ad954603295a435e47701326c8c912d1.ppt
- Количество слайдов: 44
 Chapter 11 DNA Polymorphisms and Human Identification
Chapter 11 DNA Polymorphisms and Human Identification
 Objectives § Compare and contrast different types of polymorphisms. § Define restriction fragment length polymorphisms. § Describe short tandem repeat structure and nomenclature. § Describe gender identification using the amelogenin locus. § Illustrate the use of STR for bone marrow engraftment monitoring. § Define single nucleotide polymorphisms. § Discuss mitochondrial DNA typing.
Objectives § Compare and contrast different types of polymorphisms. § Define restriction fragment length polymorphisms. § Describe short tandem repeat structure and nomenclature. § Describe gender identification using the amelogenin locus. § Illustrate the use of STR for bone marrow engraftment monitoring. § Define single nucleotide polymorphisms. § Discuss mitochondrial DNA typing.
 Polymorphism § A DNA polymorphism is a sequence difference compared to a reference standard that is present in at least 1– 2% of a population. § Polymorphisms can be single bases or thousands of bases. § Polymorphisms may or may not have phenotypic effects.
Polymorphism § A DNA polymorphism is a sequence difference compared to a reference standard that is present in at least 1– 2% of a population. § Polymorphisms can be single bases or thousands of bases. § Polymorphisms may or may not have phenotypic effects.
 Polymorphic DNA Sequences § Polymorphisms are found throughout the genome. § If the location of a polymorphic sequence is known, it can serve as a landmark or marker for locating other genes or genetics regions. § Each polymorphic marker has different versions or alleles.
Polymorphic DNA Sequences § Polymorphisms are found throughout the genome. § If the location of a polymorphic sequence is known, it can serve as a landmark or marker for locating other genes or genetics regions. § Each polymorphic marker has different versions or alleles.
 Types of Polymorphic DNA Sequences § RFLP: restriction fragment length polymorphisms § VNTR: variable number tandem repeats (8 to >50 base pairs) § STR: short tandem repeats (1– 8 base pairs) § SNP: single-nucleotide polymorphisms
Types of Polymorphic DNA Sequences § RFLP: restriction fragment length polymorphisms § VNTR: variable number tandem repeats (8 to >50 base pairs) § STR: short tandem repeats (1– 8 base pairs) § SNP: single-nucleotide polymorphisms
