13_рус.ppt
- Количество слайдов: 51
 Chapter 13 The Structure of Genomes Variations in genome anatomy in different organisms © 2005 Prentice Hall Inc. / A Pearson Education Company / Upper Saddle River, New Jersey 07458
Chapter 13 The Structure of Genomes Variations in genome anatomy in different organisms © 2005 Prentice Hall Inc. / A Pearson Education Company / Upper Saddle River, New Jersey 07458
 Contents · · · Differences in gene structure among the life domains Variations in genome size Isochores Homology in noncoding regions Noncoding DNA · Pseudogenes · Repeats · Transposons · Function of noncoding DNA © 2005 Prentice Hall Inc. / A Pearson Education Company / Upper Saddle River, New Jersey 07458
Contents · · · Differences in gene structure among the life domains Variations in genome size Isochores Homology in noncoding regions Noncoding DNA · Pseudogenes · Repeats · Transposons · Function of noncoding DNA © 2005 Prentice Hall Inc. / A Pearson Education Company / Upper Saddle River, New Jersey 07458
 Variation in genome structure · Genetic code is almost universal, but genome structure varies considerably from organism to organism · Genomics reveals whole-genome view of organisms · Sources of genome variation · Duplication events (pseudogenes, genome duplications) · Transposons (sequence elements that jump around genome) · Mutations (e. g. , microsatellites) · Biophysical constraints (repeats at centromeres, telomeres) © 2005 Prentice Hall Inc. / A Pearson Education Company / Upper Saddle River, New Jersey 07458
Variation in genome structure · Genetic code is almost universal, but genome structure varies considerably from organism to organism · Genomics reveals whole-genome view of organisms · Sources of genome variation · Duplication events (pseudogenes, genome duplications) · Transposons (sequence elements that jump around genome) · Mutations (e. g. , microsatellites) · Biophysical constraints (repeats at centromeres, telomeres) © 2005 Prentice Hall Inc. / A Pearson Education Company / Upper Saddle River, New Jersey 07458
 Comparison of genome structure between life domains · Bacteria · No introns · Circular chromosomes w/ plasmids · Archaea · Some introns · TATA box–like binding sites (like Eukarya, unlike Bacteria) · Circular chromosomes with plasmids · Eukarya · Many introns and exons · Chromosomes located inside nucleus · Chromosomal DNA tightly bound to histones © 2005 Prentice Hall Inc. / A Pearson Education Company / Upper Saddle River, New Jersey 07458
Comparison of genome structure between life domains · Bacteria · No introns · Circular chromosomes w/ plasmids · Archaea · Some introns · TATA box–like binding sites (like Eukarya, unlike Bacteria) · Circular chromosomes with plasmids · Eukarya · Many introns and exons · Chromosomes located inside nucleus · Chromosomal DNA tightly bound to histones © 2005 Prentice Hall Inc. / A Pearson Education Company / Upper Saddle River, New Jersey 07458
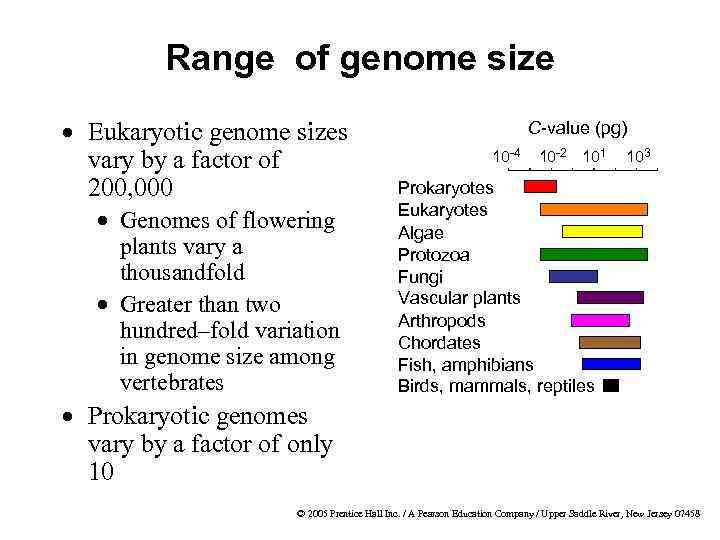 Range of genome size · Eukaryotic genome sizes vary by a factor of 200, 000 · Genomes of flowering plants vary a thousandfold · Greater than two hundred–fold variation in genome size among vertebrates C-value (pg) 10 -4 10 -2 101 103 Prokaryotes Eukaryotes Algae Protozoa Fungi Vascular plants Arthropods Chordates Fish, amphibians Birds, mammals, reptiles · Prokaryotic genomes vary by a factor of only 10 © 2005 Prentice Hall Inc. / A Pearson Education Company / Upper Saddle River, New Jersey 07458
Range of genome size · Eukaryotic genome sizes vary by a factor of 200, 000 · Genomes of flowering plants vary a thousandfold · Greater than two hundred–fold variation in genome size among vertebrates C-value (pg) 10 -4 10 -2 101 103 Prokaryotes Eukaryotes Algae Protozoa Fungi Vascular plants Arthropods Chordates Fish, amphibians Birds, mammals, reptiles · Prokaryotic genomes vary by a factor of only 10 © 2005 Prentice Hall Inc. / A Pearson Education Company / Upper Saddle River, New Jersey 07458
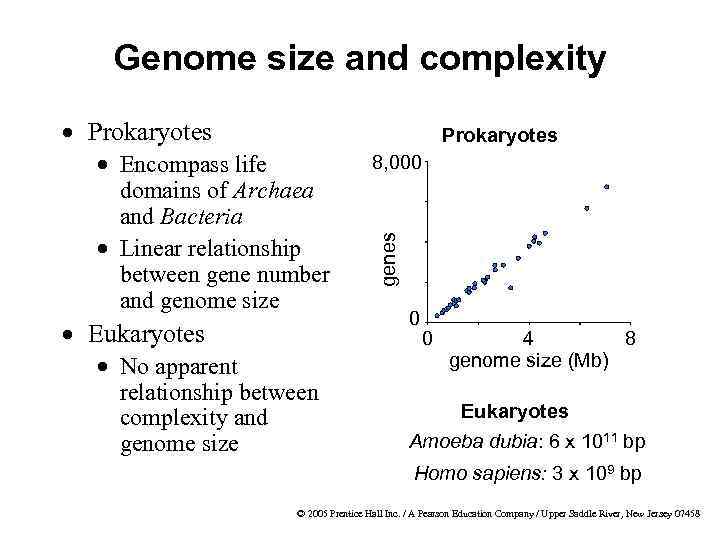 Genome size and complexity · Prokaryotes · Eukaryotes · No apparent relationship between complexity and genome size 8, 000 genes · Encompass life domains of Archaea and Bacteria · Linear relationship between gene number and genome size 0 0 4 8 genome size (Mb) Eukaryotes Amoeba dubia: 6 x 1011 bp Homo sapiens: 3 x 109 bp © 2005 Prentice Hall Inc. / A Pearson Education Company / Upper Saddle River, New Jersey 07458
Genome size and complexity · Prokaryotes · Eukaryotes · No apparent relationship between complexity and genome size 8, 000 genes · Encompass life domains of Archaea and Bacteria · Linear relationship between gene number and genome size 0 0 4 8 genome size (Mb) Eukaryotes Amoeba dubia: 6 x 1011 bp Homo sapiens: 3 x 109 bp © 2005 Prentice Hall Inc. / A Pearson Education Company / Upper Saddle River, New Jersey 07458
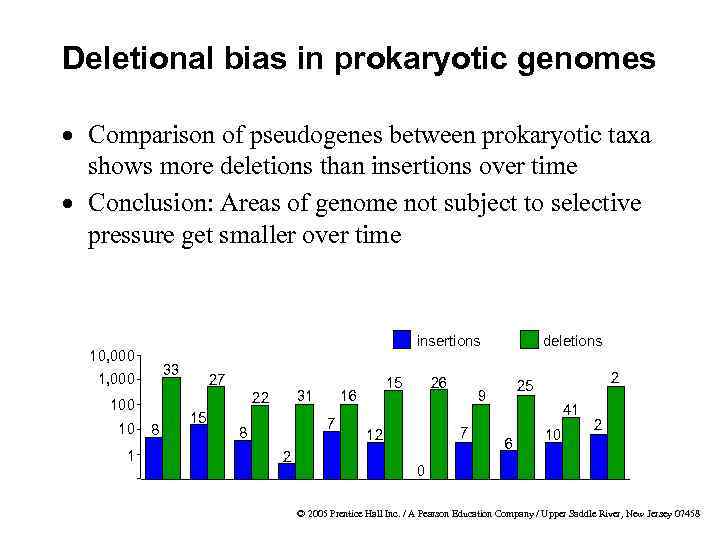 Deletional bias in prokaryotic genomes · Comparison of pseudogenes between prokaryotic taxa shows more deletions than insertions over time · Conclusion: Areas of genome not subject to selective pressure get smaller over time insertions 10, 000 33 1, 000 1 8 15 7 8 2 15 16 31 22 100 10 27 26 2 25 9 7 12 deletions 41 6 10 2 0 © 2005 Prentice Hall Inc. / A Pearson Education Company / Upper Saddle River, New Jersey 07458
