6756a6edac26e63fd00d1f822dc94841.ppt
- Количество слайдов: 28
 Chapter 11 Molecular Mechanisms of Gene regulation Jones and Bartlett Publishers © 2005
Chapter 11 Molecular Mechanisms of Gene regulation Jones and Bartlett Publishers © 2005
 Outline of topics • Transcriptional regulation in eukaryotes • Epigenetics • Regulation through RNA processing and decay • Translational control • Programmed DNA rearrangements
Outline of topics • Transcriptional regulation in eukaryotes • Epigenetics • Regulation through RNA processing and decay • Translational control • Programmed DNA rearrangements
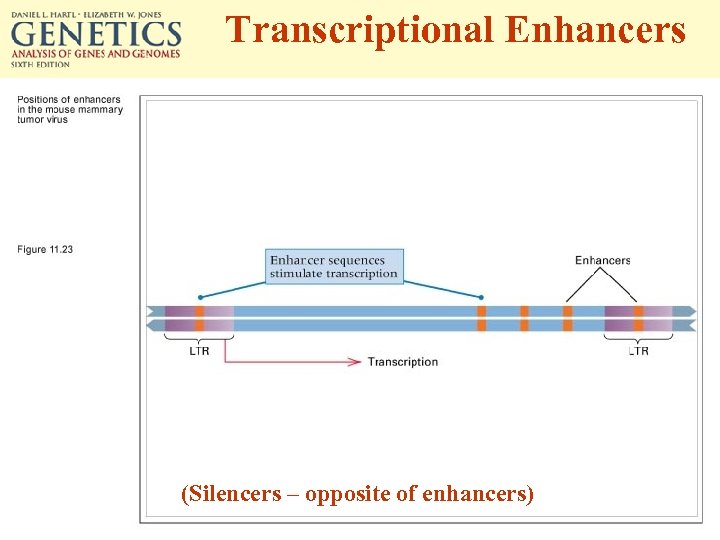 Transcriptional Enhancers (Silencers – opposite of enhancers)
Transcriptional Enhancers (Silencers – opposite of enhancers)
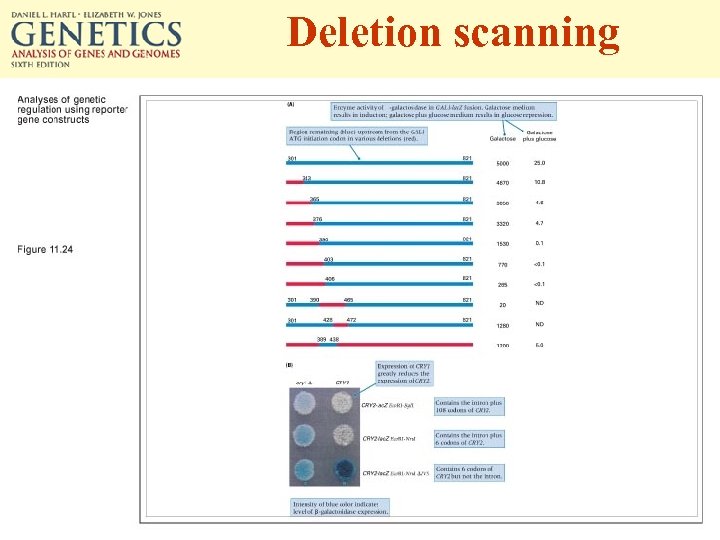 Deletion scanning
Deletion scanning
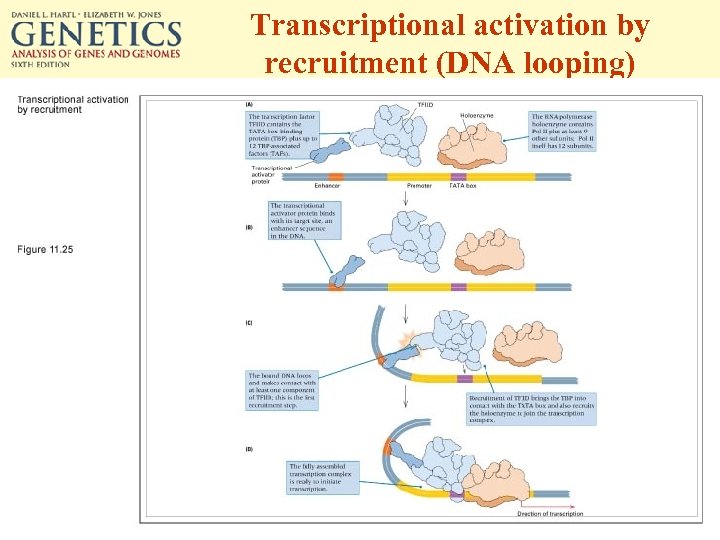 Transcriptional activation by recruitment (DNA looping)
Transcriptional activation by recruitment (DNA looping)
