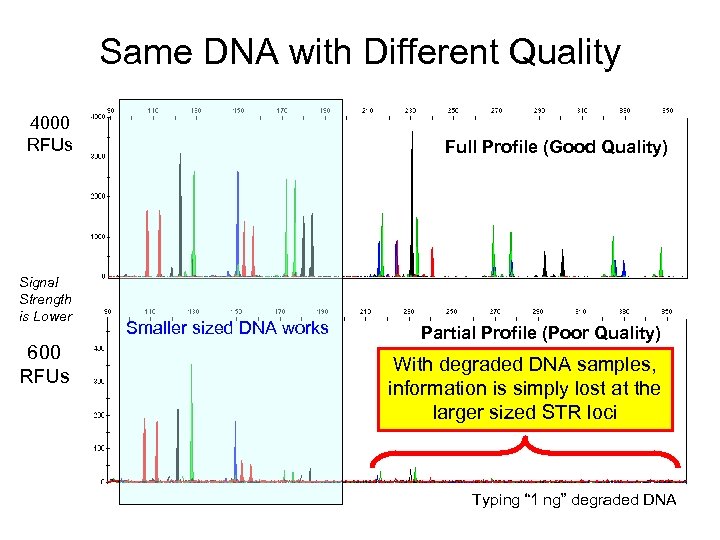 Same DNA with Different Quality 4000 RFUs Signal Strength is Lower 600 RFUs Full Profile (Good Quality) Smaller sized DNA works Partial Profile (Poor Quality) With degraded DNA samples, information is simply lost at the larger sized STR loci Typing “ 1 ng” degraded DNA
Same DNA with Different Quality 4000 RFUs Signal Strength is Lower 600 RFUs Full Profile (Good Quality) Smaller sized DNA works Partial Profile (Poor Quality) With degraded DNA samples, information is simply lost at the larger sized STR loci Typing “ 1 ng” degraded DNA
 Impact of Degraded DNA Samples • Comparison to a phone number (string of 13 numbers) 001 -301 -975 -4049 • If you only had “ 4049”…this information would be of limited value since it is not as specific (and could match other phone numbers from different area codes) • DNA profiles are essentially a string of numbers – if the DNA is damaged, then the string of numbers is shorter and less informative… ------4049 or ----301 -9 -------
Impact of Degraded DNA Samples • Comparison to a phone number (string of 13 numbers) 001 -301 -975 -4049 • If you only had “ 4049”…this information would be of limited value since it is not as specific (and could match other phone numbers from different area codes) • DNA profiles are essentially a string of numbers – if the DNA is damaged, then the string of numbers is shorter and less informative… ------4049 or ----301 -9 -------
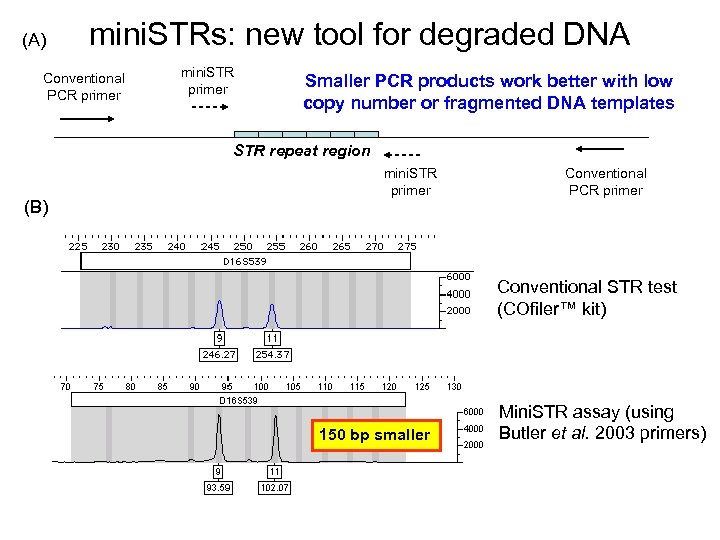 (A) mini. STRs: new tool for degraded DNA Conventional PCR primer mini. STR primer Smaller PCR products work better with low copy number or fragmented DNA templates STR repeat region (B) mini. STR primer Conventional PCR primer Conventional STR test (COfiler™ kit) 150 bp smaller Mini. STR assay (using Butler et al. 2003 primers)