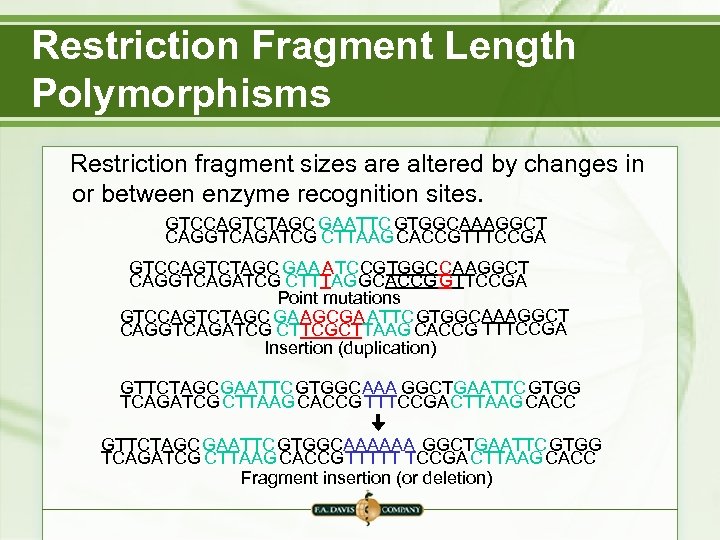 Restriction Fragment Length Polymorphisms Restriction fragment sizes are altered by changes in or between enzyme recognition sites. GTCCAGTCTAGC GAATTC GTGGCAAAGGCT CAGGTCAGATCG CTTAAG CACCGTTTCCGA GTCCAGTCTAGC GAA ATCCGTGGCCAAGGCT CAGGTCAGATCG CTTTAGGCACCGGTTCCGA Point mutations GTCCAGTCTAGC GAAGCGA ATTCGTGGCAAAGGCT CAGGTCAGATCG CTTCGCTTAAG CACCG TTTCCGA Insertion (duplication) GTTCTAGCGAATTC GTGGCAAA GGCTGAATTC GTGG TCAGATCG CTTAAG CACCGTTTCCGACTTAAG CACC GTTCTAGCGAATTC GTGGCAAAAAA GGCTGAATTC GTGG TCAGATCG CTTAAG CACCGTTTTT TCCGACTTAAG CACC Fragment insertion (or deletion)
Restriction Fragment Length Polymorphisms Restriction fragment sizes are altered by changes in or between enzyme recognition sites. GTCCAGTCTAGC GAATTC GTGGCAAAGGCT CAGGTCAGATCG CTTAAG CACCGTTTCCGA GTCCAGTCTAGC GAA ATCCGTGGCCAAGGCT CAGGTCAGATCG CTTTAGGCACCGGTTCCGA Point mutations GTCCAGTCTAGC GAAGCGA ATTCGTGGCAAAGGCT CAGGTCAGATCG CTTCGCTTAAG CACCG TTTCCGA Insertion (duplication) GTTCTAGCGAATTC GTGGCAAA GGCTGAATTC GTGG TCAGATCG CTTAAG CACCGTTTCCGACTTAAG CACC GTTCTAGCGAATTC GTGGCAAAAAA GGCTGAATTC GTGG TCAGATCG CTTAAG CACCGTTTTT TCCGACTTAAG CACC Fragment insertion (or deletion)
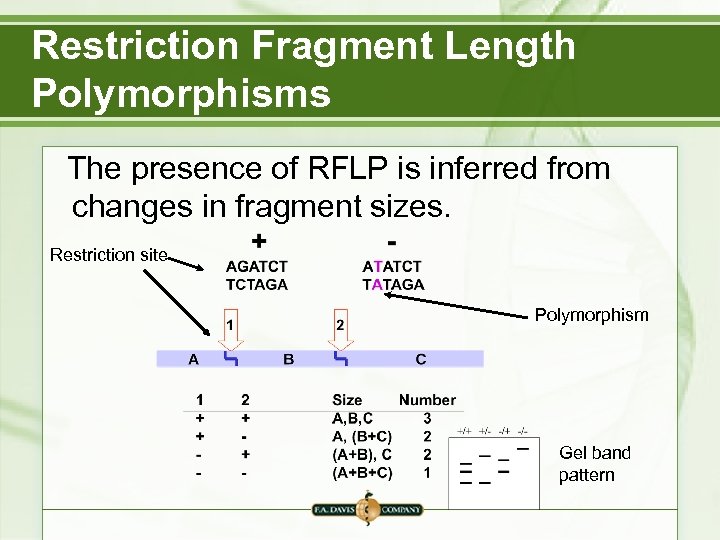 Restriction Fragment Length Polymorphisms The presence of RFLP is inferred from changes in fragment sizes. Restriction site Polymorphism Gel band pattern
Restriction Fragment Length Polymorphisms The presence of RFLP is inferred from changes in fragment sizes. Restriction site Polymorphism Gel band pattern
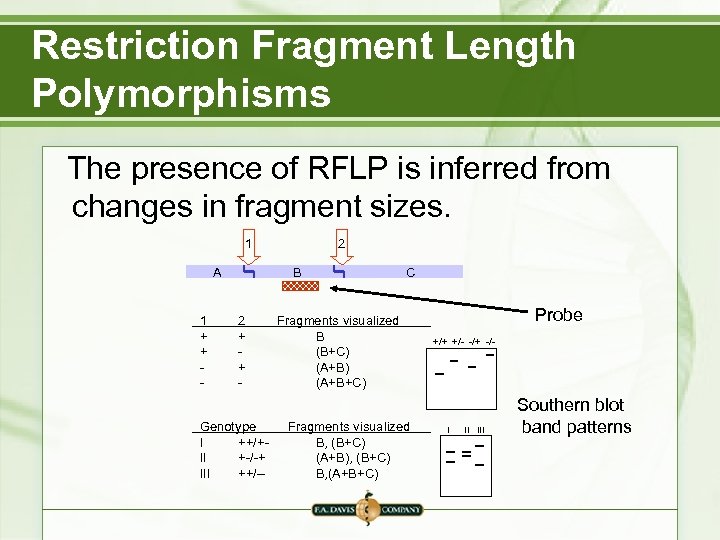 Restriction Fragment Length Polymorphisms The presence of RFLP is inferred from changes in fragment sizes. 1 A 1 + + - 2 B 2 + + - Genotype I ++/+II +-/-+ III ++/-- C Fragments visualized B (B+C) (A+B+C) Fragments visualized B, (B+C) (A+B), (B+C) B, (A+B+C) Probe +/+ +/- -/+ -/- I II III Southern blot band patterns
Restriction Fragment Length Polymorphisms The presence of RFLP is inferred from changes in fragment sizes. 1 A 1 + + - 2 B 2 + + - Genotype I ++/+II +-/-+ III ++/-- C Fragments visualized B (B+C) (A+B+C) Fragments visualized B, (B+C) (A+B), (B+C) B, (A+B+C) Probe +/+ +/- -/+ -/- I II III Southern blot band patterns
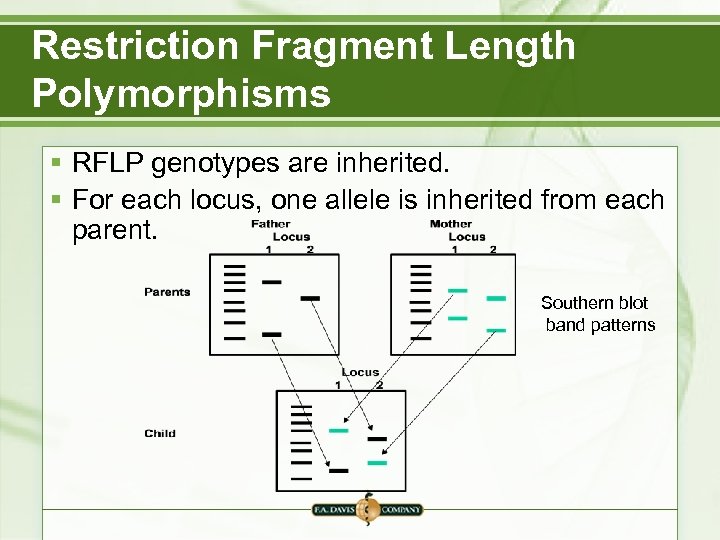 Restriction Fragment Length Polymorphisms § RFLP genotypes are inherited. § For each locus, one allele is inherited from each parent. Southern blot band patterns
Restriction Fragment Length Polymorphisms § RFLP genotypes are inherited. § For each locus, one allele is inherited from each parent. Southern blot band patterns
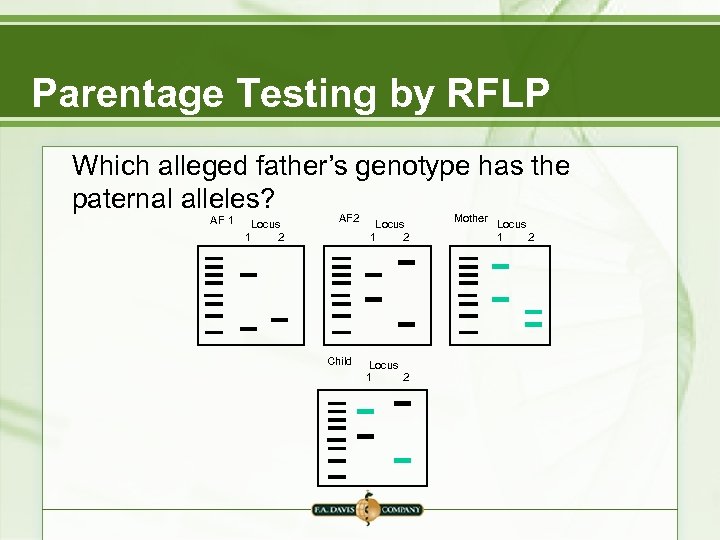 Parentage Testing by RFLP Which alleged father’s genotype has the paternal alleles? AF 1 Locus 1 2 AF 2 Child Locus 1 2 Mother Locus 1 2
Parentage Testing by RFLP Which alleged father’s genotype has the paternal alleles? AF 1 Locus 1 2 AF 2 Child Locus 1 2 Mother Locus 1 2
