68ca7760cf75b19a8fbd9412490ef507.ppt
- Количество слайдов: 75
 Chapter 10 STR Typing and Data Interpretation Fundamentals of Forensic DNA Typing Slides prepared by John M. Butler June 2009
Chapter 10 STR Typing and Data Interpretation Fundamentals of Forensic DNA Typing Slides prepared by John M. Butler June 2009
 Chapter 10 – Data Interpretation Chapter Summary Once peaks are produced in a multi-colored electropherogram, STR genotyping and data interpretation involves deciphering true STR alleles from possible instrumental or biological artifacts. Peaks are typically sized using a “Local Southern” algorithm that uses two peaks from the internal size standard on either side of the sample peak. Only peaks above a detection threshold set by the user will be recognized and recorded in the genotyping software output. Resultant peaks that are sized in comparison to the internal size standard and above the detection threshold are then converted to STR repeat numbers through comparison to a sequenced allelic ladder provided with each STR kit. A high degree of precision must exist from sample-tosample so that sample alleles can be reliably compared to the allele rungs in the allelic ladder. Typically a sizing bin of +/- 0. 5 bp is used around each allele in the STR allelic ladder. Off-ladder alleles, also known as microvariants, that contain nucleotide changes or insertions or deletions in the STR repeat region or immediate flanking regions are known to exist and can be determined with a high precision CE instrument. Other biological artifacts that can complicate STR data interpretation include stutter products, non-template addition, tri-allelic patterns, allele dropout due to “null alleles”, and STR allele mutation. Instrumental or technology-related artifact peaks can arise from sample contaminants, electrical spikes, dye blobs, and matrix (color separation) failure due to off-scale data. Partial profiles or DNA mixtures can further complicate data interpretation. Ultimately, an analyst must decide whether or not two DNA profiles match or can be excluded from coming from the same biological source.
Chapter 10 – Data Interpretation Chapter Summary Once peaks are produced in a multi-colored electropherogram, STR genotyping and data interpretation involves deciphering true STR alleles from possible instrumental or biological artifacts. Peaks are typically sized using a “Local Southern” algorithm that uses two peaks from the internal size standard on either side of the sample peak. Only peaks above a detection threshold set by the user will be recognized and recorded in the genotyping software output. Resultant peaks that are sized in comparison to the internal size standard and above the detection threshold are then converted to STR repeat numbers through comparison to a sequenced allelic ladder provided with each STR kit. A high degree of precision must exist from sample-tosample so that sample alleles can be reliably compared to the allele rungs in the allelic ladder. Typically a sizing bin of +/- 0. 5 bp is used around each allele in the STR allelic ladder. Off-ladder alleles, also known as microvariants, that contain nucleotide changes or insertions or deletions in the STR repeat region or immediate flanking regions are known to exist and can be determined with a high precision CE instrument. Other biological artifacts that can complicate STR data interpretation include stutter products, non-template addition, tri-allelic patterns, allele dropout due to “null alleles”, and STR allele mutation. Instrumental or technology-related artifact peaks can arise from sample contaminants, electrical spikes, dye blobs, and matrix (color separation) failure due to off-scale data. Partial profiles or DNA mixtures can further complicate data interpretation. Ultimately, an analyst must decide whether or not two DNA profiles match or can be excluded from coming from the same biological source.