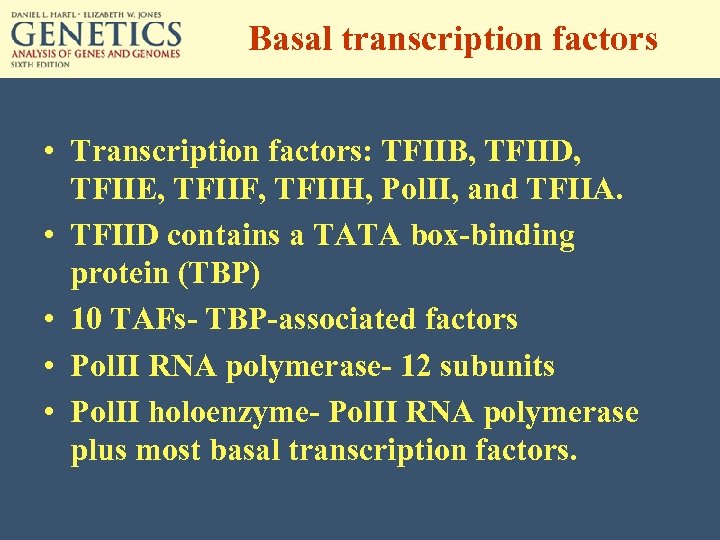 Basal transcription factors • Transcription factors: TFIIB, TFIID, TFIIE, TFIIF, TFIIH, Pol. II, and TFIIA. • TFIID contains a TATA box-binding protein (TBP) • 10 TAFs- TBP-associated factors • Pol. II RNA polymerase- 12 subunits • Pol. II holoenzyme- Pol. II RNA polymerase plus most basal transcription factors.
Basal transcription factors • Transcription factors: TFIIB, TFIID, TFIIE, TFIIF, TFIIH, Pol. II, and TFIIA. • TFIID contains a TATA box-binding protein (TBP) • 10 TAFs- TBP-associated factors • Pol. II RNA polymerase- 12 subunits • Pol. II holoenzyme- Pol. II RNA polymerase plus most basal transcription factors.
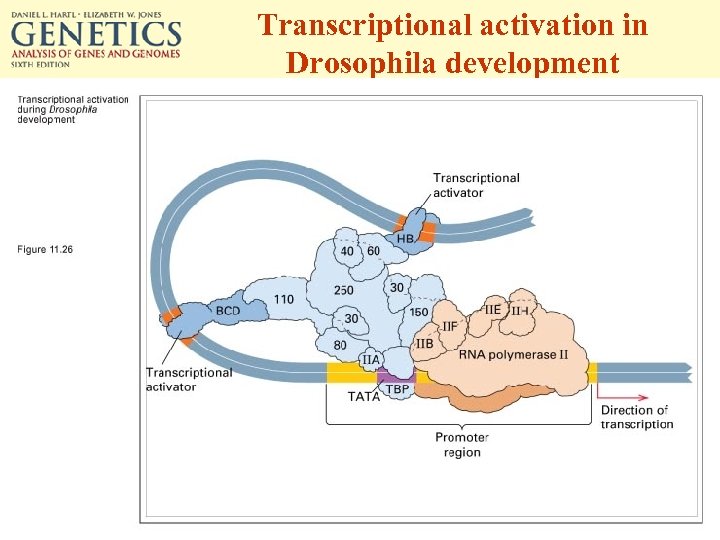 Transcriptional activation in Drosophila development
Transcriptional activation in Drosophila development
 Chromatin remodeling complexes • Purified chromatin + components of transcription don’t result in transcription. • CRCs: SWI-SNF, RSC (remodels the structure of chromatin), NURF (nucleosome remodeling factor), CHRAC (chromatin accessibility complex, and ACF (ATP-dependent chromatin assembly and remodeling factor). • CRCs are part of Pol. II holoenzyme.