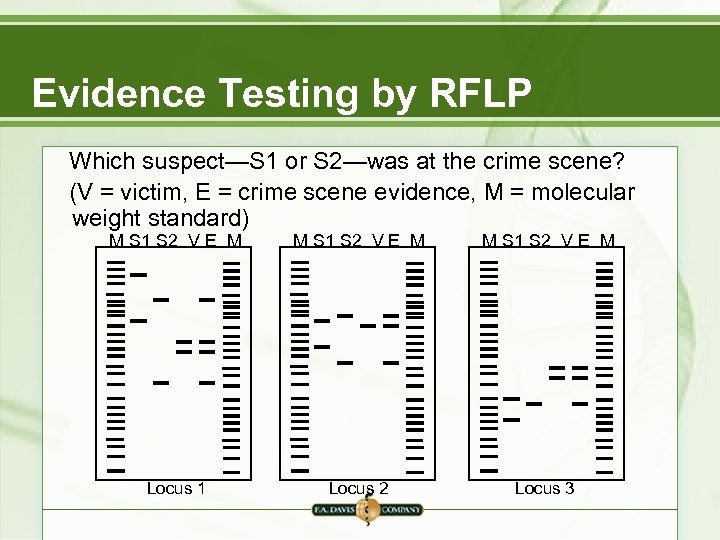 Evidence Testing by RFLP Which suspect—S 1 or S 2—was at the crime scene? (V = victim, E = crime scene evidence, M = molecular weight standard) M S 1 S 2 V E M Locus 1 Locus 2 Locus 3
Evidence Testing by RFLP Which suspect—S 1 or S 2—was at the crime scene? (V = victim, E = crime scene evidence, M = molecular weight standard) M S 1 S 2 V E M Locus 1 Locus 2 Locus 3
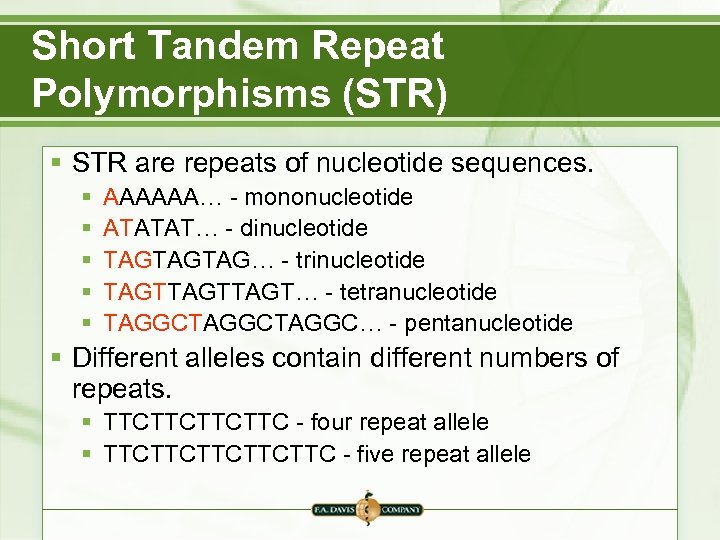 Short Tandem Repeat Polymorphisms (STR) § STR are repeats of nucleotide sequences. § § § AAAAAA… - mononucleotide ATATAT… - dinucleotide TAGTAGTAG… - trinucleotide TAGTTAGT… - tetranucleotide TAGGCTAGGC… - pentanucleotide § Different alleles contain different numbers of repeats. § TTCTTC - four repeat allele § TTCTTCTTC - five repeat allele
Short Tandem Repeat Polymorphisms (STR) § STR are repeats of nucleotide sequences. § § § AAAAAA… - mononucleotide ATATAT… - dinucleotide TAGTAGTAG… - trinucleotide TAGTTAGT… - tetranucleotide TAGGCTAGGC… - pentanucleotide § Different alleles contain different numbers of repeats. § TTCTTC - four repeat allele § TTCTTCTTC - five repeat allele
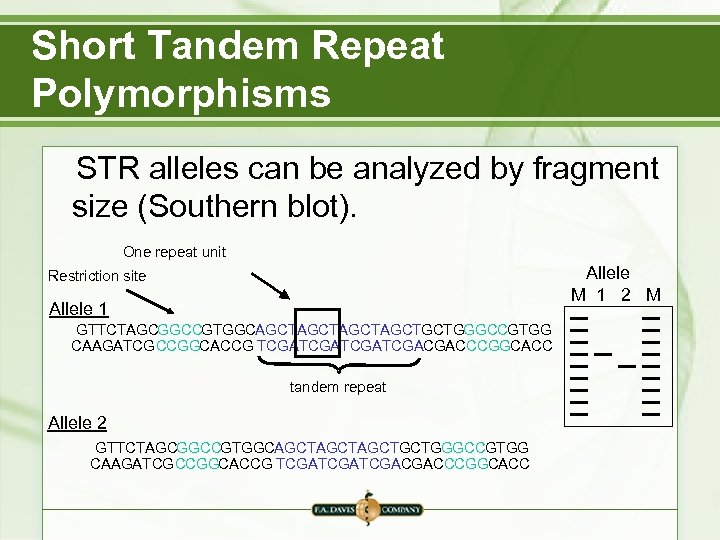 Short Tandem Repeat Polymorphisms STR alleles can be analyzed by fragment size (Southern blot). One repeat unit Allele M 1 2 M Restriction site Allele 1 GTTCTAGCGGCCGTGGCAGCTAGCTGCTGGGCCGTGG CAAGATCGCCGGCACCG TCGATCGACGACCCGGCACC tandem repeat Allele 2 GTTCTAGCGGCCGTGGCAGCTAGCTGCTGGGCCGTGG CAAGATCGCCGGCACCG TCGATCGACGACCCGGCACC
Short Tandem Repeat Polymorphisms STR alleles can be analyzed by fragment size (Southern blot). One repeat unit Allele M 1 2 M Restriction site Allele 1 GTTCTAGCGGCCGTGGCAGCTAGCTGCTGGGCCGTGG CAAGATCGCCGGCACCG TCGATCGACGACCCGGCACC tandem repeat Allele 2 GTTCTAGCGGCCGTGGCAGCTAGCTGCTGGGCCGTGG CAAGATCGCCGGCACCG TCGATCGACGACCCGGCACC
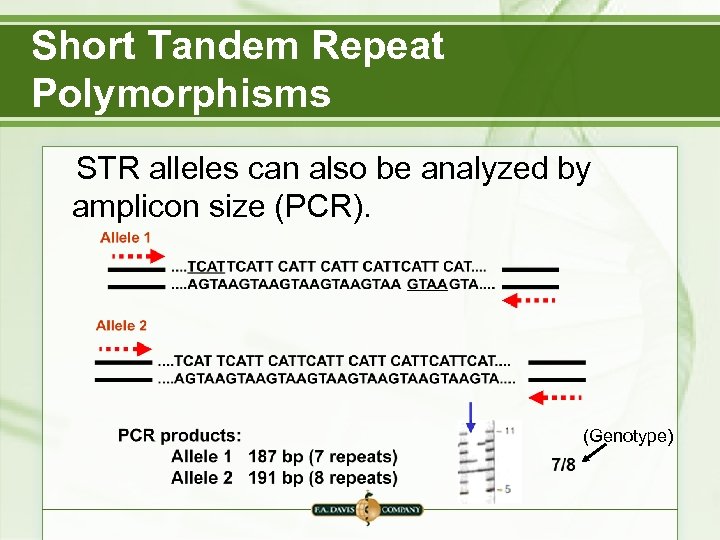 Short Tandem Repeat Polymorphisms STR alleles can also be analyzed by amplicon size (PCR). (Genotype)
Short Tandem Repeat Polymorphisms STR alleles can also be analyzed by amplicon size (PCR). (Genotype)
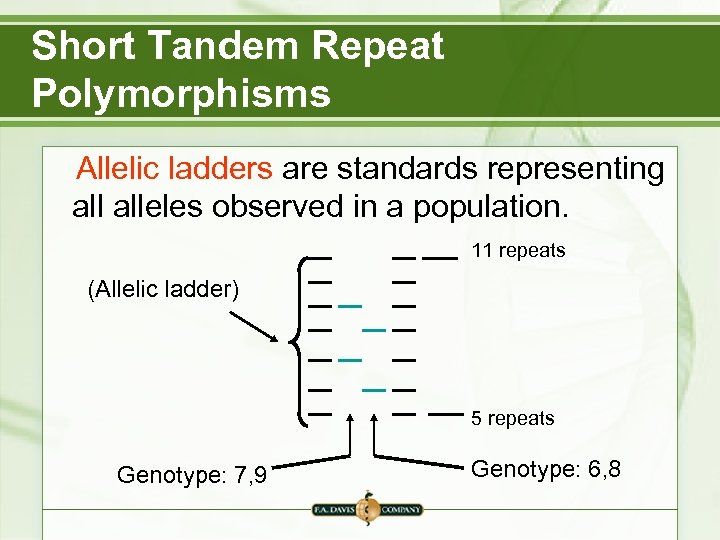 Short Tandem Repeat Polymorphisms Allelic ladders are standards representing alleles observed in a population. 11 repeats (Allelic ladder) 5 repeats Genotype: 7, 9 Genotype: 6, 8
Short Tandem Repeat Polymorphisms Allelic ladders are standards representing alleles observed in a population. 11 repeats (Allelic ladder) 5 repeats Genotype: 7, 9 Genotype: 6, 8
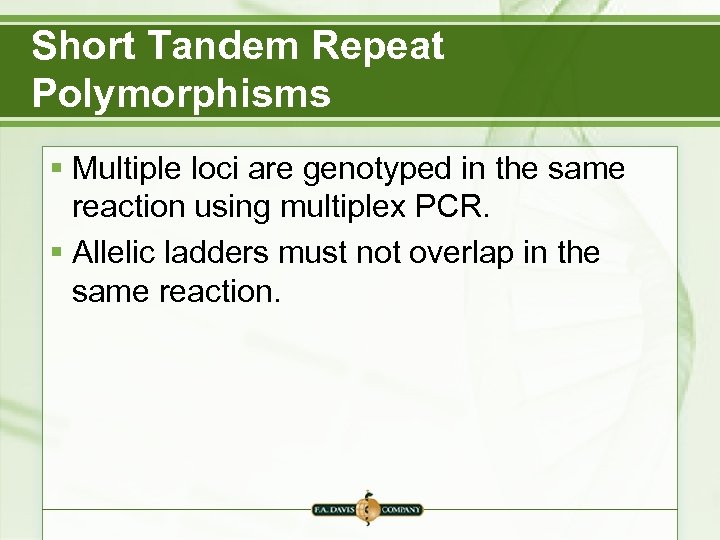 Short Tandem Repeat Polymorphisms § Multiple loci are genotyped in the same reaction using multiplex PCR. § Allelic ladders must not overlap in the same reaction.