 Data Transfer • Following data collection, the data (. fsa) files are typically transferred from the lab computer to one in an office where data analysis is performed • A USB thumb drive permits rapid and easy transfer of data files
Data Transfer • Following data collection, the data (. fsa) files are typically transferred from the lab computer to one in an office where data analysis is performed • A USB thumb drive permits rapid and easy transfer of data files
 Data Analysis • The analyst carefully reviews the DNA data (electropherogram) and checks software genotype calls and edits out artifacts • Software designates sample genotypes via comparison to an allelic ladder (mixture of common allele possibilities)
Data Analysis • The analyst carefully reviews the DNA data (electropherogram) and checks software genotype calls and edits out artifacts • Software designates sample genotypes via comparison to an allelic ladder (mixture of common allele possibilities)
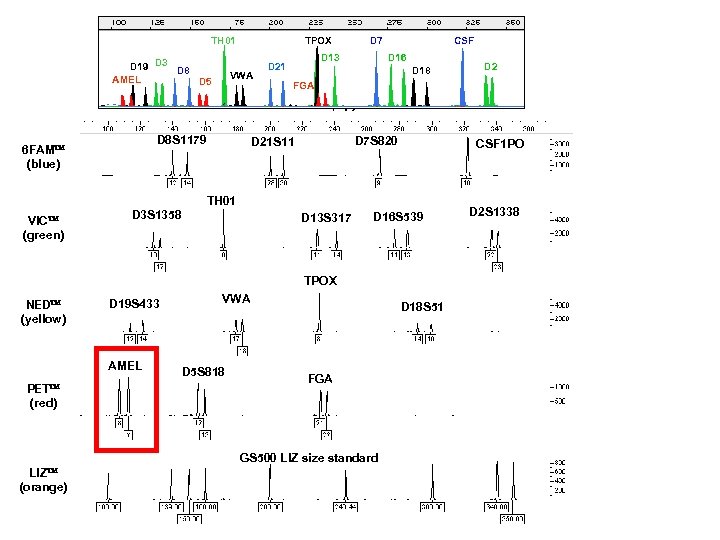 TH 01 D 19 D 3 D 8 AMEL D 5 TPOX VWA D 7 D 13 D 21 CSF D 16 D 18 D 2 FGA DNA Size (bp) D 8 S 1179 6 FAM (blue) D 7 S 820 D 21 S 11 CSF 1 PO TH 01 VIC (green) D 3 S 1358 D 13 S 317 D 16 S 539 TPOX NED (yellow) D 19 S 433 AMEL PET (red) VWA D 5 S 818 D 18 S 51 FGA GS 500 LIZ size standard LIZ (orange) D 2 S 1338
TH 01 D 19 D 3 D 8 AMEL D 5 TPOX VWA D 7 D 13 D 21 CSF D 16 D 18 D 2 FGA DNA Size (bp) D 8 S 1179 6 FAM (blue) D 7 S 820 D 21 S 11 CSF 1 PO TH 01 VIC (green) D 3 S 1358 D 13 S 317 D 16 S 539 TPOX NED (yellow) D 19 S 433 AMEL PET (red) VWA D 5 S 818 D 18 S 51 FGA GS 500 LIZ size standard LIZ (orange) D 2 S 1338