Chromatin remodeling complexes • Purified chromatin + components of transcription don’t result in transcription. • CRCs: SWI-SNF, RSC (remodels the structure of chromatin), NURF (nucleosome remodeling factor), CHRAC (chromatin accessibility complex, and ACF (ATP-dependent chromatin assembly and remodeling factor). • CRCs are part of Pol. II holoenzyme.
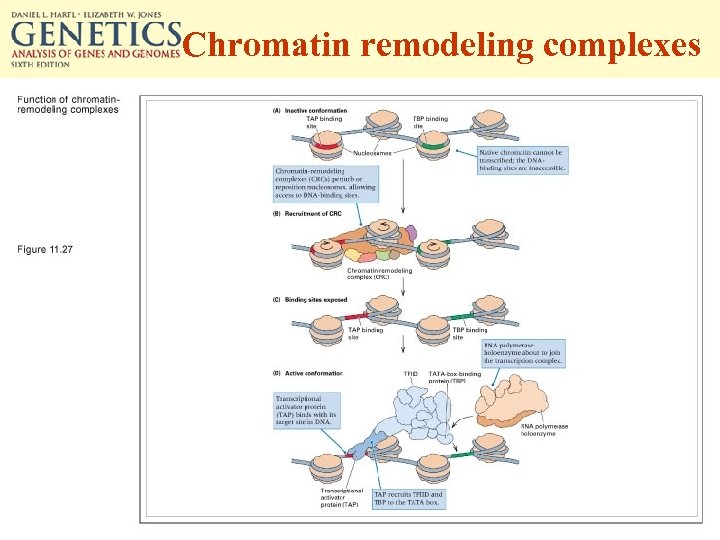 Chromatin remodeling complexes
Chromatin remodeling complexes
 • How are genes regulated during different stages of development in an organism? – Use of alternative promoter