(A) mini. STRs: new tool for degraded DNA Conventional PCR primer mini. STR primer Smaller PCR products work better with low copy number or fragmented DNA templates STR repeat region (B) mini. STR primer Conventional PCR primer Conventional STR test (COfiler™ kit) 150 bp smaller Mini. STR assay (using Butler et al. 2003 primers)
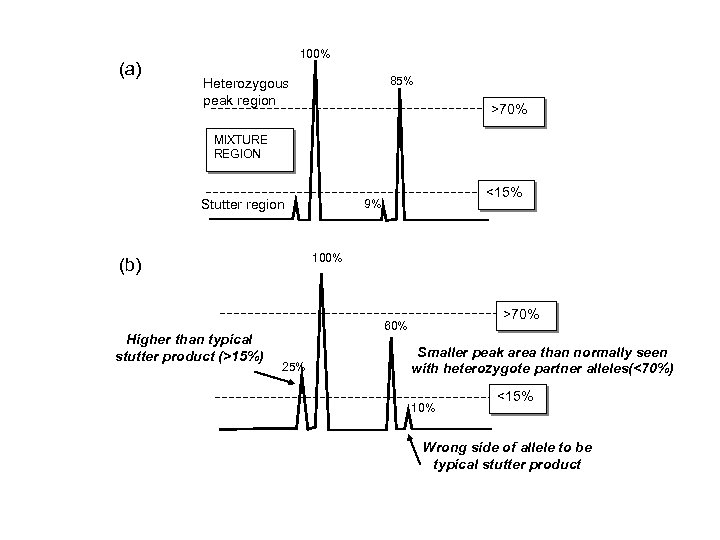 (a) 100% 85% Heterozygous peak region >70% MIXTURE REGION Stutter region 100% (b) Higher than typical stutter product (>15%) <15% 9% >70% 60% 25% Smaller peak area than normally seen with heterozygote partner alleles(<70%) 10% <15% Wrong side of allele to be typical stutter product
(a) 100% 85% Heterozygous peak region >70% MIXTURE REGION Stutter region 100% (b) Higher than typical stutter product (>15%) <15% 9% >70% 60% 25% Smaller peak area than normally seen with heterozygote partner alleles(<70%) 10% <15% Wrong side of allele to be typical stutter product
 Mixture Basics From J. M. Butler (2005) Forensic DNA Typing, 2 nd Edition, p. 154 • Mixtures arise when two or more individuals contribute to the sample being tested. • Mixtures can be challenging to detect and interpret without extensive experience and careful training. Even more challenging with poor quality data when degraded DNA is present… • Differential extraction can help distinguish male and female components of many sexual assault mixtures. Y-chromosome markers can help here in some cases…
Mixture Basics From J. M. Butler (2005) Forensic DNA Typing, 2 nd Edition, p. 154 • Mixtures arise when two or more individuals contribute to the sample being tested. • Mixtures can be challenging to detect and interpret without extensive experience and careful training. Even more challenging with poor quality data when degraded DNA is present… • Differential extraction can help distinguish male and female components of many sexual assault mixtures. Y-chromosome markers can help here in some cases…