Deletional bias in prokaryotic genomes · Comparison of pseudogenes between prokaryotic taxa shows more deletions than insertions over time · Conclusion: Areas of genome not subject to selective pressure get smaller over time insertions 10, 000 33 1, 000 1 8 15 7 8 2 15 16 31 22 100 10 27 26 2 25 9 7 12 deletions 41 6 10 2 0 © 2005 Prentice Hall Inc. / A Pearson Education Company / Upper Saddle River, New Jersey 07458
 The C-value paradox · Why is there a lack of correlation between genome size and complexity in eukaryotes? · Some things do correlate to genome size · Duration of the cell cycle · Minimum cell volume · Presence of LTR transposon sequences in various species of grasses © 2005 Prentice Hall Inc. / A Pearson Education Company / Upper Saddle River, New Jersey 07458
The C-value paradox · Why is there a lack of correlation between genome size and complexity in eukaryotes? · Some things do correlate to genome size · Duration of the cell cycle · Minimum cell volume · Presence of LTR transposon sequences in various species of grasses © 2005 Prentice Hall Inc. / A Pearson Education Company / Upper Saddle River, New Jersey 07458
 Hypotheses to explain the C-value paradox · Some hypotheses to explain the paradox · · Junk DNA Selfish DNA Nucleoskeletal hypothesis Nucleotypic hypothesis · Developmental control · Transposon hypothesis © 2005 Prentice Hall Inc. / A Pearson Education Company / Upper Saddle River, New Jersey 07458
Hypotheses to explain the C-value paradox · Some hypotheses to explain the paradox · · Junk DNA Selfish DNA Nucleoskeletal hypothesis Nucleotypic hypothesis · Developmental control · Transposon hypothesis © 2005 Prentice Hall Inc. / A Pearson Education Company / Upper Saddle River, New Jersey 07458
 Caveats · Junk DNA and selfish DNA · Lack of constant accumulation in many organisms with large genomes · Nucleoskeletal hypothesis · Not compatible with sudden changes in genome size · Nucleotypic hypothesis · Not always true across taxa · Transposons · Relationship not quite linear © 2005 Prentice Hall Inc. / A Pearson Education Company / Upper Saddle River, New Jersey 07458
Caveats · Junk DNA and selfish DNA · Lack of constant accumulation in many organisms with large genomes · Nucleoskeletal hypothesis · Not compatible with sudden changes in genome size · Nucleotypic hypothesis · Not always true across taxa · Transposons · Relationship not quite linear © 2005 Prentice Hall Inc. / A Pearson Education Company / Upper Saddle River, New Jersey 07458
 Genome features · Variations in base-pair content · Isochores · Duplicated genes · Paralogous genes · Pseudogenes · Repetitive sequences · Minisatellites and microsatellites · Transposon sequences · Foreign DNA · DNA from other free-living organisms · Viral DNA © 2005 Prentice Hall Inc. / A Pearson Education Company / Upper Saddle River, New Jersey 07458
Genome features · Variations in base-pair content · Isochores · Duplicated genes · Paralogous genes · Pseudogenes · Repetitive sequences · Minisatellites and microsatellites · Transposon sequences · Foreign DNA · DNA from other free-living organisms · Viral DNA © 2005 Prentice Hall Inc. / A Pearson Education Company / Upper Saddle River, New Jersey 07458
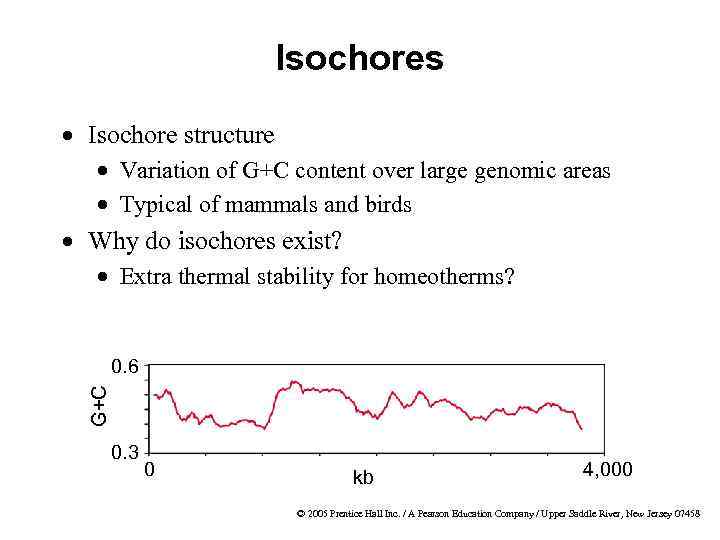 Isochores · Isochore structure · Variation of G+C content over large genomic areas · Typical of mammals and birds · Why do isochores exist? · Extra thermal stability for homeotherms? G+C 0. 6 0. 3 0 kb 4, 000 © 2005 Prentice Hall Inc. / A Pearson Education Company / Upper Saddle River, New Jersey 07458
Isochores · Isochore structure · Variation of G+C content over large genomic areas · Typical of mammals and birds · Why do isochores exist? · Extra thermal stability for homeotherms? G+C 0. 6 0. 3 0 kb 4, 000 © 2005 Prentice Hall Inc. / A Pearson Education Company / Upper Saddle River, New Jersey 07458
 Genome duplications · A driving force in evolution? · Large-scale duplication of genes · Differentiation of function in duplicated genes over time · Evidence · Polyploidy · More than two copies per chromosome · Endopolyploidy · Evidence of past genome duplications in diploid organisms © 2005 Prentice Hall Inc. / A Pearson Education Company / Upper Saddle River, New Jersey 07458
Genome duplications · A driving force in evolution? · Large-scale duplication of genes · Differentiation of function in duplicated genes over time · Evidence · Polyploidy · More than two copies per chromosome · Endopolyploidy · Evidence of past genome duplications in diploid organisms © 2005 Prentice Hall Inc. / A Pearson Education Company / Upper Saddle River, New Jersey 07458
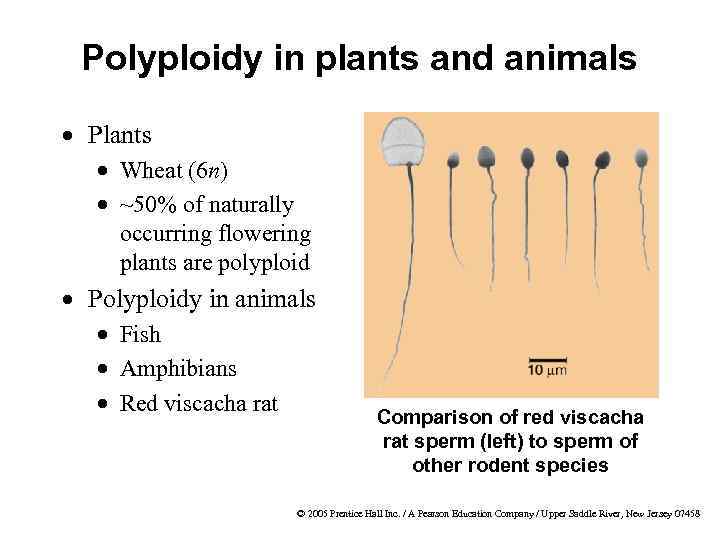 Polyploidy in plants and animals · Plants · Wheat (6 n) · ~50% of naturally occurring flowering plants are polyploid · Polyploidy in animals · Fish · Amphibians · Red viscacha rat 10 mm Comparison of red viscacha rat sperm (left) to sperm of other rodent species © 2005 Prentice Hall Inc. / A Pearson Education Company / Upper Saddle River, New Jersey 07458