Short Tandem Repeat Polymorphisms § Multiple loci are genotyped in the same reaction using multiplex PCR. § Allelic ladders must not overlap in the same reaction.
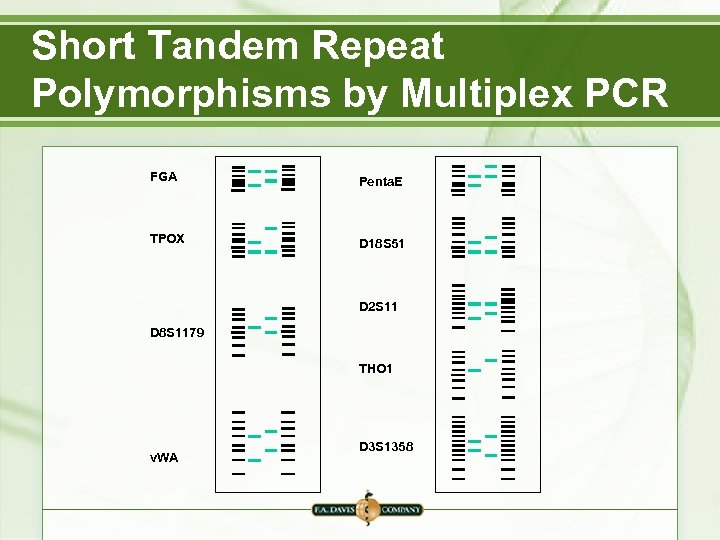 Short Tandem Repeat Polymorphisms by Multiplex PCR FGA Penta. E TPOX D 18 S 51 D 2 S 11 D 8 S 1179 THO 1 v. WA D 3 S 1358
Short Tandem Repeat Polymorphisms by Multiplex PCR FGA Penta. E TPOX D 18 S 51 D 2 S 11 D 8 S 1179 THO 1 v. WA D 3 S 1358
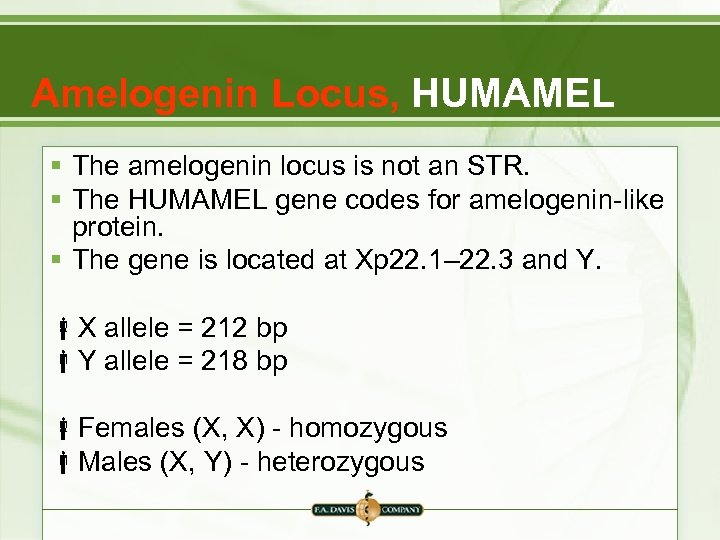 Amelogenin Locus, HUMAMEL § The amelogenin locus is not an STR. § The HUMAMEL gene codes for amelogenin-like protein. § The gene is located at Xp 22. 1– 22. 3 and Y. X allele = 212 bp Y allele = 218 bp Females (X, X) - homozygous Males (X, Y) - heterozygous
Amelogenin Locus, HUMAMEL § The amelogenin locus is not an STR. § The HUMAMEL gene codes for amelogenin-like protein. § The gene is located at Xp 22. 1– 22. 3 and Y. X allele = 212 bp Y allele = 218 bp Females (X, X) - homozygous Males (X, Y) - heterozygous
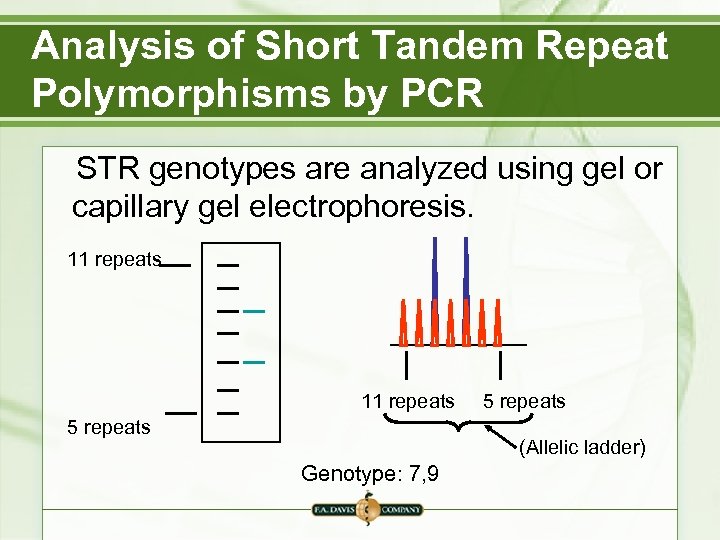 Analysis of Short Tandem Repeat Polymorphisms by PCR STR genotypes are analyzed using gel or capillary gel electrophoresis. 11 repeats 5 repeats (Allelic ladder) Genotype: 7, 9
Analysis of Short Tandem Repeat Polymorphisms by PCR STR genotypes are analyzed using gel or capillary gel electrophoresis. 11 repeats 5 repeats (Allelic ladder) Genotype: 7, 9
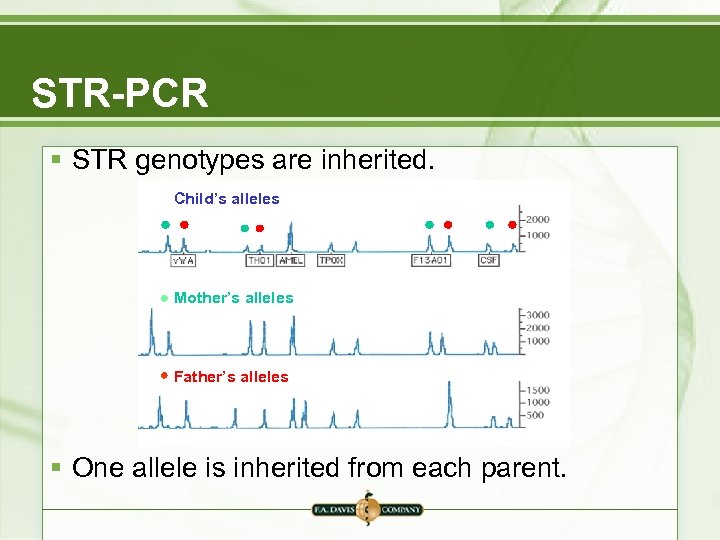 STR-PCR § STR genotypes are inherited. Child’s alleles Mother’s alleles Father’s alleles § One allele is inherited from each parent.