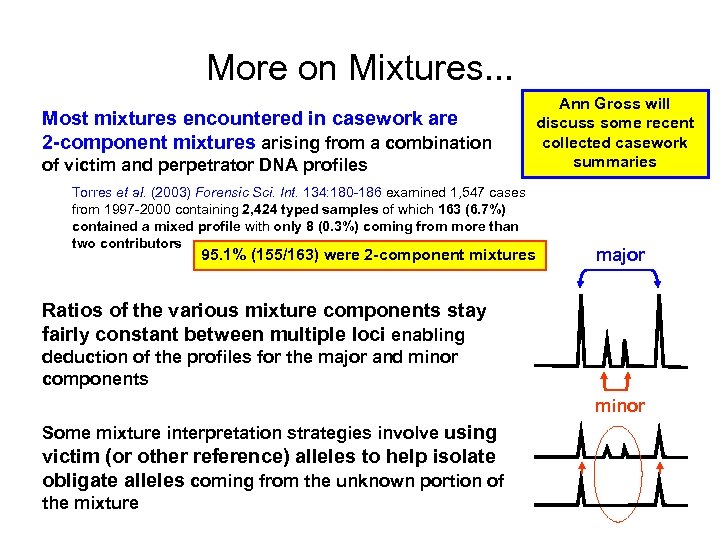 More on Mixtures. . . Most mixtures encountered in casework are 2 -component mixtures arising from a combination of victim and perpetrator DNA profiles Torres et al. (2003) Forensic Sci. Int. 134: 180 -186 examined 1, 547 cases from 1997 -2000 containing 2, 424 typed samples of which 163 (6. 7%) contained a mixed profile with only 8 (0. 3%) coming from more than two contributors Ann Gross will discuss some recent collected casework summaries 95. 1% (155/163) were 2 -component mixtures major Ratios of the various mixture components stay fairly constant between multiple loci enabling deduction of the profiles for the major and minor components minor Some mixture interpretation strategies involve using victim (or other reference) alleles to help isolate obligate alleles coming from the unknown portion of the mixture
More on Mixtures. . . Most mixtures encountered in casework are 2 -component mixtures arising from a combination of victim and perpetrator DNA profiles Torres et al. (2003) Forensic Sci. Int. 134: 180 -186 examined 1, 547 cases from 1997 -2000 containing 2, 424 typed samples of which 163 (6. 7%) contained a mixed profile with only 8 (0. 3%) coming from more than two contributors Ann Gross will discuss some recent collected casework summaries 95. 1% (155/163) were 2 -component mixtures major Ratios of the various mixture components stay fairly constant between multiple loci enabling deduction of the profiles for the major and minor components minor Some mixture interpretation strategies involve using victim (or other reference) alleles to help isolate obligate alleles coming from the unknown portion of the mixture