Polyploidy in plants and animals · Plants · Wheat (6 n) · ~50% of naturally occurring flowering plants are polyploid · Polyploidy in animals · Fish · Amphibians · Red viscacha rat 10 mm Comparison of red viscacha rat sperm (left) to sperm of other rodent species © 2005 Prentice Hall Inc. / A Pearson Education Company / Upper Saddle River, New Jersey 07458
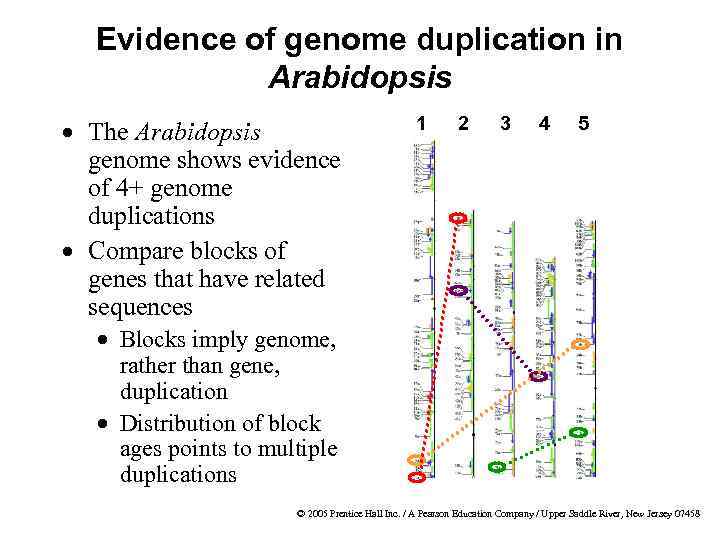 Evidence of genome duplication in Arabidopsis · The Arabidopsis genome shows evidence of 4+ genome duplications · Compare blocks of genes that have related sequences 1 2 3 4 5 · Blocks imply genome, rather than gene, duplication · Distribution of block ages points to multiple duplications © 2005 Prentice Hall Inc. / A Pearson Education Company / Upper Saddle River, New Jersey 07458
Evidence of genome duplication in Arabidopsis · The Arabidopsis genome shows evidence of 4+ genome duplications · Compare blocks of genes that have related sequences 1 2 3 4 5 · Blocks imply genome, rather than gene, duplication · Distribution of block ages points to multiple duplications © 2005 Prentice Hall Inc. / A Pearson Education Company / Upper Saddle River, New Jersey 07458
 Junk DNA? · Junk DNA · Sequences that serve no apparent evolutionary function · Is noncoding DNA junk? · Evidence against junk DNA · Statistical evidence of selection acting on noncoding regions · Biological functionality for noncoding regions AGACCAGGAACTTACAGCGACCTTGAACTGTTCCATTGCTCTTTTCCTGGGGCGG-GGGC |||||| |||||||||||||||| ||| AGACCAGGAACTCGTGGCGGCCGTGAACTGTTCCATTGCTCTTTTCCTGGGGCGGAGGG Comparison of human and mouse intergenic regions © 2005 Prentice Hall Inc. / A Pearson Education Company / Upper Saddle River, New Jersey 07458
Junk DNA? · Junk DNA · Sequences that serve no apparent evolutionary function · Is noncoding DNA junk? · Evidence against junk DNA · Statistical evidence of selection acting on noncoding regions · Biological functionality for noncoding regions AGACCAGGAACTTACAGCGACCTTGAACTGTTCCATTGCTCTTTTCCTGGGGCGG-GGGC |||||| |||||||||||||||| ||| AGACCAGGAACTCGTGGCGGCCGTGAACTGTTCCATTGCTCTTTTCCTGGGGCGGAGGG Comparison of human and mouse intergenic regions © 2005 Prentice Hall Inc. / A Pearson Education Company / Upper Saddle River, New Jersey 07458
 Variations in the amount of noncoding DNA · Prokaryotes · Bacteria: ~15% · Eukaryotes · · · Yeast (S. cerevisiae): 30% Malarial parasite (P. falciparum): 50% Flowering plant (A. thaliana): 70% Nematode worm (C. elegans): 70% Human (H. sapiens): 95% © 2005 Prentice Hall Inc. / A Pearson Education Company / Upper Saddle River, New Jersey 07458
Variations in the amount of noncoding DNA · Prokaryotes · Bacteria: ~15% · Eukaryotes · · · Yeast (S. cerevisiae): 30% Malarial parasite (P. falciparum): 50% Flowering plant (A. thaliana): 70% Nematode worm (C. elegans): 70% Human (H. sapiens): 95% © 2005 Prentice Hall Inc. / A Pearson Education Company / Upper Saddle River, New Jersey 07458
 Classification of noncoding DNA · Pseudogenes · Duplicated genes that have accumulated too many deleterious mutations to function · Repeats · Repeated units of DNA are 1– 200 bp long · Transposable elements · Pieces of DNA that have the ability to jump from place to place in the genome © 2005 Prentice Hall Inc. / A Pearson Education Company / Upper Saddle River, New Jersey 07458
Classification of noncoding DNA · Pseudogenes · Duplicated genes that have accumulated too many deleterious mutations to function · Repeats · Repeated units of DNA are 1– 200 bp long · Transposable elements · Pieces of DNA that have the ability to jump from place to place in the genome © 2005 Prentice Hall Inc. / A Pearson Education Company / Upper Saddle River, New Jersey 07458
 Pseudogenes · Pseudogenes can exist because a single functioning copy of a gene is sufficient · Processed pseudogenes lack a promoter and introns · Believed to derive from m. RNA copy · Reverse transcribed into c. DNA · Reintroduced into genome · Pseudogenes can be quite common · In C. elegans, there is one pseudogene for every eight genes © 2005 Prentice Hall Inc. / A Pearson Education Company / Upper Saddle River, New Jersey 07458
Pseudogenes · Pseudogenes can exist because a single functioning copy of a gene is sufficient · Processed pseudogenes lack a promoter and introns · Believed to derive from m. RNA copy · Reverse transcribed into c. DNA · Reintroduced into genome · Pseudogenes can be quite common · In C. elegans, there is one pseudogene for every eight genes © 2005 Prentice Hall Inc. / A Pearson Education Company / Upper Saddle River, New Jersey 07458
 Minisatellites · Tandem repeats 7 -100 bp long · Generally GC rich · Highly polymorphic · Minisatellites in coding regions · Example: apolipoprotein family · Association with specific lipoprotein depends on number of repeats in coding minisatellite · Minisatellites can also affect gene regulation · Found in wide range of eukaryotic organisms © 2005 Prentice Hall Inc. / A Pearson Education Company / Upper Saddle River, New Jersey 07458
Minisatellites · Tandem repeats 7 -100 bp long · Generally GC rich · Highly polymorphic · Minisatellites in coding regions · Example: apolipoprotein family · Association with specific lipoprotein depends on number of repeats in coding minisatellite · Minisatellites can also affect gene regulation · Found in wide range of eukaryotic organisms © 2005 Prentice Hall Inc. / A Pearson Education Company / Upper Saddle River, New Jersey 07458