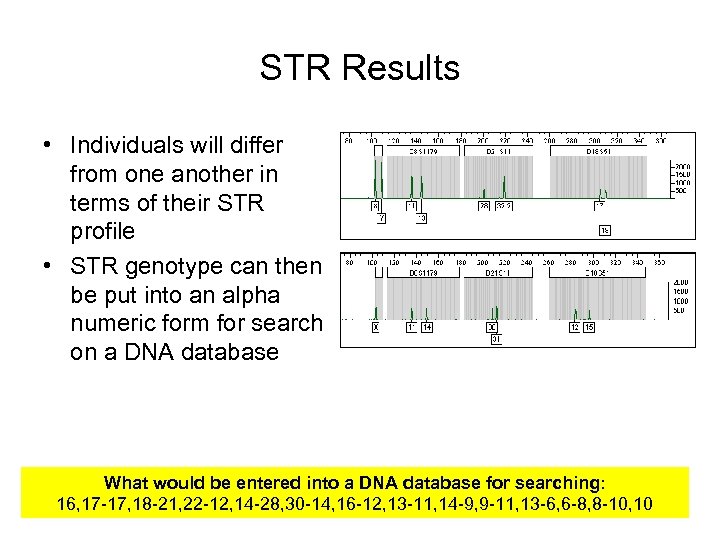 STR Results • Individuals will differ from one another in terms of their STR profile • STR genotype can then be put into an alpha numeric form for search on a DNA database What would be entered into a DNA database for searching: 16, 17 -17, 18 -21, 22 -12, 14 -28, 30 -14, 16 -12, 13 -11, 14 -9, 9 -11, 13 -6, 6 -8, 8 -10, 10
STR Results • Individuals will differ from one another in terms of their STR profile • STR genotype can then be put into an alpha numeric form for search on a DNA database What would be entered into a DNA database for searching: 16, 17 -17, 18 -21, 22 -12, 14 -28, 30 -14, 16 -12, 13 -11, 14 -9, 9 -11, 13 -6, 6 -8, 8 -10, 10
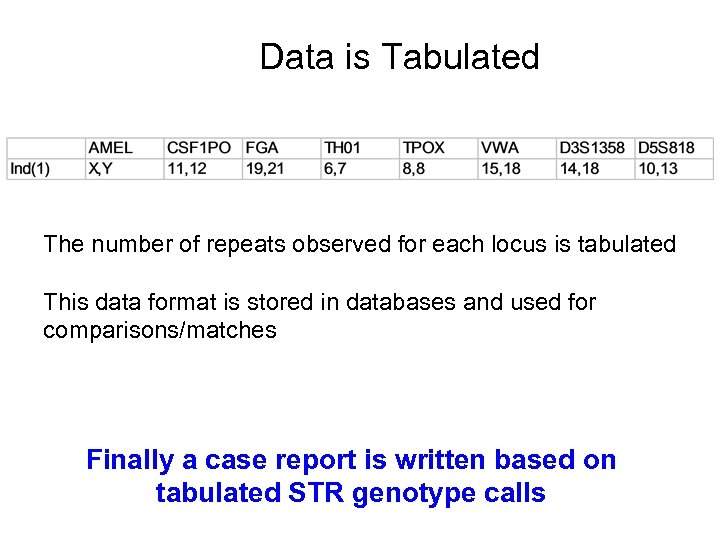 Data is Tabulated The number of repeats observed for each locus is tabulated This data format is stored in databases and used for comparisons/matches Finally a case report is written based on tabulated STR genotype calls
Data is Tabulated The number of repeats observed for each locus is tabulated This data format is stored in databases and used for comparisons/matches Finally a case report is written based on tabulated STR genotype calls
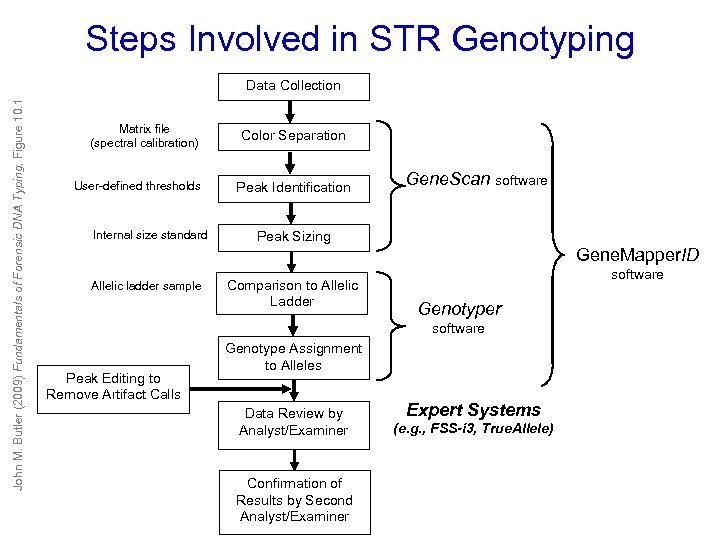 Steps Involved in STR Genotyping John M. Butler (2009) Fundamentals of Forensic DNA Typing, Figure 10. 1 Data Collection Matrix file (spectral calibration) User-defined thresholds Internal size standard Color Separation Peak Identification Gene. Scan software Peak Sizing Gene. Mapper. ID Allelic ladder sample Comparison to Allelic Ladder software Genotyper software Peak Editing to Remove Artifact Calls Genotype Assignment to Alleles Data Review by Analyst/Examiner Confirmation of Results by Second Analyst/Examiner Expert Systems (e. g. , FSS-i 3, True. Allele)