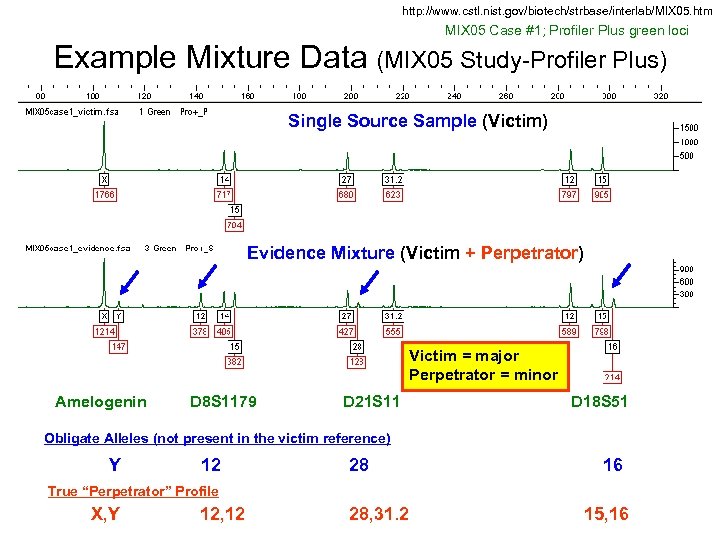 http: //www. cstl. nist. gov/biotech/strbase/interlab/MIX 05. htm MIX 05 Case #1; Profiler Plus green loci Example Mixture Data (MIX 05 Study-Profiler Plus) Single Source Sample (Victim) Evidence Mixture (Victim + Perpetrator) Victim = major Perpetrator = minor Amelogenin D 8 S 1179 D 21 S 11 D 18 S 51 Obligate Alleles (not present in the victim reference) Y 12 28 16 True “Perpetrator” Profile X, Y 12, 12 28, 31. 2 15, 16
http: //www. cstl. nist. gov/biotech/strbase/interlab/MIX 05. htm MIX 05 Case #1; Profiler Plus green loci Example Mixture Data (MIX 05 Study-Profiler Plus) Single Source Sample (Victim) Evidence Mixture (Victim + Perpetrator) Victim = major Perpetrator = minor Amelogenin D 8 S 1179 D 21 S 11 D 18 S 51 Obligate Alleles (not present in the victim reference) Y 12 28 16 True “Perpetrator” Profile X, Y 12, 12 28, 31. 2 15, 16
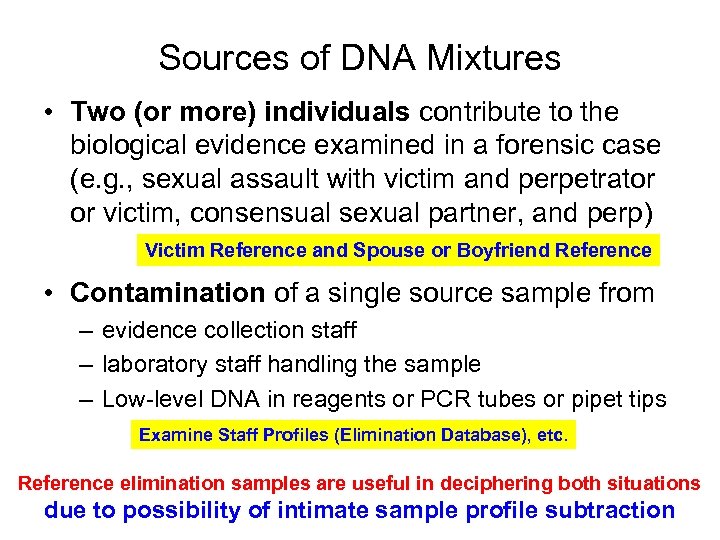 Sources of DNA Mixtures • Two (or more) individuals contribute to the biological evidence examined in a forensic case (e. g. , sexual assault with victim and perpetrator or victim, consensual sexual partner, and perp) Victim Reference and Spouse or Boyfriend Reference • Contamination of a single source sample from – evidence collection staff – laboratory staff handling the sample – Low-level DNA in reagents or PCR tubes or pipet tips Examine Staff Profiles (Elimination Database), etc. Reference elimination samples are useful in deciphering both situations due to possibility of intimate sample profile subtraction
Sources of DNA Mixtures • Two (or more) individuals contribute to the biological evidence examined in a forensic case (e. g. , sexual assault with victim and perpetrator or victim, consensual sexual partner, and perp) Victim Reference and Spouse or Boyfriend Reference • Contamination of a single source sample from – evidence