 Instability in human minisatellites · Minisatellites in humans are hypermutable · Mutation rate > 0. 5% per sperm per allele · Not the case in mouse, rat, or pig genome · Insertion of human minisatellites into mouse genome does not increase germ line mutation rate · Differences between minisatellites in humans and other mammals · 90% of minisatellites are in highly recombinant regions · 66% of pig, 30% of rat, and 15% of mouse minisatellites are in similar regions © 2005 Prentice Hall Inc. / A Pearson Education Company / Upper Saddle River, New Jersey 07458
Instability in human minisatellites · Minisatellites in humans are hypermutable · Mutation rate > 0. 5% per sperm per allele · Not the case in mouse, rat, or pig genome · Insertion of human minisatellites into mouse genome does not increase germ line mutation rate · Differences between minisatellites in humans and other mammals · 90% of minisatellites are in highly recombinant regions · 66% of pig, 30% of rat, and 15% of mouse minisatellites are in similar regions © 2005 Prentice Hall Inc. / A Pearson Education Company / Upper Saddle River, New Jersey 07458
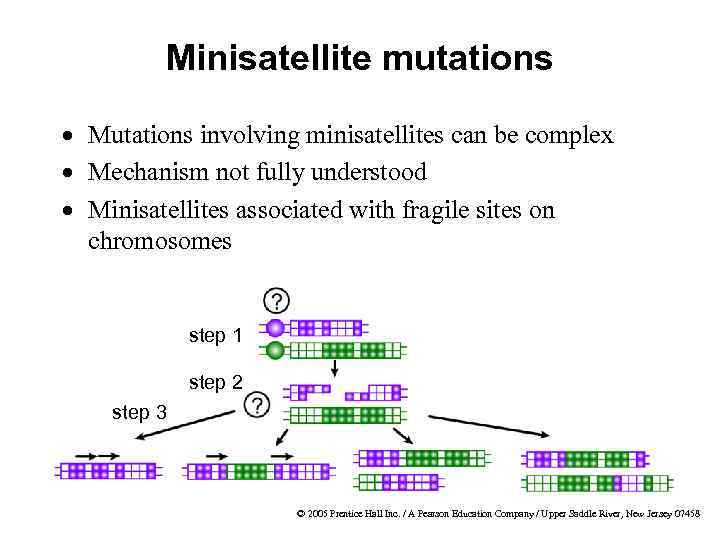 Minisatellite mutations · Mutations involving minisatellites can be complex · Mechanism not fully understood · Minisatellites associated with fragile sites on chromosomes step 1 step 2 step 3 © 2005 Prentice Hall Inc. / A Pearson Education Company / Upper Saddle River, New Jersey 07458
Minisatellite mutations · Mutations involving minisatellites can be complex · Mechanism not fully understood · Minisatellites associated with fragile sites on chromosomes step 1 step 2 step 3 © 2005 Prentice Hall Inc. / A Pearson Education Company / Upper Saddle River, New Jersey 07458
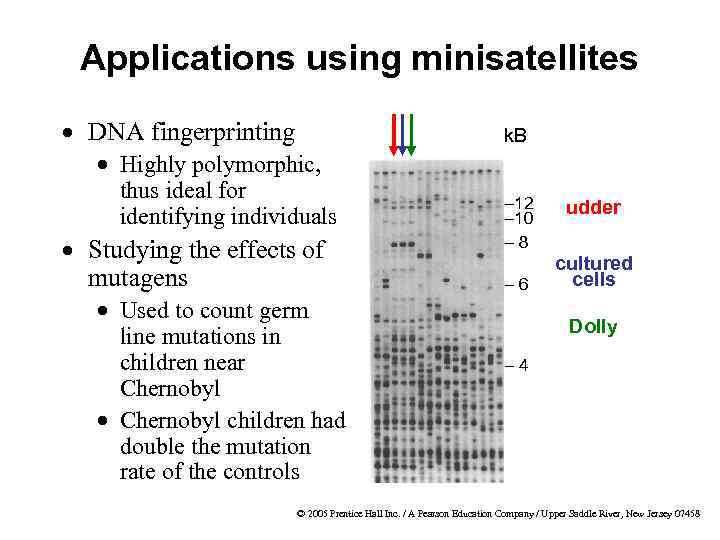 Applications using minisatellites · DNA fingerprinting k. B · Highly polymorphic, thus ideal for identifying individuals · Studying the effects of mutagens · Used to count germ line mutations in children near Chernobyl · Chernobyl children had double the mutation rate of the controls – 12 – 10 – 8 – 6 udder cultured cells Dolly – 4 © 2005 Prentice Hall Inc. / A Pearson Education Company / Upper Saddle River, New Jersey 07458
Applications using minisatellites · DNA fingerprinting k. B · Highly polymorphic, thus ideal for identifying individuals · Studying the effects of mutagens · Used to count germ line mutations in children near Chernobyl · Chernobyl children had double the mutation rate of the controls – 12 – 10 – 8 – 6 udder cultured cells Dolly – 4 © 2005 Prentice Hall Inc. / A Pearson Education Company / Upper Saddle River, New Jersey 07458
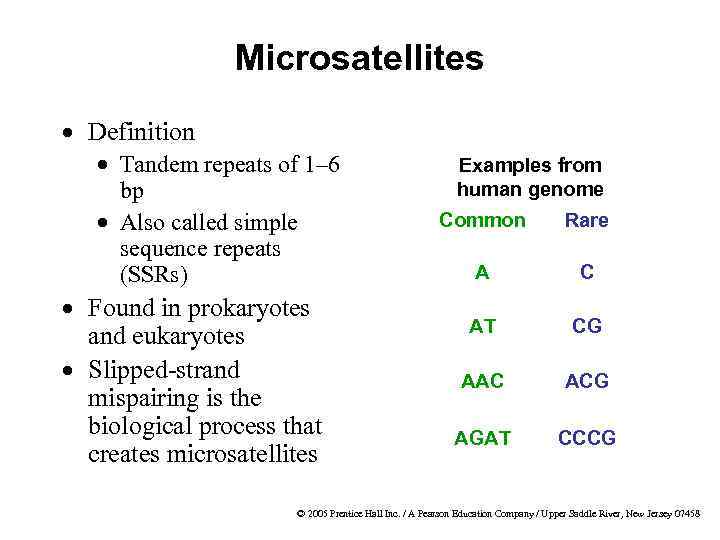 Microsatellites · Definition · Tandem repeats of 1– 6 bp · Also called simple sequence repeats (SSRs) · Found in prokaryotes and eukaryotes · Slipped-strand mispairing is the biological process that creates microsatellites Examples from human genome Common Rare A C AT CG AAC ACG AGAT CCCG © 2005 Prentice Hall Inc. / A Pearson Education Company / Upper Saddle River, New Jersey 07458
Microsatellites · Definition · Tandem repeats of 1– 6 bp · Also called simple sequence repeats (SSRs) · Found in prokaryotes and eukaryotes · Slipped-strand mispairing is the biological process that creates microsatellites Examples from human genome Common Rare A C AT CG AAC ACG AGAT CCCG © 2005 Prentice Hall Inc. / A Pearson Education Company / Upper Saddle River, New Jersey 07458
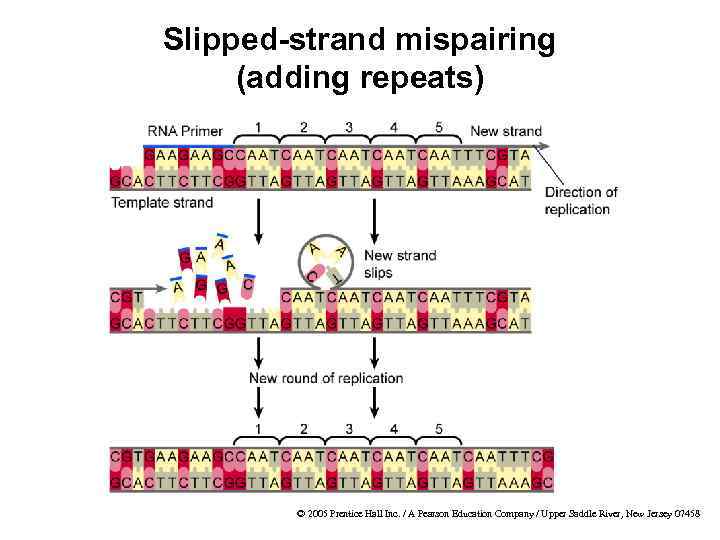 Slipped-strand mispairing (adding repeats) © 2005 Prentice Hall Inc. / A Pearson Education Company / Upper Saddle River, New Jersey 07458
Slipped-strand mispairing (adding repeats) © 2005 Prentice Hall Inc. / A Pearson Education Company / Upper Saddle River, New Jersey 07458
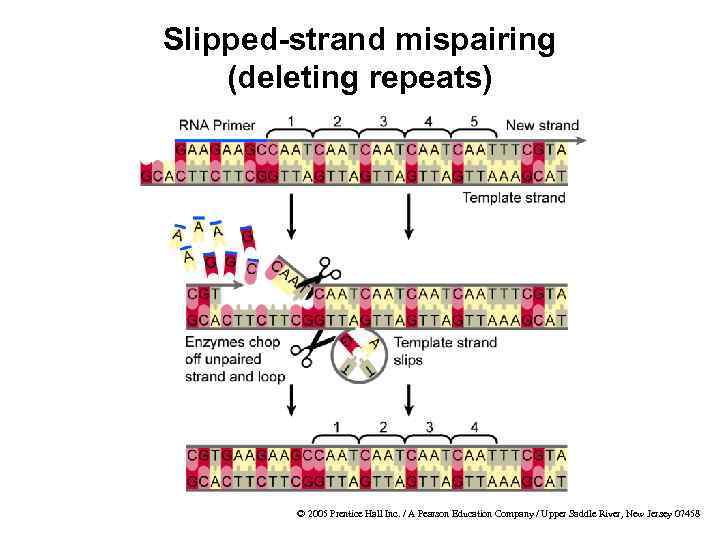 Slipped-strand mispairing (deleting repeats) © 2005 Prentice Hall Inc. / A Pearson Education Company / Upper Saddle River, New Jersey 07458
Slipped-strand mispairing (deleting repeats) © 2005 Prentice Hall Inc. / A Pearson Education Company / Upper Saddle River, New Jersey 07458
 Microsatellite function in bacteria · Microsatellites are unstable because of susceptibility to slipped-strand mispairing · Slipped-strand mispairing causes mutations that alter protein function · Virulent bacteria have microsatellites associated with multiple genes · By maintaining genetic diversity within a small population, bacteria can increase adaptability © 2005 Prentice Hall Inc. / A Pearson Education Company / Upper Saddle River, New Jersey 07458
Microsatellite function in bacteria · Microsatellites are unstable because of susceptibility to slipped-strand mispairing · Slipped-strand mispairing causes mutations that alter protein function · Virulent bacteria have microsatellites associated with multiple genes · By maintaining genetic diversity within a small population, bacteria can increase adaptability © 2005 Prentice Hall Inc. / A Pearson Education Company / Upper Saddle River, New Jersey 07458