• How are genes regulated during different stages of development in an organism? – Use of alternative promoter
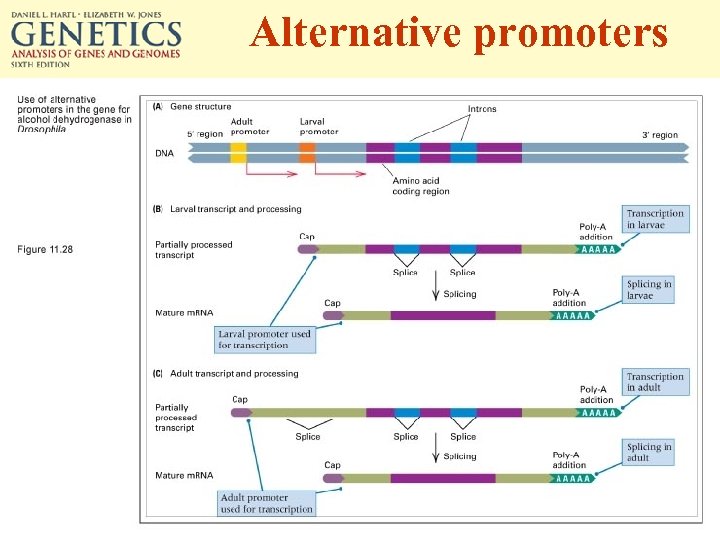 Alternative promoters
Alternative promoters
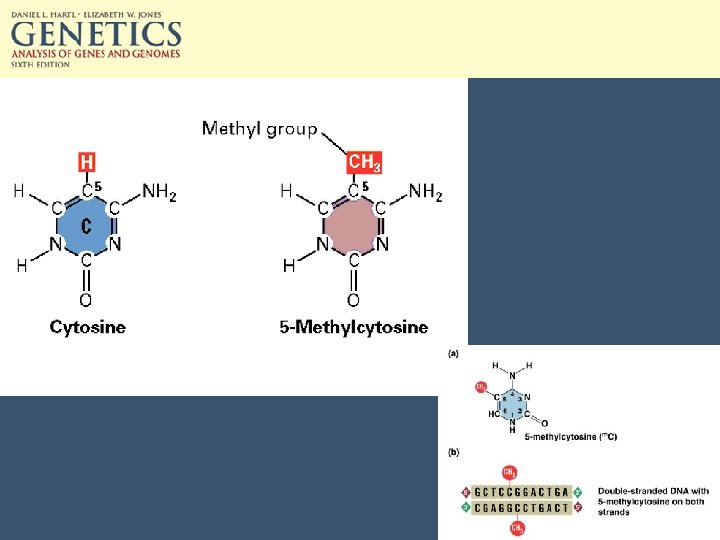
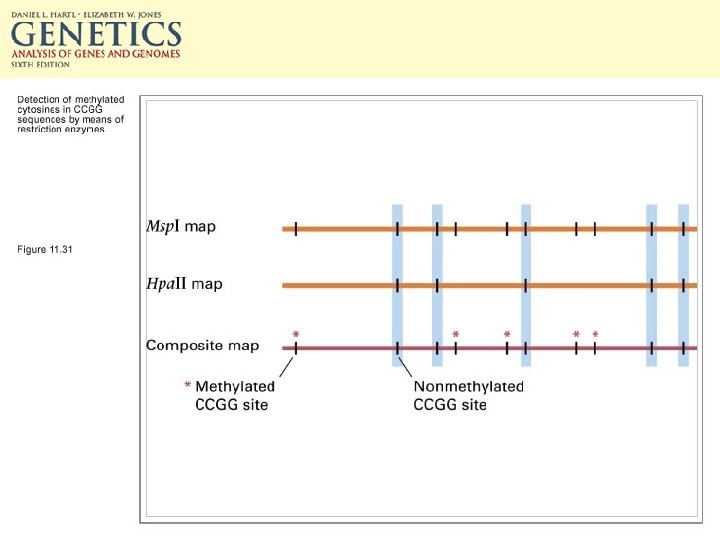
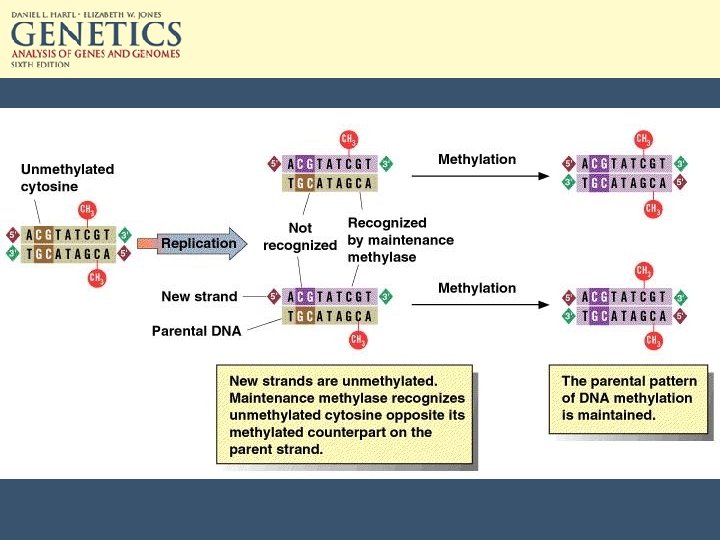
 Epigenetic methods of transcriptional regulation • CG doublets can be methylated. • 5 -methylcytosine can make up 2 -7% of C in vertebrate DNA and up to 30% of plant DNA. • Both Cs in the CG doublet are methylated. • Maintenance methylase preserves this.