STR-PCR § STR genotypes are inherited. Child’s alleles Mother’s alleles Father’s alleles § One allele is inherited from each parent.
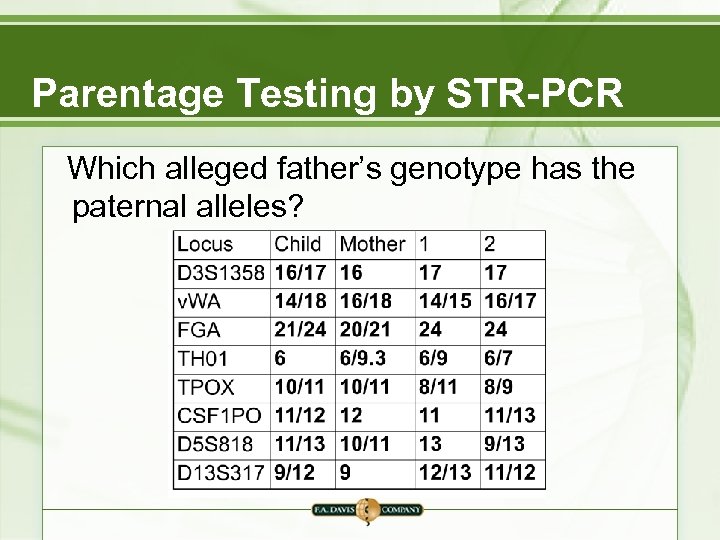 Parentage Testing by STR-PCR Which alleged father’s genotype has the paternal alleles?
Parentage Testing by STR-PCR Which alleged father’s genotype has the paternal alleles?
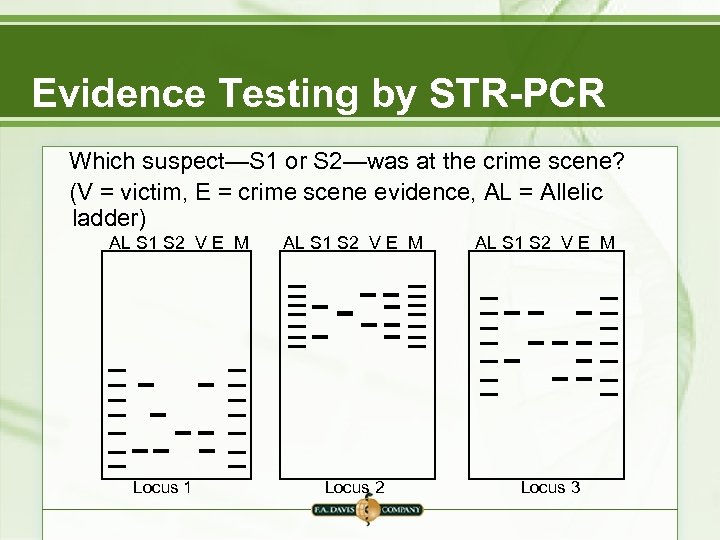 Evidence Testing by STR-PCR Which suspect—S 1 or S 2—was at the crime scene? (V = victim, E = crime scene evidence, AL = Allelic ladder) AL S 1 S 2 V E M Locus 1 AL S 1 S 2 V E M Locus 2 AL S 1 S 2 V E M Locus 3
Evidence Testing by STR-PCR Which suspect—S 1 or S 2—was at the crime scene? (V = victim, E = crime scene evidence, AL = Allelic ladder) AL S 1 S 2 V E M Locus 1 AL S 1 S 2 V E M Locus 2 AL S 1 S 2 V E M Locus 3
 Short Tandem Repeat Polymorphisms: Y-STR § The Y chromosome is inherited in a block without recombination. § STR on the Y chromosome are inherited paternally as a haplotype. § Y haplotypes are used for exclusion and paternal lineage analysis.
Short Tandem Repeat Polymorphisms: Y-STR § The Y chromosome is inherited in a block without recombination. § STR on the Y chromosome are inherited paternally as a haplotype. § Y haplotypes are used for exclusion and paternal lineage analysis.
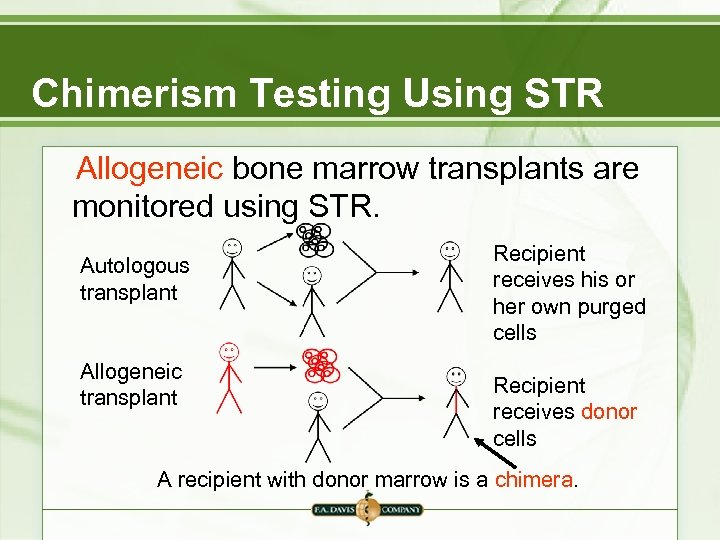 Chimerism Testing Using STR Allogeneic bone marrow transplants are monitored using STR. Autologous transplant Allogeneic transplant Recipient receives his or her own purged cells Recipient receives donor cells A recipient with donor marrow is a chimera.
Chimerism Testing Using STR Allogeneic bone marrow transplants are monitored using STR. Autologous transplant Allogeneic transplant Recipient receives his or her own purged cells Recipient receives donor cells A recipient with donor marrow is a chimera.