 An example of contingency genes · Bacterium N. gonorrhoeae causes gonorrhea · Opa genes code for surface proteins · Allow infection by facilitating adhesion to epithelial cells · But also make bacteria susceptible to phagocytes · Contain microsatellites with CTCTT repeats · Repeat deletions cause frame-shift mutations · Surface proteins no longer “sticky” · N. gonorrhoeae can thereby avoid lethal phagocytes · 1 in 100– 1, 000 new cells will have CTCTT mutation · Therefore, there is always a population of evasive and infectious cells © 2005 Prentice Hall Inc. / A Pearson Education Company / Upper Saddle River, New Jersey 07458
An example of contingency genes · Bacterium N. gonorrhoeae causes gonorrhea · Opa genes code for surface proteins · Allow infection by facilitating adhesion to epithelial cells · But also make bacteria susceptible to phagocytes · Contain microsatellites with CTCTT repeats · Repeat deletions cause frame-shift mutations · Surface proteins no longer “sticky” · N. gonorrhoeae can thereby avoid lethal phagocytes · 1 in 100– 1, 000 new cells will have CTCTT mutation · Therefore, there is always a population of evasive and infectious cells © 2005 Prentice Hall Inc. / A Pearson Education Company / Upper Saddle River, New Jersey 07458
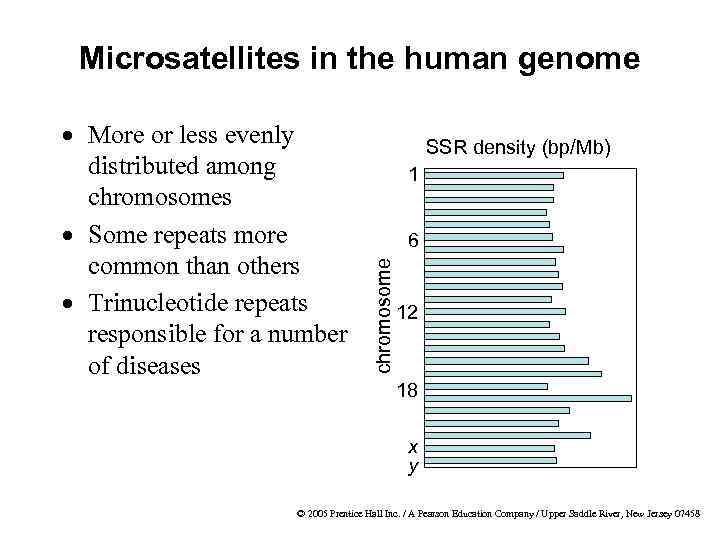 Microsatellites in the human genome SSR density (bp/Mb) 1 6 chromosome · More or less evenly distributed among chromosomes · Some repeats more common than others · Trinucleotide repeats responsible for a number of diseases 12 18 x y © 2005 Prentice Hall Inc. / A Pearson Education Company / Upper Saddle River, New Jersey 07458
Microsatellites in the human genome SSR density (bp/Mb) 1 6 chromosome · More or less evenly distributed among chromosomes · Some repeats more common than others · Trinucleotide repeats responsible for a number of diseases 12 18 x y © 2005 Prentice Hall Inc. / A Pearson Education Company / Upper Saddle River, New Jersey 07458
 Trinucleotide repeats and human disease · Expansion of trinucleotide repeats responsible for over a dozen neurological diseases · Examples: Huntington’s disease, fragile X syndrome · Repeat motifs · Mutations involving CGG, GCC, GAA, CTG, and CAG account for 14 diseases · Some located in noncoding areas · Others located in coding regions © 2005 Prentice Hall Inc. / A Pearson Education Company / Upper Saddle River, New Jersey 07458
Trinucleotide repeats and human disease · Expansion of trinucleotide repeats responsible for over a dozen neurological diseases · Examples: Huntington’s disease, fragile X syndrome · Repeat motifs · Mutations involving CGG, GCC, GAA, CTG, and CAG account for 14 diseases · Some located in noncoding areas · Others located in coding regions © 2005 Prentice Hall Inc. / A Pearson Education Company / Upper Saddle River, New Jersey 07458
 Trinucleotide expansion in Huntington’s disease · Hutington’s disease · Midlife onset of dementia, followed by death · Caused by expansion of CAG repeats in the coding region of Huntingtin · CAG repeats translated into polyglutamine tract · 6– 35 repeats: no disease · 36– 121 repeats: Huntington’s disease · > 70 repeats: Huntington’s disease with juvenile onset · Other diseases also caused by CAG expansion in coding region · Polyglutamine tract believed to cause neurotoxic aggregates © 2005 Prentice Hall Inc. / A Pearson Education Company / Upper Saddle River, New Jersey 07458
Trinucleotide expansion in Huntington’s disease · Hutington’s disease · Midlife onset of dementia, followed by death · Caused by expansion of CAG repeats in the coding region of Huntingtin · CAG repeats translated into polyglutamine tract · 6– 35 repeats: no disease · 36– 121 repeats: Huntington’s disease · > 70 repeats: Huntington’s disease with juvenile onset · Other diseases also caused by CAG expansion in coding region · Polyglutamine tract believed to cause neurotoxic aggregates © 2005 Prentice Hall Inc. / A Pearson Education Company / Upper Saddle River, New Jersey 07458
 Viral remnants in the genome · Human endogenous retroviruses (HERVs) constitute ~7% of the human genome · Retroviruses infect only a specific cell type and do not incorporate viral genetic material into the germ line · Endogenous retroviruses present challenges for xenotransplantation · Pig endogenous retroviruses (PERVs) can infect cultured human cells · PERVs can also infect mouse cells through transplants · So far, no evidence of infection in pig-to-human transplants © 2005 Prentice Hall Inc. / A Pearson Education Company / Upper Saddle River, New Jersey 07458
Viral remnants in the genome · Human endogenous retroviruses (HERVs) constitute ~7% of the human genome · Retroviruses infect only a specific cell type and do not incorporate viral genetic material into the germ line · Endogenous retroviruses present challenges for xenotransplantation · Pig endogenous retroviruses (PERVs) can infect cultured human cells · PERVs can also infect mouse cells through transplants · So far, no evidence of infection in pig-to-human transplants © 2005 Prentice Hall Inc. / A Pearson Education Company / Upper Saddle River, New Jersey 07458
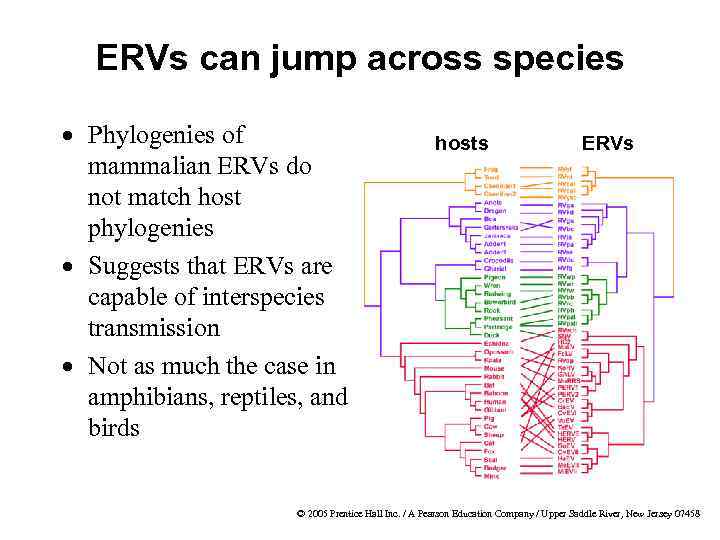 ERVs can jump across species · Phylogenies of mammalian ERVs do not match host phylogenies · Suggests that ERVs are capable of interspecies transmission · Not as much the case in amphibians, reptiles, and birds hosts ERVs © 2005 Prentice Hall Inc. / A Pearson Education Company / Upper Saddle River, New Jersey 07458