Epigenetic methods of transcriptional regulation • CG doublets can be methylated. • 5 -methylcytosine can make up 2 -7% of C in vertebrate DNA and up to 30% of plant DNA. • Both Cs in the CG doublet are methylated. • Maintenance methylase preserves this.
 Transcriptional cosuppression • Inserted gene copies can result in transcriptional silencing. • Chalcone synthase gene in petunia. • Methylation is involved here. • Drosophila have no methylation (no DNA methylase), but still have transcriptional silencing.
Transcriptional cosuppression • Inserted gene copies can result in transcriptional silencing. • Chalcone synthase gene in petunia. • Methylation is involved here. • Drosophila have no methylation (no DNA methylase), but still have transcriptional silencing.
 Genomic Imprinting • • Imprinting occurs in the germ line. Affects a few hundred genes, in clusters. Heavy methylation is involved. Imprinted genes are methylated differently in female and male germ lines. • Once imprinted and methylated, a silenced gene remains transcriptionally inactive during embryogenesis. • Imprints are erased early in germ-line development, then re-established according to sexspecific pattern.
Genomic Imprinting • • Imprinting occurs in the germ line. Affects a few hundred genes, in clusters. Heavy methylation is involved. Imprinted genes are methylated differently in female and male germ lines. • Once imprinted and methylated, a silenced gene remains transcriptionally inactive during embryogenesis. • Imprints are erased early in germ-line development, then re-established according to sexspecific pattern.
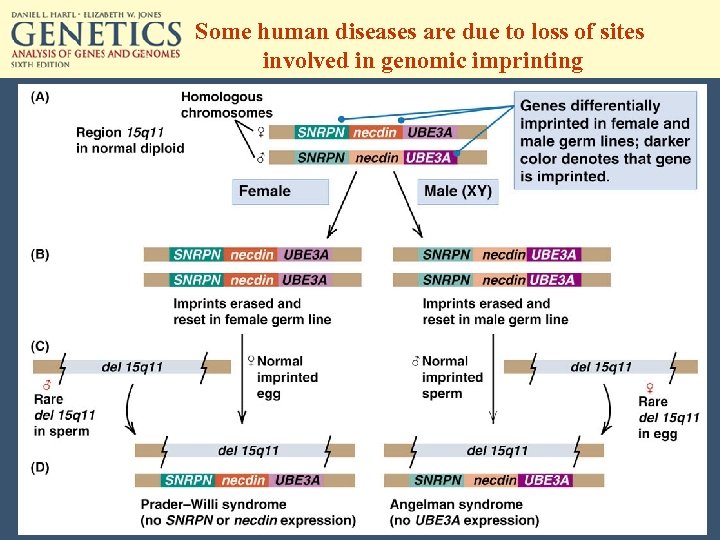 Some human diseases are due to loss of sites involved in genomic imprinting
Some human diseases are due to loss of sites involved in genomic imprinting
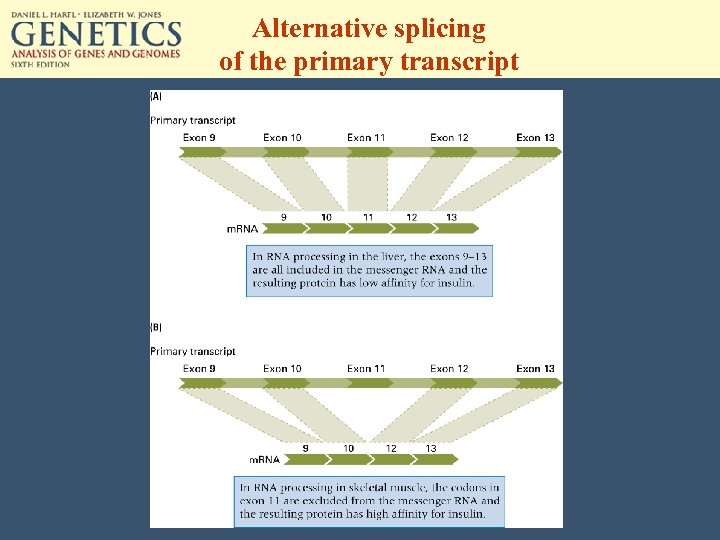 Alternative splicing of the primary transcript
Alternative splicing of the primary transcript
 Regulation through RNA processing and decay • • Alternative splicing m. RNA stability (poly-A tail is trimmed) Posttranscriptional cosuppression RNA interference: a few hundred pairs of double stranded RNA triggers RNA transcript degradation. – RISC (RNA-induced silencing complex) – Plus 21 - or 22 -mer oligonucleotides (chopped ds. RNA) as templates to recognize complementary sequences.