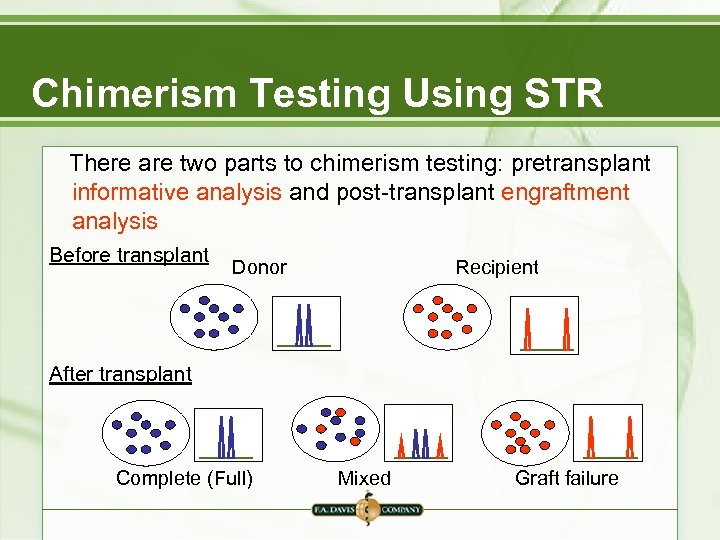 Chimerism Testing Using STR There are two parts to chimerism testing: pretransplant informative analysis and post-transplant engraftment analysis Before transplant Donor Recipient After transplant Complete (Full) Mixed Graft failure
Chimerism Testing Using STR There are two parts to chimerism testing: pretransplant informative analysis and post-transplant engraftment analysis Before transplant Donor Recipient After transplant Complete (Full) Mixed Graft failure
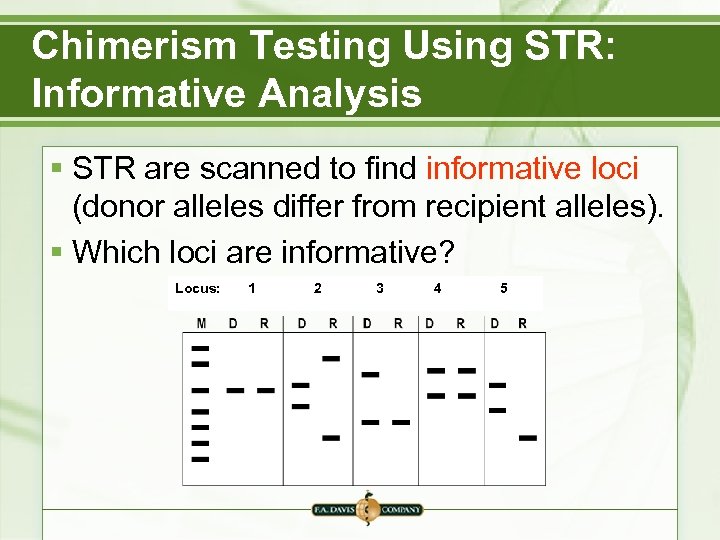 Chimerism Testing Using STR: Informative Analysis § STR are scanned to find informative loci (donor alleles differ from recipient alleles). § Which loci are informative? Locus: 1 2 3 4 5
Chimerism Testing Using STR: Informative Analysis § STR are scanned to find informative loci (donor alleles differ from recipient alleles). § Which loci are informative? Locus: 1 2 3 4 5
 Chimerism Testing Using STR: Informative Analysis § There are different degrees of informativity. § With the most informative loci, recipient bands or peaks do not overlap stutter in donor bands or peaks. § Stutter is a technical artifact of the PCR reaction in which a minor product of n-1 repeat units is produced.
Chimerism Testing Using STR: Informative Analysis § There are different degrees of informativity. § With the most informative loci, recipient bands or peaks do not overlap stutter in donor bands or peaks. § Stutter is a technical artifact of the PCR reaction in which a minor product of n-1 repeat units is produced.
![Examples of Informative Loci (Type 5) [Thiede et al. , Leukemia 18: 248 (2004)] Examples of Informative Loci (Type 5) [Thiede et al. , Leukemia 18: 248 (2004)]](https://present5.com/presentation/ad954603295a435e47701326c8c912d1/image-28.jpg) Examples of Informative Loci (Type 5) [Thiede et al. , Leukemia 18: 248 (2004)] Recipient Stutter Donor Recipient Donor
Examples of Informative Loci (Type 5) [Thiede et al. , Leukemia 18: 248 (2004)] Recipient Stutter Donor Recipient Donor
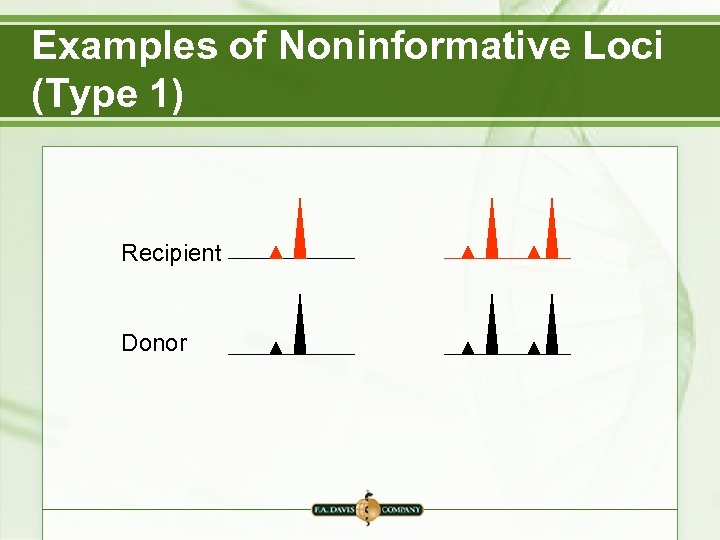 Examples of Noninformative Loci (Type 1) Recipient Donor
Examples of Noninformative Loci (Type 1) Recipient Donor
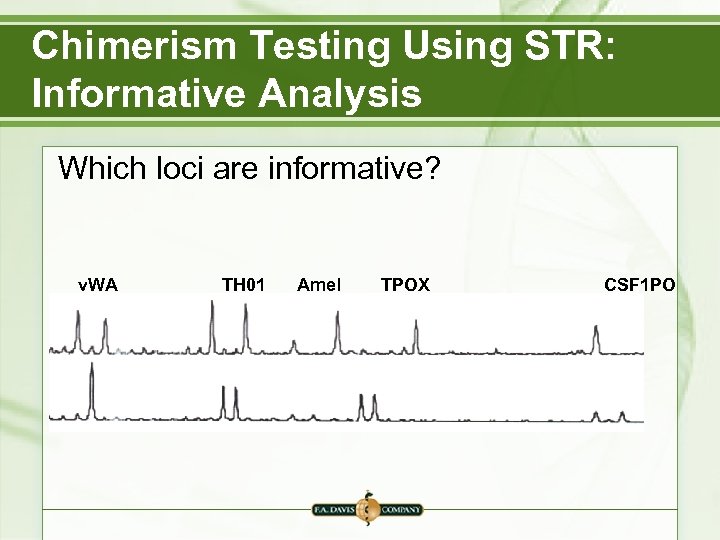 Chimerism Testing Using STR: Informative Analysis Which loci are informative? v. WA TH 01 Amel TPOX CSF 1 PO
Chimerism Testing Using STR: Informative Analysis Which loci are informative? v. WA TH 01 Amel TPOX CSF 1 PO
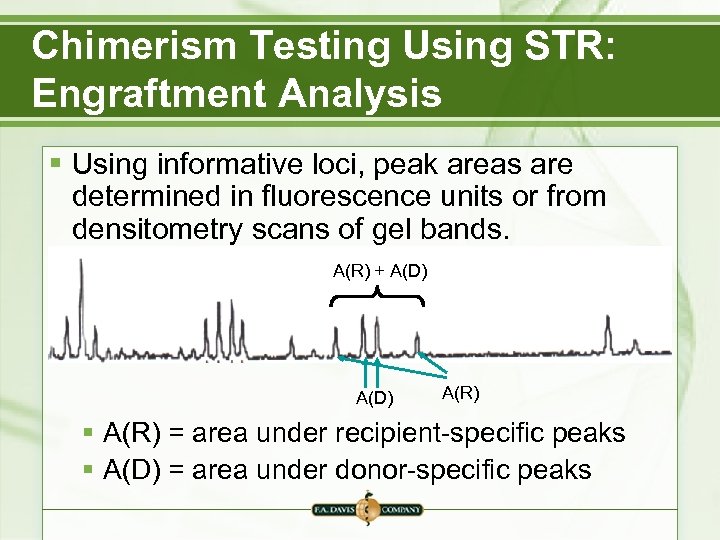 Chimerism Testing Using STR: Engraftment Analysis § Using informative loci, peak areas are determined in fluorescence units or from densitometry scans of gel bands. A(R) + A(D) A(R) § A(R) = area under recipient-specific peaks § A(D) = area under donor-specific peaks
Chimerism Testing Using STR: Engraftment Analysis § Using informative loci, peak areas are determined in fluorescence units or from densitometry scans of gel bands. A(R) + A(D) A(R) § A(R) = area under recipient-specific peaks § A(D) = area under donor-specific peaks