ERVs can jump across species · Phylogenies of mammalian ERVs do not match host phylogenies · Suggests that ERVs are capable of interspecies transmission · Not as much the case in amphibians, reptiles, and birds hosts ERVs © 2005 Prentice Hall Inc. / A Pearson Education Company / Upper Saddle River, New Jersey 07458
 The effect of ERVs on the genome · Insertion into a coding region can destroy gene function · Insertion of LTRs near preexisting genes can alter gene regulation · Gene duplication via reverse transcription of m. RNA · Processed pseudogene · Duplicated gene that evolves new function · An ERV sequence can evolve into a gene beneficial to the host · e. g. , HERV-derived gene in the human placenta © 2005 Prentice Hall Inc. / A Pearson Education Company / Upper Saddle River, New Jersey 07458
The effect of ERVs on the genome · Insertion into a coding region can destroy gene function · Insertion of LTRs near preexisting genes can alter gene regulation · Gene duplication via reverse transcription of m. RNA · Processed pseudogene · Duplicated gene that evolves new function · An ERV sequence can evolve into a gene beneficial to the host · e. g. , HERV-derived gene in the human placenta © 2005 Prentice Hall Inc. / A Pearson Education Company / Upper Saddle River, New Jersey 07458
 Transposons and ERVs · Some ERV sequences have genes capable of producing infectious viruses · Others are degraded to some extent · Transposons are sequences that can copy and reinsert themselves into the genome · LTR retrotransposons are related to ERVs · Both have long terminal repeats (LTRs) · LTR retrotransposons no longer capable of creating full-fledged virus © 2005 Prentice Hall Inc. / A Pearson Education Company / Upper Saddle River, New Jersey 07458
Transposons and ERVs · Some ERV sequences have genes capable of producing infectious viruses · Others are degraded to some extent · Transposons are sequences that can copy and reinsert themselves into the genome · LTR retrotransposons are related to ERVs · Both have long terminal repeats (LTRs) · LTR retrotransposons no longer capable of creating full-fledged virus © 2005 Prentice Hall Inc. / A Pearson Education Company / Upper Saddle River, New Jersey 07458
 Transposable elements · DNA regions flanked by repeats that can jump around the genome · Transposable-element (TE) insertion can be faster than chromosome replication · Allows TEs to rapidly accumulate in genome · 44% of human genome sequence is TEs · TEs can have different effects on the host · · Insertion into gene can destroy function TE can evolve into beneficial gene TE can affect gene expression But most TEs will have a neutral effect © 2005 Prentice Hall Inc. / A Pearson Education Company / Upper Saddle River, New Jersey 07458
Transposable elements · DNA regions flanked by repeats that can jump around the genome · Transposable-element (TE) insertion can be faster than chromosome replication · Allows TEs to rapidly accumulate in genome · 44% of human genome sequence is TEs · TEs can have different effects on the host · · Insertion into gene can destroy function TE can evolve into beneficial gene TE can affect gene expression But most TEs will have a neutral effect © 2005 Prentice Hall Inc. / A Pearson Education Company / Upper Saddle River, New Jersey 07458
 TE content among species · TE content varies greatly among species · · Humans: 44% Maize: > 50% Drosophila: 15% Arabidopsis, C. elegans, yeast: < 5% · TEs tend to be concentrated in particular genomic regions · In maize, TEs have doubled the size of the genome over the last few million years © 2005 Prentice Hall Inc. / A Pearson Education Company / Upper Saddle River, New Jersey 07458
TE content among species · TE content varies greatly among species · · Humans: 44% Maize: > 50% Drosophila: 15% Arabidopsis, C. elegans, yeast: < 5% · TEs tend to be concentrated in particular genomic regions · In maize, TEs have doubled the size of the genome over the last few million years © 2005 Prentice Hall Inc. / A Pearson Education Company / Upper Saddle River, New Jersey 07458
 Intraspecies variations in TE content · Wild barley plants examined in Israeli canyon containing distinct microclimates · Copies of BARE-1 transposon vary in individual wild barley plants · 8, 300– 22, 000 copies per plant · BARE-1 copy number (and genome size) found to positively correlate with dryness and altitude · Biological significance · Large genome correlates with increased cell volume · Large cell volumes make growth more efficient in cooler weather of higher altitudes © 2005 Prentice Hall Inc. / A Pearson Education Company / Upper Saddle River, New Jersey 07458
Intraspecies variations in TE content · Wild barley plants examined in Israeli canyon containing distinct microclimates · Copies of BARE-1 transposon vary in individual wild barley plants · 8, 300– 22, 000 copies per plant · BARE-1 copy number (and genome size) found to positively correlate with dryness and altitude · Biological significance · Large genome correlates with increased cell volume · Large cell volumes make growth more efficient in cooler weather of higher altitudes © 2005 Prentice Hall Inc. / A Pearson Education Company / Upper Saddle River, New Jersey 07458