Regulation through RNA processing and decay • • Alternative splicing m. RNA stability (poly-A tail is trimmed) Posttranscriptional cosuppression RNA interference: a few hundred pairs of double stranded RNA triggers RNA transcript degradation. – RISC (RNA-induced silencing complex) – Plus 21 - or 22 -mer oligonucleotides (chopped ds. RNA) as templates to recognize complementary sequences.
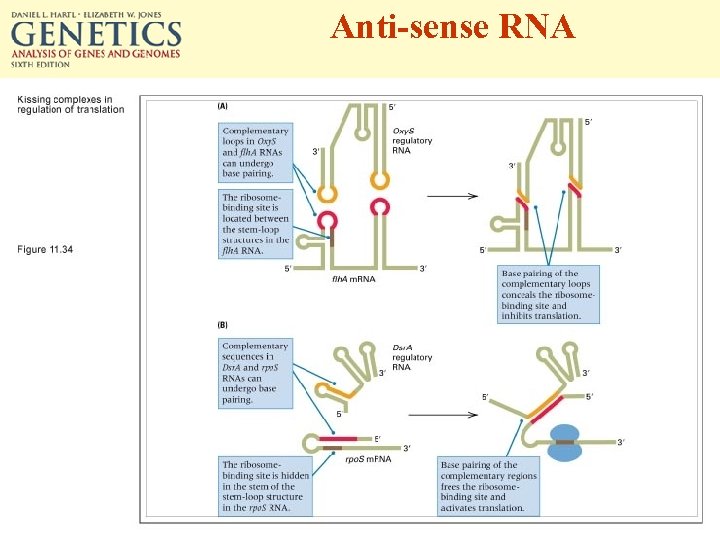 Anti-sense RNA
Anti-sense RNA
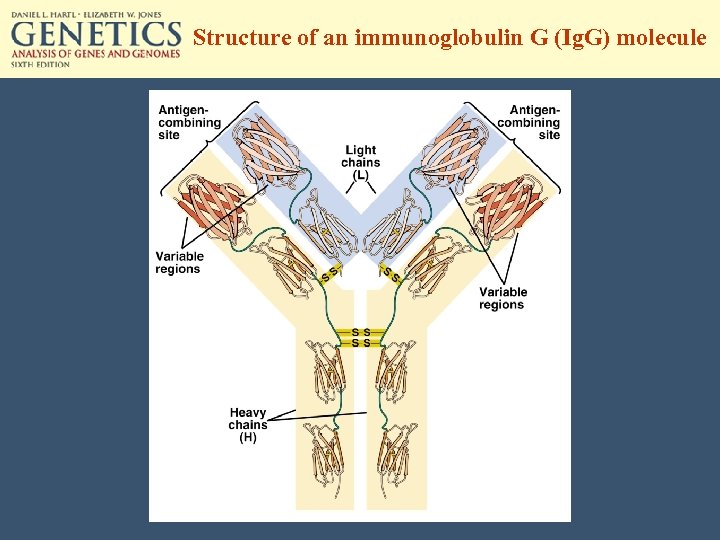 Structure of an immunoglobulin G (Ig. G) molecule
Structure of an immunoglobulin G (Ig. G) molecule