collection staff – laboratory staff handling the sample – Low-level DNA in reagents or PCR tubes or pipet tips Examine Staff Profiles (Elimination Database), etc. Reference elimination samples are useful in deciphering both situations due to possibility of intimate sample profile subtraction
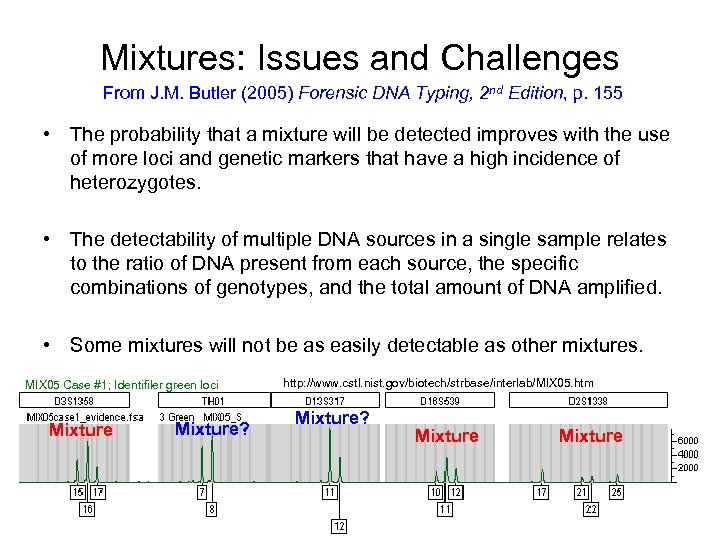 Mixtures: Issues and Challenges From J. M. Butler (2005) Forensic DNA Typing, 2 nd Edition, p. 155 • The probability that a mixture will be detected improves with the use of more loci and genetic markers that have a high incidence of heterozygotes. • The detectability of multiple DNA sources in a single sample relates to the ratio of DNA present from each source, the specific combinations of genotypes, and the total amount of DNA amplified. • Some mixtures will not be as easily detectable as other mixtures. MIX 05 Case #1; Identifiler green loci Mixture? http: //www. cstl. nist. gov/biotech/strbase/interlab/MIX 05. htm Mixture? Mixture
Mixtures: Issues and Challenges From J. M. Butler (2005) Forensic DNA Typing, 2 nd Edition, p. 155 • The probability that a mixture will be detected improves with the use of more loci and genetic markers that have a high incidence of heterozygotes. • The detectability of multiple DNA sources in a single sample relates to the ratio of DNA present from each source, the specific combinations of genotypes, and the total amount of DNA amplified. • Some mixtures will not be as easily detectable as other mixtures. MIX 05 Case #1; Identifiler green loci Mixture? http: //www. cstl. nist. gov/biotech/strbase/interlab/MIX 05. htm Mixture? Mixture
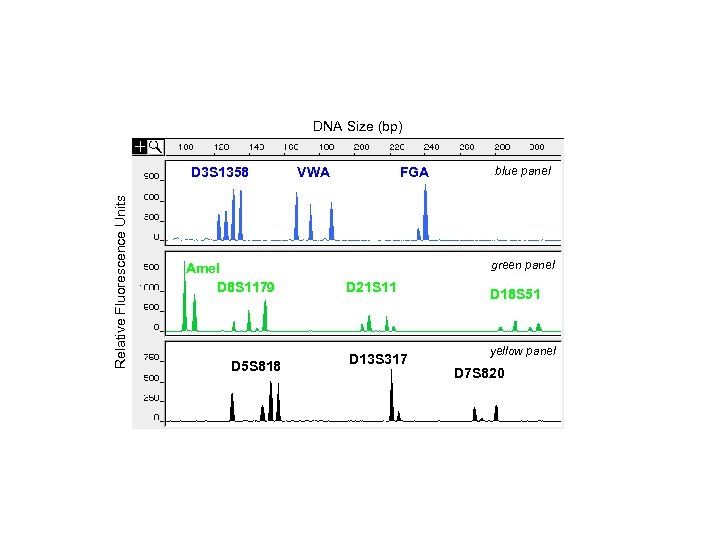 DNA Size (bp) Relative Fluorescence Units D 3 S 1358 Amel D 8 S 1179 D 5 S 818 VWA FGA blue panel green panel D 21 S 11 D 13 S 317 D 18 S 51 yellow panel D 7 S 820