Steps Involved in STR Genotyping John M. Butler (2009) Fundamentals of Forensic DNA Typing, Figure 10. 1 Data Collection Matrix file (spectral calibration) User-defined thresholds Internal size standard Color Separation Peak Identification Gene. Scan software Peak Sizing Gene. Mapper. ID Allelic ladder sample Comparison to Allelic Ladder software Genotyper software Peak Editing to Remove Artifact Calls Genotype Assignment to Alleles Data Review by Analyst/Examiner Confirmation of Results by Second Analyst/Examiner Expert Systems (e. g. , FSS-i 3, True. Allele)
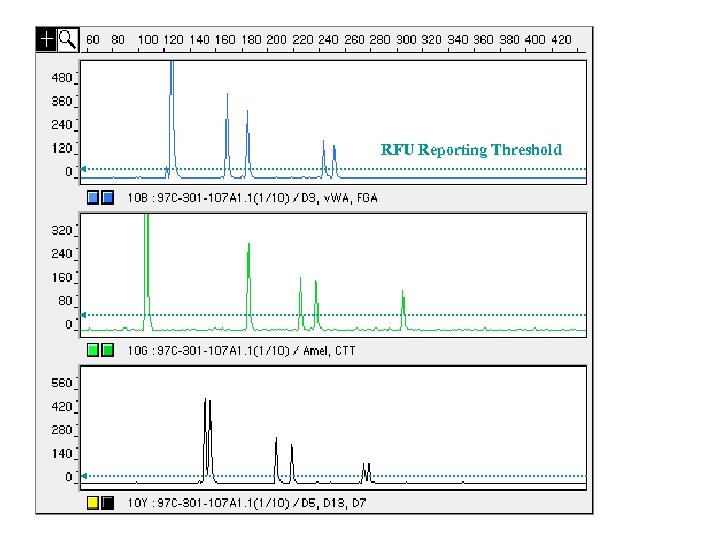
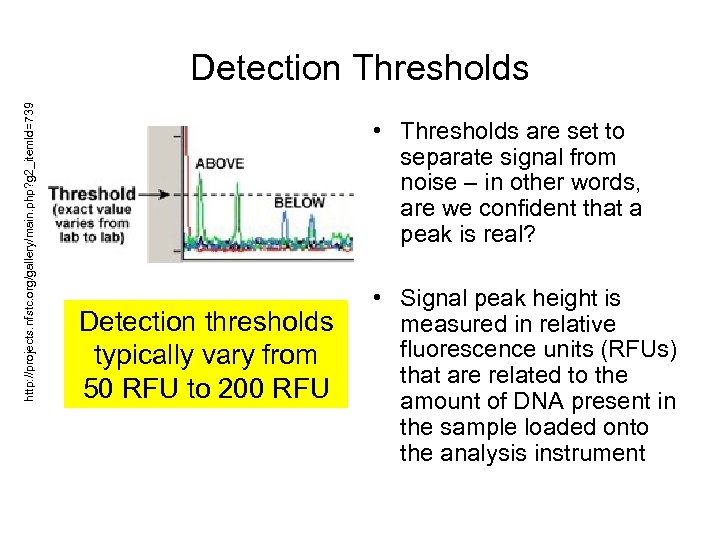 http: //projects. nfstc. org/gallery/main. php? g 2_item. Id=739 Detection Thresholds • Thresholds are set to separate signal from noise – in other words, are we confident that a peak is real? Detection thresholds typically vary from 50 RFU to 200 RFU • Signal peak height is measured in relative fluorescence units (RFUs) that are related to the amount of DNA present in the sample loaded onto the analysis instrument
http: //projects. nfstc. org/gallery/main. php? g 2_item. Id=739 Detection Thresholds • Thresholds are set to separate signal from noise – in other words, are we confident that a peak is real? Detection thresholds typically vary from 50 RFU to 200 RFU • Signal peak height is measured in relative fluorescence units (RFUs) that are related to the amount of DNA present in the sample loaded onto the analysis instrument
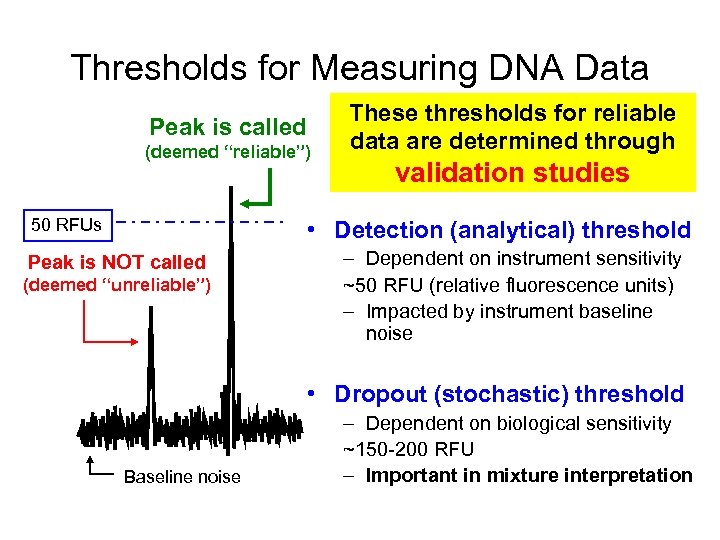 Thresholds for Measuring DNA Data Peak is called (deemed “reliable”) 50 RFUs These thresholds for reliable data are determined through validation studies • Detection (analytical) threshold Peak is NOT called (deemed “unreliable”) – Dependent on instrument sensitivity ~50 RFU (relative fluorescence units) – Impacted by instrument baseline noise • Dropout (stochastic) threshold Baseline noise – Dependent on biological sensitivity ~150 -200 RFU – Important in mixture interpretation