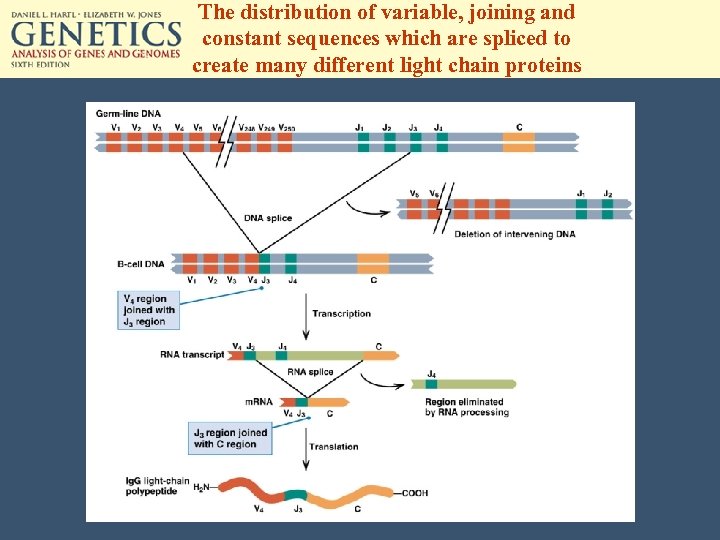 The distribution of variable, joining and constant sequences which are spliced to create many different light chain proteins
The distribution of variable, joining and constant sequences which are spliced to create many different light chain proteins
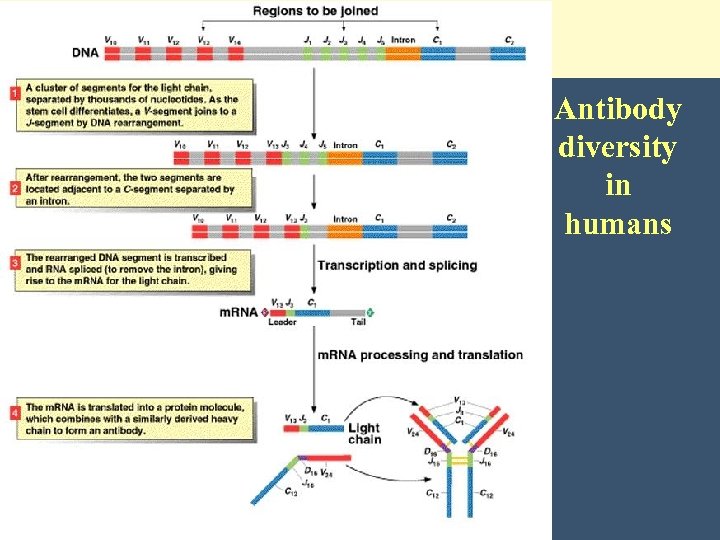 Antibody diversity in humans
Antibody diversity in humans
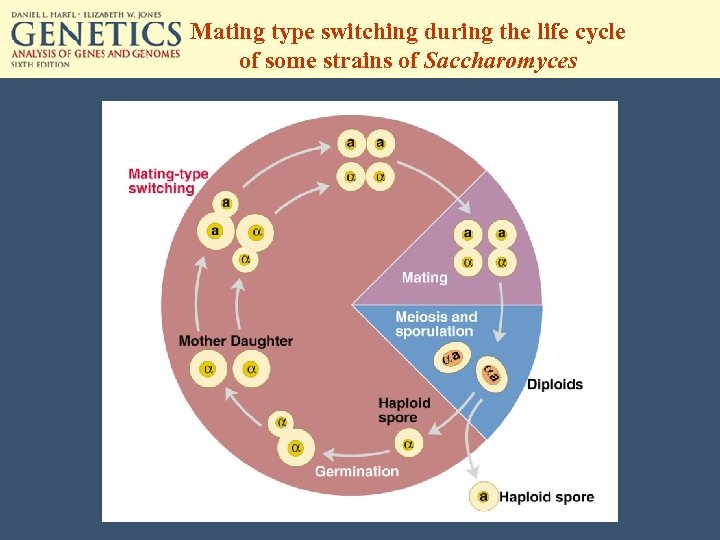 Mating type switching during the life cycle of some strains of Saccharomyces
Mating type switching during the life cycle of some strains of Saccharomyces
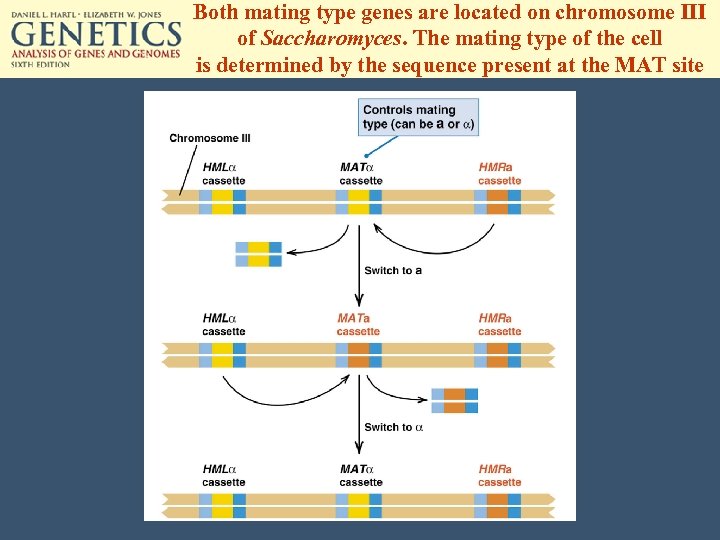 Both mating type genes are located on chromosome III of Saccharomyces. The mating type of the cell is determined by the sequence present at the MAT site