DNA Size (bp) Relative Fluorescence Units D 3 S 1358 Amel D 8 S 1179 D 5 S 818 VWA FGA blue panel green panel D 21 S 11 D 13 S 317 D 18 S 51 yellow panel D 7 S 820
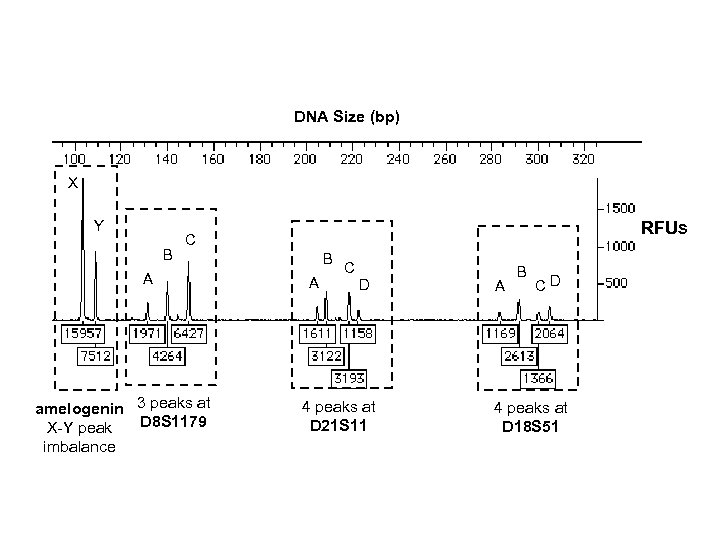 DNA Size (bp) X Y B RFUs C A amelogenin 3 peaks at D 8 S 1179 X-Y peak imbalance B A C D 4 peaks at D 21 S 11 A B CD 4 peaks at D 18 S 51
DNA Size (bp) X Y B RFUs C A amelogenin 3 peaks at D 8 S 1179 X-Y peak imbalance B A C D 4 peaks at D 21 S 11 A B CD 4 peaks at D 18 S 51
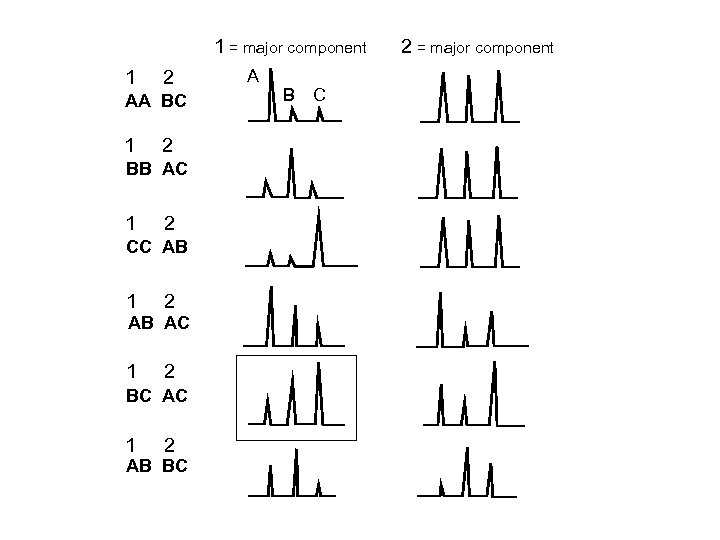 1 = major component 1 2 AA BC 1 2 BB AC 1 2 CC AB 1 2 AB AC 1 2 BC AC 1 2 AB BC A B C 2 = major component
1 = major component 1 2 AA BC 1 2 BB AC 1 2 CC AB 1 2 AB AC 1 2 BC AC 1 2 AB BC A B C 2 = major component
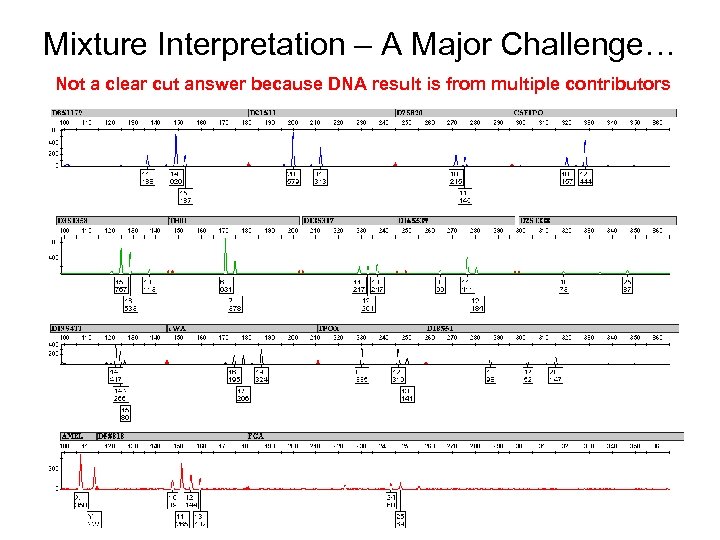 Mixture Interpretation – A Major Challenge… Not a clear cut answer because DNA result is from multiple contributors
Mixture Interpretation – A Major Challenge… Not a clear cut answer because DNA result is from multiple contributors
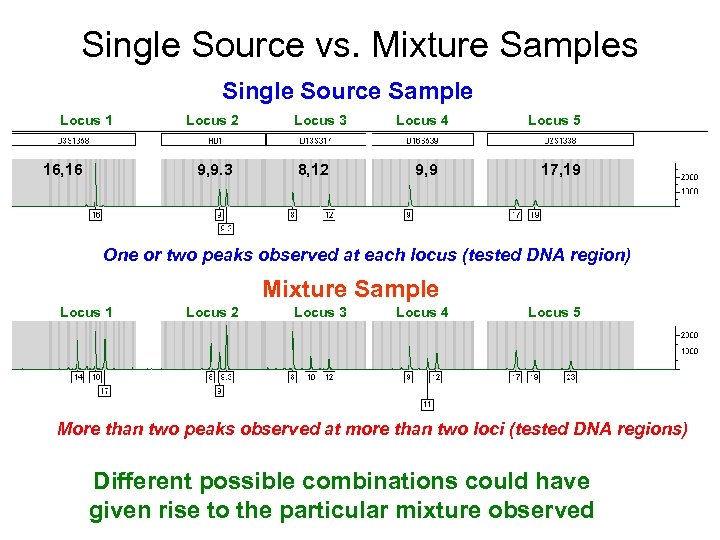 Single Source vs. Mixture Samples Single Source Sample Locus 1 16, 16 Locus 2 9, 9. 3 Locus 3 8, 12 Locus 4 9, 9 Locus 5 17, 19 One or two peaks observed at each locus (tested DNA region) Mixture Sample Locus 1 Locus 2 Locus 3 Locus 4 Locus 5 More than two peaks observed at more than two loci (tested DNA regions) Different possible combinations could have given rise to the particular mixture observed
Single Source vs. Mixture Samples Single Source Sample Locus 1 16, 16 Locus 2 9, 9. 3 Locus 3 8, 12 Locus 4 9, 9 Locus 5 17, 19 One or two peaks observed at each locus (tested DNA region) Mixture Sample Locus 1 Locus 2 Locus 3 Locus 4 Locus 5 More than two peaks observed at more than two loci (tested DNA regions) Different possible combinations could have given rise to the particular mixture observed