Thresholds for Measuring DNA Data Peak is called (deemed “reliable”) 50 RFUs These thresholds for reliable data are determined through validation studies • Detection (analytical) threshold Peak is NOT called (deemed “unreliable”) – Dependent on instrument sensitivity ~50 RFU (relative fluorescence units) – Impacted by instrument baseline noise • Dropout (stochastic) threshold Baseline noise – Dependent on biological sensitivity ~150 -200 RFU – Important in mixture interpretation
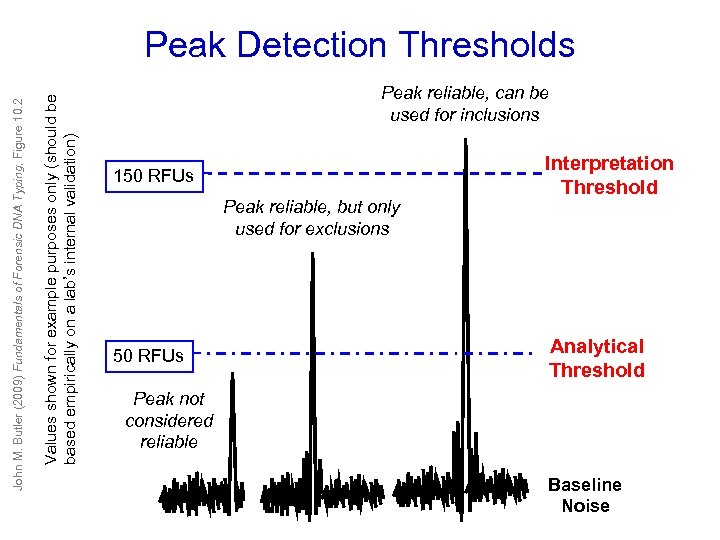 Values shown for example purposes only (should be based empirically on a lab’s internal validation) John M. Butler (2009) Fundamentals of Forensic DNA Typing, Figure 10. 2 Peak Detection Thresholds Peak reliable, can be used for inclusions 150 RFUs Peak reliable, but only used for exclusions 50 RFUs Interpretation Threshold Analytical Threshold Peak not considered reliable Baseline Noise
Values shown for example purposes only (should be based empirically on a lab’s internal validation) John M. Butler (2009) Fundamentals of Forensic DNA Typing, Figure 10. 2 Peak Detection Thresholds Peak reliable, can be used for inclusions 150 RFUs Peak reliable, but only used for exclusions 50 RFUs Interpretation Threshold Analytical Threshold Peak not considered reliable Baseline Noise
 DNA Data Quality • The raw DNA data itself does not have quality scores directly attached to it. • Only the STR allele designations are stored without an indication of data quality. • Checks and balances exist in the entire system to try and ensure good quality data. • Retesting of offender database sample is performed when a DNA database hit is observed.