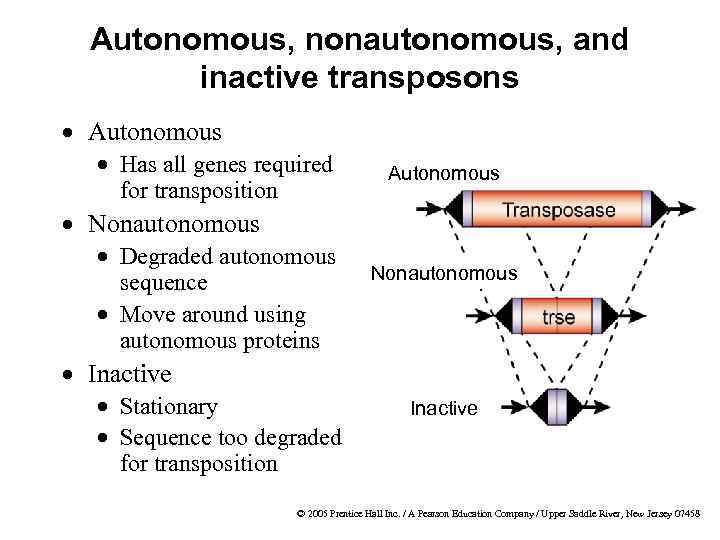 Autonomous, nonautonomous, and inactive transposons · Autonomous · Has all genes required for transposition Autonomous · Nonautonomous · Degraded autonomous sequence · Move around using autonomous proteins Nonautonomous · Inactive · Stationary · Sequence too degraded for transposition Inactive © 2005 Prentice Hall Inc. / A Pearson Education Company / Upper Saddle River, New Jersey 07458
Autonomous, nonautonomous, and inactive transposons · Autonomous · Has all genes required for transposition Autonomous · Nonautonomous · Degraded autonomous sequence · Move around using autonomous proteins Nonautonomous · Inactive · Stationary · Sequence too degraded for transposition Inactive © 2005 Prentice Hall Inc. / A Pearson Education Company / Upper Saddle River, New Jersey 07458
 Class I transposable elements · Use RNA to transcribe themselves · Types · Long terminal repeat (LTR) retrotransposons · Non-LTR retrotransposons · Class I transposable elements in the human genome account for ~42% of the total genome sequence © 2005 Prentice Hall Inc. / A Pearson Education Company / Upper Saddle River, New Jersey 07458
Class I transposable elements · Use RNA to transcribe themselves · Types · Long terminal repeat (LTR) retrotransposons · Non-LTR retrotransposons · Class I transposable elements in the human genome account for ~42% of the total genome sequence © 2005 Prentice Hall Inc. / A Pearson Education Company / Upper Saddle River, New Jersey 07458
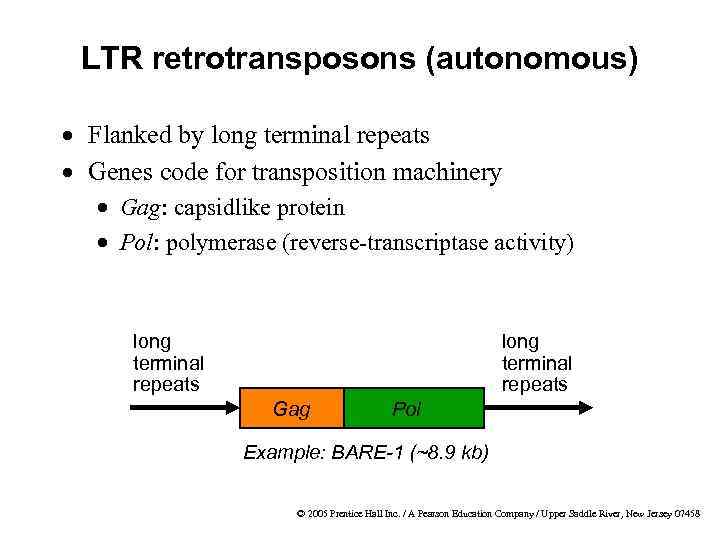 LTR retrotransposons (autonomous) · Flanked by long terminal repeats · Genes code for transposition machinery · Gag: capsidlike protein · Pol: polymerase (reverse-transcriptase activity) long terminal repeats Gag Pol Example: BARE-1 (~8. 9 kb) © 2005 Prentice Hall Inc. / A Pearson Education Company / Upper Saddle River, New Jersey 07458
LTR retrotransposons (autonomous) · Flanked by long terminal repeats · Genes code for transposition machinery · Gag: capsidlike protein · Pol: polymerase (reverse-transcriptase activity) long terminal repeats Gag Pol Example: BARE-1 (~8. 9 kb) © 2005 Prentice Hall Inc. / A Pearson Education Company / Upper Saddle River, New Jersey 07458
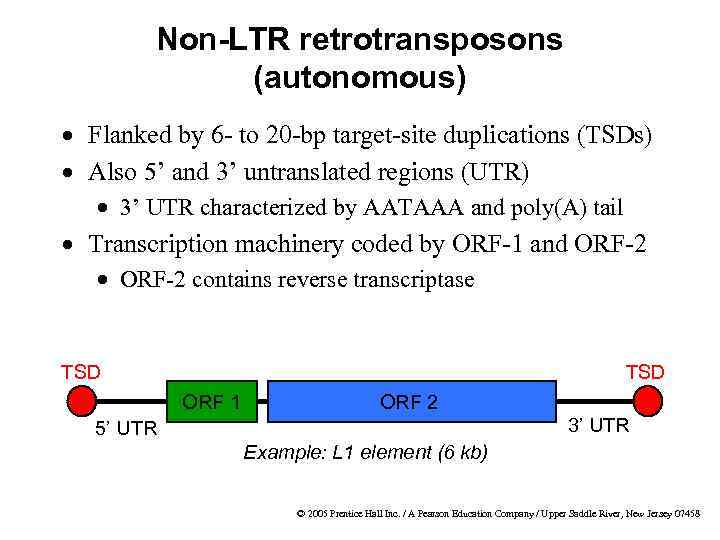 Non-LTR retrotransposons (autonomous) · Flanked by 6 - to 20 -bp target-site duplications (TSDs) · Also 5’ and 3’ untranslated regions (UTR) · 3’ UTR characterized by AATAAA and poly(A) tail · Transcription machinery coded by ORF-1 and ORF-2 · ORF-2 contains reverse transcriptase TSD ORF 1 ORF 2 3’ UTR 5’ UTR Example: L 1 element (6 kb) © 2005 Prentice Hall Inc. / A Pearson Education Company / Upper Saddle River, New Jersey 07458
Non-LTR retrotransposons (autonomous) · Flanked by 6 - to 20 -bp target-site duplications (TSDs) · Also 5’ and 3’ untranslated regions (UTR) · 3’ UTR characterized by AATAAA and poly(A) tail · Transcription machinery coded by ORF-1 and ORF-2 · ORF-2 contains reverse transcriptase TSD ORF 1 ORF 2 3’ UTR 5’ UTR Example: L 1 element (6 kb) © 2005 Prentice Hall Inc. / A Pearson Education Company / Upper Saddle River, New Jersey 07458
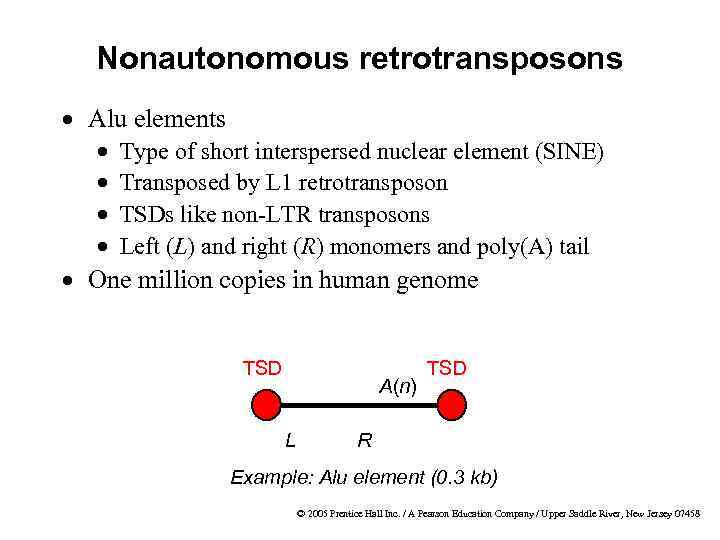 Nonautonomous retrotransposons · Alu elements · · Type of short interspersed nuclear element (SINE) Transposed by L 1 retrotransposon TSDs like non-LTR transposons Left (L) and right (R) monomers and poly(A) tail · One million copies in human genome TSD A(n) L TSD R Example: Alu element (0. 3 kb) © 2005 Prentice Hall Inc. / A Pearson Education Company / Upper Saddle River, New Jersey 07458
Nonautonomous retrotransposons · Alu elements · · Type of short interspersed nuclear element (SINE) Transposed by L 1 retrotransposon TSDs like non-LTR transposons Left (L) and right (R) monomers and poly(A) tail · One million copies in human genome TSD A(n) L TSD R Example: Alu element (0. 3 kb) © 2005 Prentice Hall Inc. / A Pearson Education Company / Upper Saddle River, New Jersey 07458
 Effects of active retrotransposons · Cis retrotransposition · Trans retrotransposition · L 1 retrotransposons replicate Alu elements · Insertional mutagenesis · Retrotransposon inserts into coding region of gene · Unequal homologous recombination · Duplications and deletions · Effects on gene expression · Some L 1 sequences have acquired enhancer function © 2005 Prentice Hall Inc. / A Pearson Education Company / Upper Saddle River, New Jersey 07458
Effects of active retrotransposons · Cis retrotransposition · Trans retrotransposition · L 1 retrotransposons replicate Alu elements · Insertional mutagenesis · Retrotransposon inserts into coding region of gene · Unequal homologous recombination · Duplications and deletions · Effects on gene expression · Some L 1 sequences have acquired enhancer function © 2005 Prentice Hall Inc. / A Pearson Education Company / Upper Saddle River, New Jersey 07458