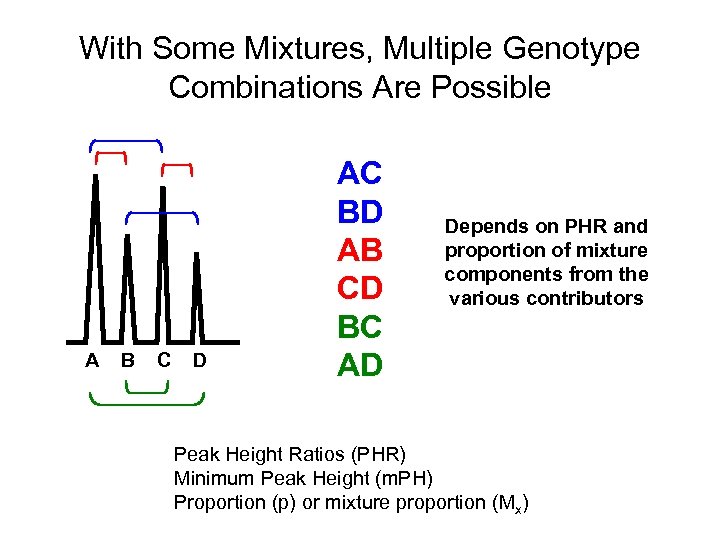 With Some Mixtures, Multiple Genotype Combinations Are Possible A B C D AC BD AB CD BC AD Depends on PHR and proportion of mixture components from the various contributors Peak Height Ratios (PHR) Minimum Peak Height (m. PH) Proportion (p) or mixture proportion (Mx)
With Some Mixtures, Multiple Genotype Combinations Are Possible A B C D AC BD AB CD BC AD Depends on PHR and proportion of mixture components from the various contributors Peak Height Ratios (PHR) Minimum Peak Height (m. PH) Proportion (p) or mixture proportion (Mx)
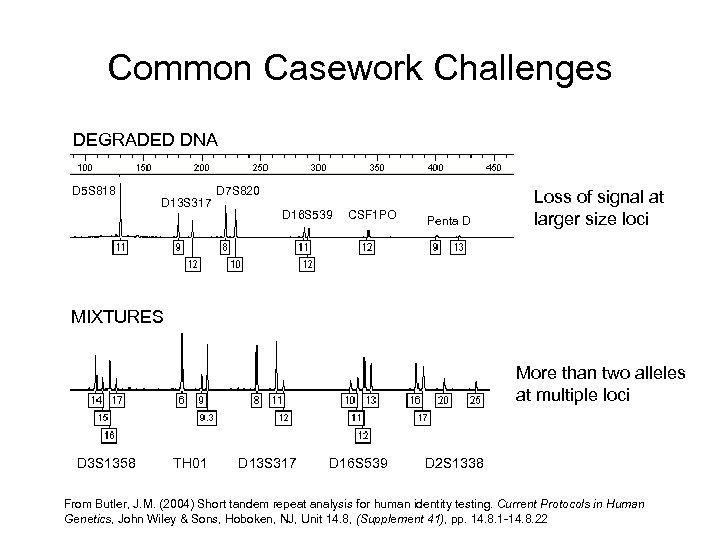 Common Casework Challenges DEGRADED DNA D 5 S 818 D 13 S 317 D 7 S 820 D 16 S 539 CSF 1 PO Penta D Loss of signal at larger size loci MIXTURES More than two alleles at multiple loci D 3 S 1358 TH 01 D 13 S 317 D 16 S 539 D 2 S 1338 From Butler, J. M. (2004) Short tandem repeat analysis for human identity testing. Current Protocols in Human Genetics, John Wiley & Sons, Hoboken, NJ, Unit 14. 8, (Supplement 41), pp. 14. 8. 1 -14. 8. 22
Common Casework Challenges DEGRADED DNA D 5 S 818 D 13 S 317 D 7 S 820 D 16 S 539 CSF 1 PO Penta D Loss of signal at larger size loci MIXTURES More than two alleles at multiple loci D 3 S 1358 TH 01 D 13 S 317 D 16 S 539 D 2 S 1338 From Butler, J. M. (2004) Short tandem repeat analysis for human identity testing. Current Protocols in Human Genetics, John Wiley & Sons, Hoboken, NJ, Unit 14. 8, (Supplement 41), pp. 14. 8. 1 -14. 8. 22
 DNA Testing Has Become Extremely Sensitive… • What does it mean to obtain a DNA match between a suspect and material from a crime scene? • Is the fact that a DNA profile obtained mean that this information is probative? • More complicated samples (mixtures) and more items per case being submitted to labs