DNA Data Quality • The raw DNA data itself does not have quality scores directly attached to it. • Only the STR allele designations are stored without an indication of data quality. • Checks and balances exist in the entire system to try and ensure good quality data. • Retesting of offender database sample is performed when a DNA database hit is observed.
 Data Interpretation Issues • Artifact Peaks vs. Allele Peaks • • • Pull-Up Stutter “N” Peaks Off Ladder Alleles Tri-Alleles • Allelic Drop-Out • Degradation • Inhibition/Primer Binding Site Mutation • Mixture • Mutation
Data Interpretation Issues • Artifact Peaks vs. Allele Peaks • • • Pull-Up Stutter “N” Peaks Off Ladder Alleles Tri-Alleles • Allelic Drop-Out • Degradation • Inhibition/Primer Binding Site Mutation • Mixture • Mutation
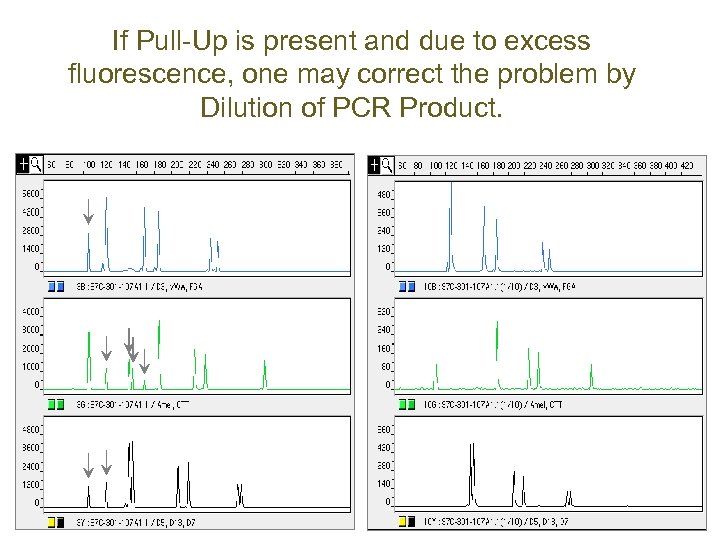 If Pull-Up is present and due to excess fluorescence, one may correct the problem by Dilution of PCR Product.
If Pull-Up is present and due to excess fluorescence, one may correct the problem by Dilution of PCR Product.