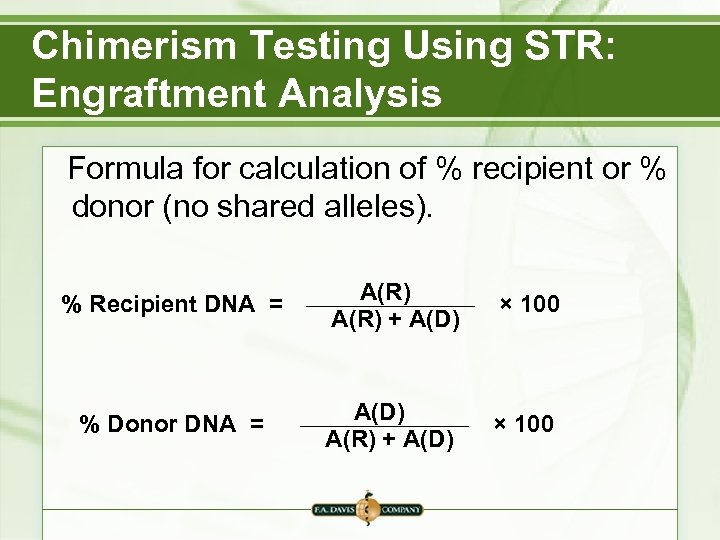 Chimerism Testing Using STR: Engraftment Analysis Formula for calculation of % recipient or % donor (no shared alleles). % Recipient DNA = A(R) + A(D) × 100 % Donor DNA = A(D) A(R) + A(D) × 100
Chimerism Testing Using STR: Engraftment Analysis Formula for calculation of % recipient or % donor (no shared alleles). % Recipient DNA = A(R) + A(D) × 100 % Donor DNA = A(D) A(R) + A(D) × 100
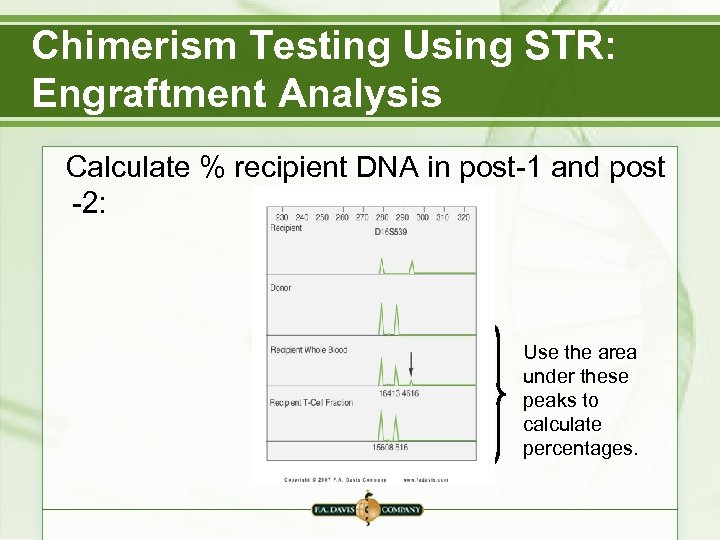 Chimerism Testing Using STR: Engraftment Analysis Calculate % recipient DNA in post-1 and post -2: Use the area under these peaks to calculate percentages.
Chimerism Testing Using STR: Engraftment Analysis Calculate % recipient DNA in post-1 and post -2: Use the area under these peaks to calculate percentages.
 Chimerism Analysis of Cellular Subsets § Cell subsets (T cells, granulocytes, NK cells, etc. ) engraft with different kinetics. § Analysis of cellular subsets provides a more detailed description of the engrafting cell population. § Analysis of cellular subsets also increases the sensitivity of the engraftment assay.
Chimerism Analysis of Cellular Subsets § Cell subsets (T cells, granulocytes, NK cells, etc. ) engraft with different kinetics. § Analysis of cellular subsets provides a more detailed description of the engrafting cell population. § Analysis of cellular subsets also increases the sensitivity of the engraftment assay.
 Chimerism Analysis of Cellular Subsets § T cells (CD 3), NK cells (CD 56), granulocytes, myeloid cells (CD 13, CD 33), myelomonocytic cells (CD 14), B cells (CD 19), stem cells (CD 34) § Methods § Flow cytometric sorting § Immunomagnetic cell sorting § Immunohistochemistry + XY FISH
Chimerism Analysis of Cellular Subsets § T cells (CD 3), NK cells (CD 56), granulocytes, myeloid cells (CD 13, CD 33), myelomonocytic cells (CD 14), B cells (CD 19), stem cells (CD 34) § Methods § Flow cytometric sorting § Immunomagnetic cell sorting § Immunohistochemistry + XY FISH
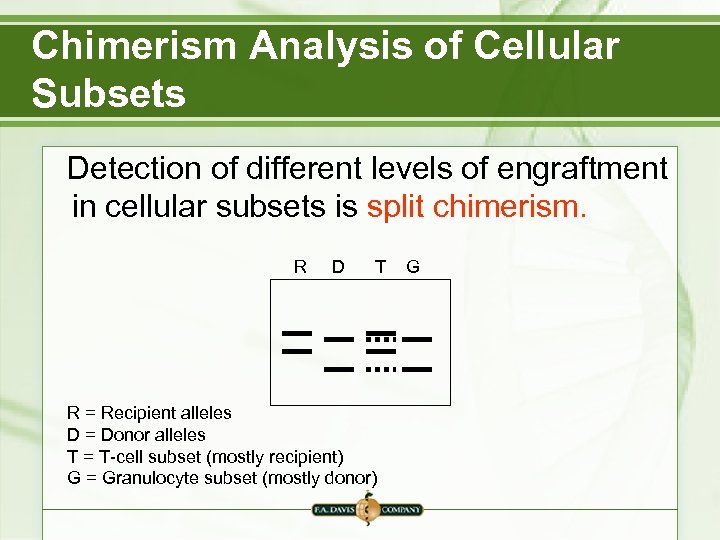 Chimerism Analysis of Cellular Subsets Detection of different levels of engraftment in cellular subsets is split chimerism. R D T R = Recipient alleles D = Donor alleles T = T-cell subset (mostly recipient) G = Granulocyte subset (mostly donor) G
Chimerism Analysis of Cellular Subsets Detection of different levels of engraftment in cellular subsets is split chimerism. R D T R = Recipient alleles D = Donor alleles T = T-cell subset (mostly recipient) G = Granulocyte subset (mostly donor) G
 Single Nucleotide Polymorphisms (SNP) § Single-nucleotide differences between DNA sequences. § One SNP occurs approximately every 1, 250 base pairs in human DNA. § SNPs are detected by sequencing, melt curve analysis, or other methods. § 99% have no biological effect; 60, 000 are within genes.