Both mating type genes are located on chromosome III of Saccharomyces. The mating type of the cell is determined by the sequence present at the MAT site
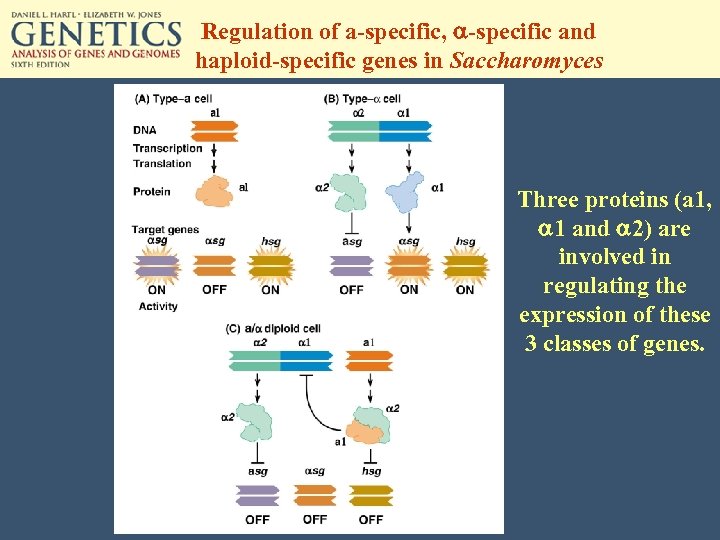 Regulation of a-specific, a-specific and haploid-specific genes in Saccharomyces Three proteins (a 1, a 1 and a 2) are involved in regulating the expression of these 3 classes of genes.
Regulation of a-specific, a-specific and haploid-specific genes in Saccharomyces Three proteins (a 1, a 1 and a 2) are involved in regulating the expression of these 3 classes of genes.