 RFU Reporting Threshold
RFU Reporting Threshold
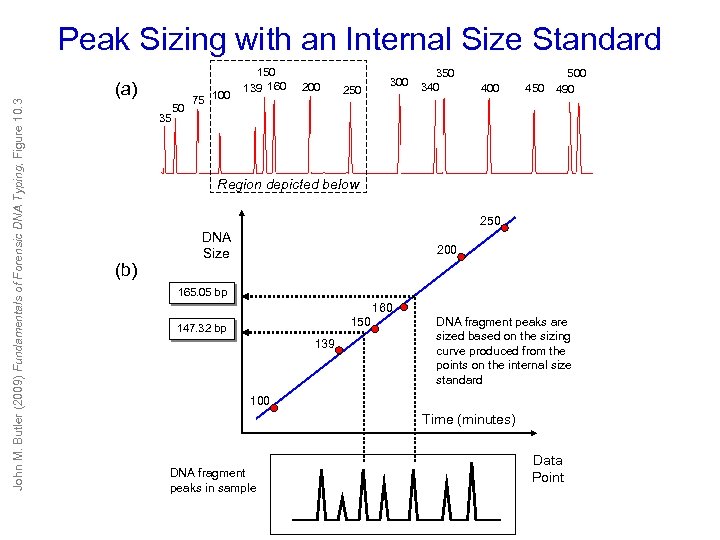 Peak Sizing with an Internal Size Standard John M. Butler (2009) Fundamentals of Forensic DNA Typing, Figure 10. 3 (a) 35 50 75 100 150 139 160 200 250 300 350 340 400 450 500 490 Region depicted below 250 (b) DNA Size 200 165. 05 bp 160 150 147. 32 bp 139 DNA fragment peaks are sized based on the sizing curve produced from the points on the internal size standard 100 Time (minutes) DNA fragment peaks in sample Data Point
Peak Sizing with an Internal Size Standard John M. Butler (2009) Fundamentals of Forensic DNA Typing, Figure 10. 3 (a) 35 50 75 100 150 139 160 200 250 300 350 340 400 450 500 490 Region depicted below 250 (b) DNA Size 200 165. 05 bp 160 150 147. 32 bp 139 DNA fragment peaks are sized based on the sizing curve produced from the points on the internal size standard 100 Time (minutes) DNA fragment peaks in sample Data Point
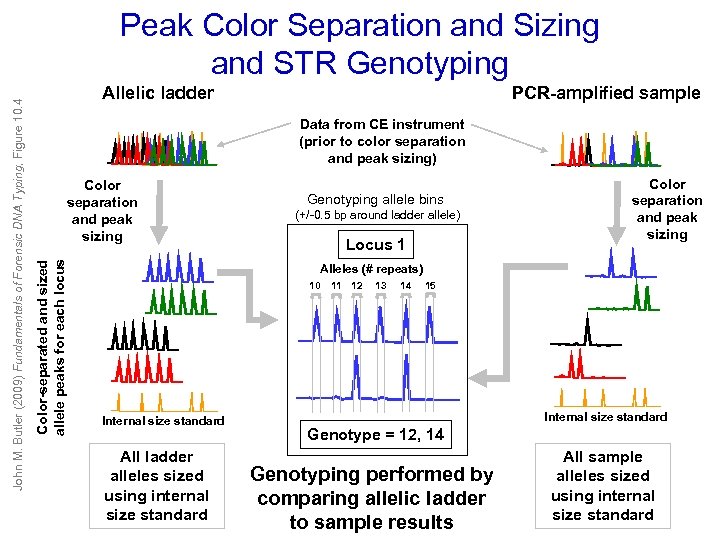 Allelic ladder PCR-amplified sample Data from CE instrument (prior to color separation and peak sizing) Color separation and peak sizing Color-separated and sized allele peaks for each locus John M. Butler (2009) Fundamentals of Forensic DNA Typing, Figure 10. 4 Peak Color Separation and Sizing and STR Genotyping allele bins (+/-0. 5 bp around ladder allele) Locus 1 Color separation and peak sizing Alleles (# repeats) 10 Internal size standard All ladder alleles sized using internal size standard 11 12 13 14 15 Internal size standard Genotype = 12, 14 Genotyping performed by comparing allelic ladder to sample results All sample alleles sized using internal size standard