Single Nucleotide Polymorphisms (SNP) § Single-nucleotide differences between DNA sequences. § One SNP occurs approximately every 1, 250 base pairs in human DNA. § SNPs are detected by sequencing, melt curve analysis, or other methods. § 99% have no biological effect; 60, 000 are within genes.
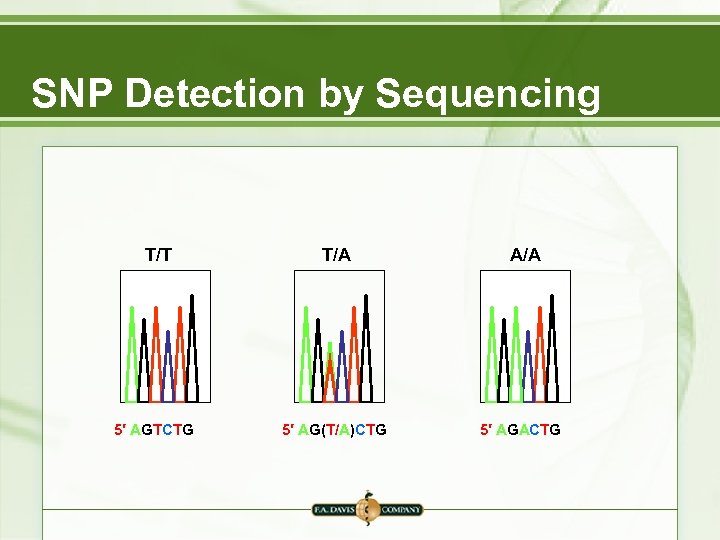 SNP Detection by Sequencing T/T 5′ AGTCTG T/A 5′ AG(T/A)CTG A/A 5′ AGACTG
SNP Detection by Sequencing T/T 5′ AGTCTG T/A 5′ AG(T/A)CTG A/A 5′ AGACTG
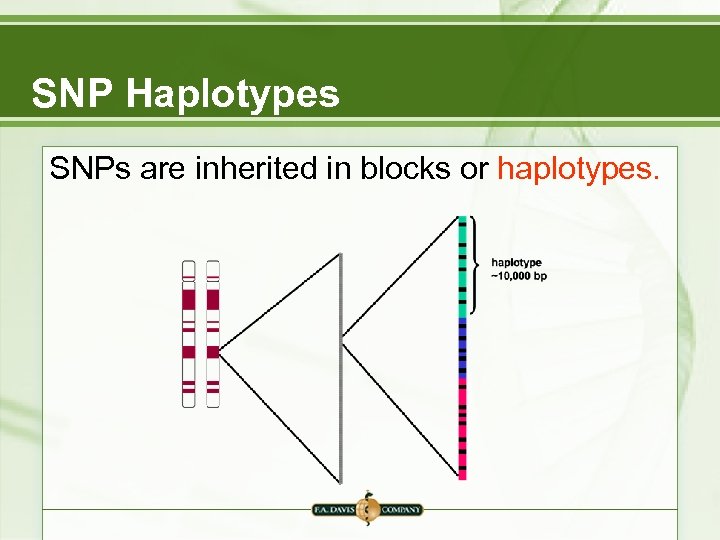 SNP Haplotypes SNPs are inherited in blocks or haplotypes.
SNP Haplotypes SNPs are inherited in blocks or haplotypes.
 Applications of SNP Analysis § SNPs can be used for mapping genes, human identification, chimerism analysis, and many other applications. § The Human Haplotype Mapping (Hap. Map) Project is aimed at identifying SNP haplotypes throughout the human genome.
Applications of SNP Analysis § SNPs can be used for mapping genes, human identification, chimerism analysis, and many other applications. § The Human Haplotype Mapping (Hap. Map) Project is aimed at identifying SNP haplotypes throughout the human genome.
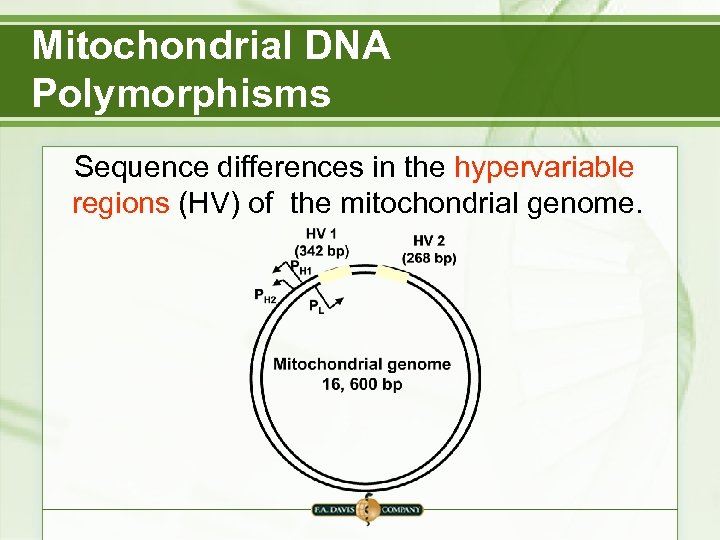 Mitochondrial DNA Polymorphisms Sequence differences in the hypervariable regions (HV) of the mitochondrial genome.
Mitochondrial DNA Polymorphisms Sequence differences in the hypervariable regions (HV) of the mitochondrial genome.
 Mitochondrial DNA Polymorphisms § Mitochondria are maternally inherited. § There an average of 8. 5 base differences in the mitochondrial HV sequences of unrelated individuals. § All maternal relatives will have the same mitochondrial sequences. § Mitochondrial typing can be used for legal exclusion of individuals or confirmation of maternal lineage.
Mitochondrial DNA Polymorphisms § Mitochondria are maternally inherited. § There an average of 8. 5 base differences in the mitochondrial HV sequences of unrelated individuals. § All maternal relatives will have the same mitochondrial sequences. § Mitochondrial typing can be used for legal exclusion of individuals or confirmation of maternal lineage.
 Summary § Four types of polymorphisms are used for a variety of purposes in the laboratory: RFLP, VNTR, STR, and SNP. § Polymorphisms are used for human identification and parentage testing. § Y-STR haplotypes are paternally inherited. § Polymorphisms are used to measure engraftment after allogeneic bone marrow transplants.
Summary § Four types of polymorphisms are used for a variety of purposes in the laboratory: RFLP, VNTR, STR, and SNP. § Polymorphisms are used for human identification and parentage testing. § Y-STR haplotypes are paternally inherited. § Polymorphisms are used to measure engraftment after allogeneic bone marrow transplants.
 Summary § Single-nucleotide polymorphisms are detected by sequencing, melt curve analysis, or other methods. § SNPs can be used for the same applications as other polymorphisms. § Mitochondrial DNA typing is performed by sequencing the mitochondrial HV regions. § Mitochondrial types are maternally inherited.
Summary § Single-nucleotide polymorphisms are detected by sequencing, melt curve analysis, or other methods. § SNPs can be used for the same applications as other polymorphisms. § Mitochondrial DNA typing is performed by sequencing the mitochondrial HV regions. § Mitochondrial types are maternally inherited.