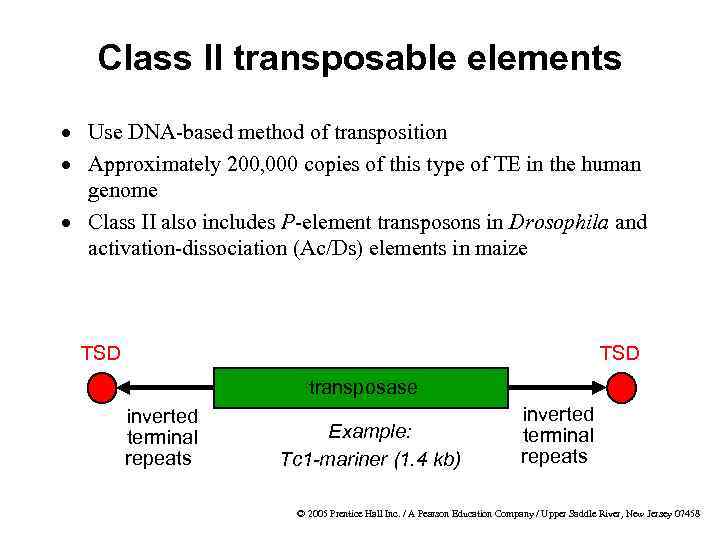 Class II transposable elements · Use DNA-based method of transposition · Approximately 200, 000 copies of this type of TE in the human genome · Class II also includes P-element transposons in Drosophila and activation-dissociation (Ac/Ds) elements in maize TSD transposase inverted terminal repeats Example: Tc 1 -mariner (1. 4 kb) inverted terminal repeats © 2005 Prentice Hall Inc. / A Pearson Education Company / Upper Saddle River, New Jersey 07458
Class II transposable elements · Use DNA-based method of transposition · Approximately 200, 000 copies of this type of TE in the human genome · Class II also includes P-element transposons in Drosophila and activation-dissociation (Ac/Ds) elements in maize TSD transposase inverted terminal repeats Example: Tc 1 -mariner (1. 4 kb) inverted terminal repeats © 2005 Prentice Hall Inc. / A Pearson Education Company / Upper Saddle River, New Jersey 07458
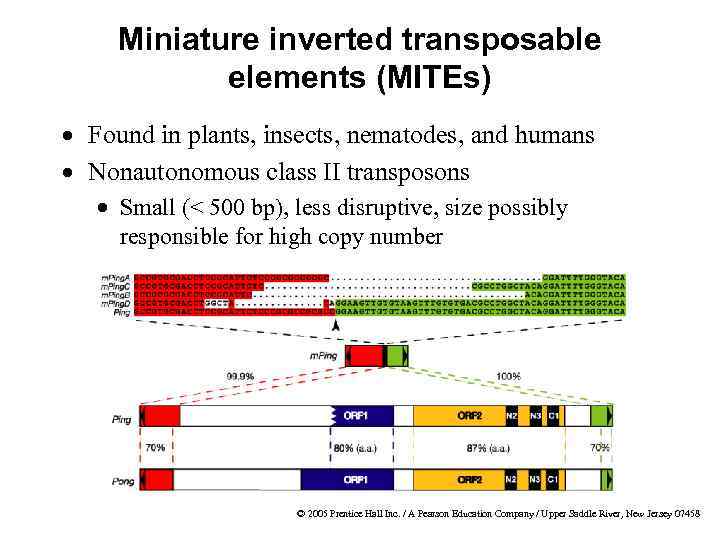 Miniature inverted transposable elements (MITEs) · Found in plants, insects, nematodes, and humans · Nonautonomous class II transposons · Small (< 500 bp), less disruptive, size possibly responsible for high copy number © 2005 Prentice Hall Inc. / A Pearson Education Company / Upper Saddle River, New Jersey 07458
Miniature inverted transposable elements (MITEs) · Found in plants, insects, nematodes, and humans · Nonautonomous class II transposons · Small (< 500 bp), less disruptive, size possibly responsible for high copy number © 2005 Prentice Hall Inc. / A Pearson Education Company / Upper Saddle River, New Jersey 07458
 TEs and X-chromosome inactivation · One X chromosome in each somatic cell inactivated · Occurs during embryonic development in female mammals · Regions of X chromosome that are inactivated are rich in L 1 elements · 10% of X chromosome that escapes inactivation is in region with low L 1 density · L 1 elements on X chromosome dated to 100 million years ago (emergence of mammals) · Correlations suggest L 1 elements might be involved in X-chromosome inactivation © 2005 Prentice Hall Inc. / A Pearson Education Company / Upper Saddle River, New Jersey 07458
TEs and X-chromosome inactivation · One X chromosome in each somatic cell inactivated · Occurs during embryonic development in female mammals · Regions of X chromosome that are inactivated are rich in L 1 elements · 10% of X chromosome that escapes inactivation is in region with low L 1 density · L 1 elements on X chromosome dated to 100 million years ago (emergence of mammals) · Correlations suggest L 1 elements might be involved in X-chromosome inactivation © 2005 Prentice Hall Inc. / A Pearson Education Company / Upper Saddle River, New Jersey 07458
 Summary I · Genome structure varies tremendously · Genome size and complexity · Linear relationship in prokaryotes · No clear relationship in eukaryotes · Genome features · Statistical features · Isochores: regions of elevated G+C content © 2005 Prentice Hall Inc. / A Pearson Education Company / Upper Saddle River, New Jersey 07458
Summary I · Genome structure varies tremendously · Genome size and complexity · Linear relationship in prokaryotes · No clear relationship in eukaryotes · Genome features · Statistical features · Isochores: regions of elevated G+C content © 2005 Prentice Hall Inc. / A Pearson Education Company / Upper Saddle River, New Jersey 07458
 Summary II · Coding regions · Bacteria: no introns · Archaea: some introns, TATA boxes · Eukarya: many introns and exons, TATA boxes · Noncoding regions · Pseudogenes · Repetitive sequences · Minisatellites · Microsatellites · Endogenous viral sequences © 2005 Prentice Hall Inc. / A Pearson Education Company / Upper Saddle River, New Jersey 07458
Summary II · Coding regions · Bacteria: no introns · Archaea: some introns, TATA boxes · Eukarya: many introns and exons, TATA boxes · Noncoding regions · Pseudogenes · Repetitive sequences · Minisatellites · Microsatellites · Endogenous viral sequences © 2005 Prentice Hall Inc. / A Pearson Education Company / Upper Saddle River, New Jersey 07458
 Summary III · Transposons · Class I · RNA intermediate · Frequent replication · Class II · DNA intermediate · Rare replication · MITEs · DNA intermediate · High copy number in plants © 2005 Prentice Hall Inc. / A Pearson Education Company / Upper Saddle River, New Jersey 07458
Summary III · Transposons · Class I · RNA intermediate · Frequent replication · Class II · DNA intermediate · Rare replication · MITEs · DNA intermediate · High copy number in plants © 2005 Prentice Hall Inc. / A Pearson Education Company / Upper Saddle River, New Jersey 07458
 Summary IV · Forces affecting genome structure · · · Point mutations Gene and genome duplications Viral infection Transposable elements Tandem repeat–associated mutations · Functions of noncoding DNA · Gene regulation · Adaptation to environmental conditions · Co-opting for new gene function © 2005 Prentice Hall Inc. / A Pearson Education Company / Upper Saddle River, New Jersey 07458
Summary IV · Forces affecting genome structure · · · Point mutations Gene and genome duplications Viral infection Transposable elements Tandem repeat–associated mutations · Functions of noncoding DNA · Gene regulation · Adaptation to environmental conditions · Co-opting for new gene function © 2005 Prentice Hall Inc. / A Pearson Education Company / Upper Saddle River, New Jersey 07458