DNA Testing Has Become Extremely Sensitive… • What does it mean to obtain a DNA match between a suspect and material from a crime scene? • Is the fact that a DNA profile obtained mean that this information is probative? • More complicated samples (mixtures) and more items per case being submitted to labs
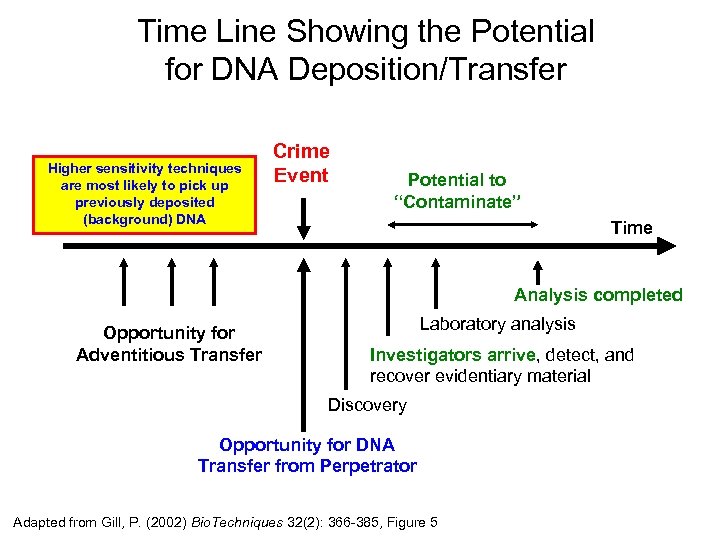 Time Line Showing the Potential for DNA Deposition/Transfer Higher sensitivity techniques are most likely to pick up previously deposited (background) DNA Crime Event Potential to “Contaminate” Time Analysis completed Opportunity for Adventitious Transfer Laboratory analysis Investigators arrive, detect, and recover evidentiary material Discovery Opportunity for DNA Transfer from Perpetrator Adapted from Gill, P. (2002) Bio. Techniques 32(2): 366 -385, Figure 5
Time Line Showing the Potential for DNA Deposition/Transfer Higher sensitivity techniques are most likely to pick up previously deposited (background) DNA Crime Event Potential to “Contaminate” Time Analysis completed Opportunity for Adventitious Transfer Laboratory analysis Investigators arrive, detect, and recover evidentiary material Discovery Opportunity for DNA Transfer from Perpetrator Adapted from Gill, P. (2002) Bio. Techniques 32(2): 366 -385, Figure 5
 Chapter 14 – Points for Discussion • Discuss advantages and disadvantages of mini. STR assays. • Name at least two ways that the presence of a mixture can be detected. • What are the causes of allele drop-in and allele drop-out in the context of amplifying low amounts of DNA template?
Chapter 14 – Points for Discussion • Discuss advantages and disadvantages of mini. STR assays. • Name at least two ways that the presence of a mixture can be detected. • What are the causes of allele drop-in and allele drop-out in the context of amplifying low amounts of DNA template?