Allelic ladder PCR-amplified sample Data from CE instrument (prior to color separation and peak sizing) Color separation and peak sizing Color-separated and sized allele peaks for each locus John M. Butler (2009) Fundamentals of Forensic DNA Typing, Figure 10. 4 Peak Color Separation and Sizing and STR Genotyping allele bins (+/-0. 5 bp around ladder allele) Locus 1 Color separation and peak sizing Alleles (# repeats) 10 Internal size standard All ladder alleles sized using internal size standard 11 12 13 14 15 Internal size standard Genotype = 12, 14 Genotyping performed by comparing allelic ladder to sample results All sample alleles sized using internal size standard
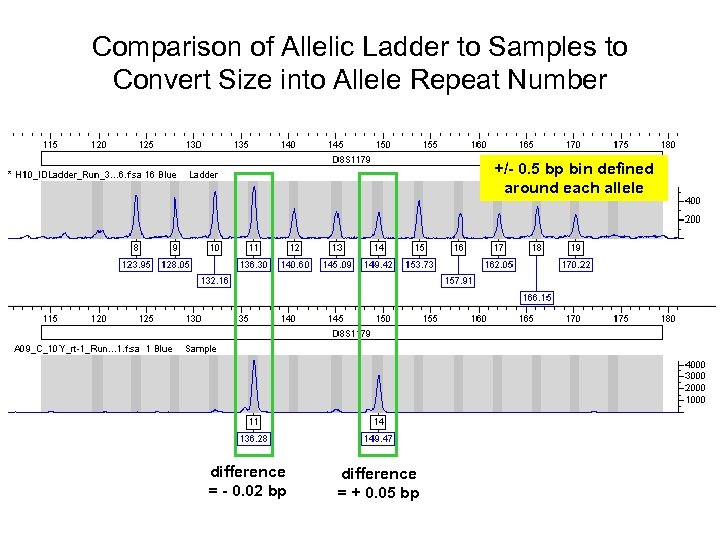 Comparison of Allelic Ladder to Samples to Convert Size into Allele Repeat Number +/- 0. 5 bp bin defined around each allele difference = - 0. 02 bp difference = + 0. 05 bp
Comparison of Allelic Ladder to Samples to Convert Size into Allele Repeat Number +/- 0. 5 bp bin defined around each allele difference = - 0. 02 bp difference = + 0. 05 bp
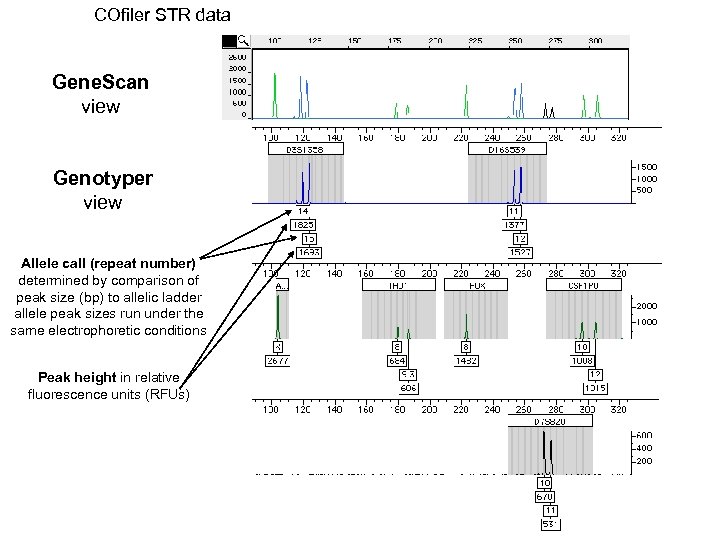 COfiler STR data Gene. Scan view Genotyper view Allele call (repeat number) determined by comparison of peak size (bp) to allelic ladder allele peak sizes run under the same electrophoretic conditions Peak height in relative fluorescence units (RFUs)
COfiler STR data Gene. Scan view Genotyper view Allele call (repeat number) determined by comparison of peak size (bp) to allelic ladder allele peak sizes run under the same electrophoretic conditions Peak height in relative fluorescence units (RFUs)
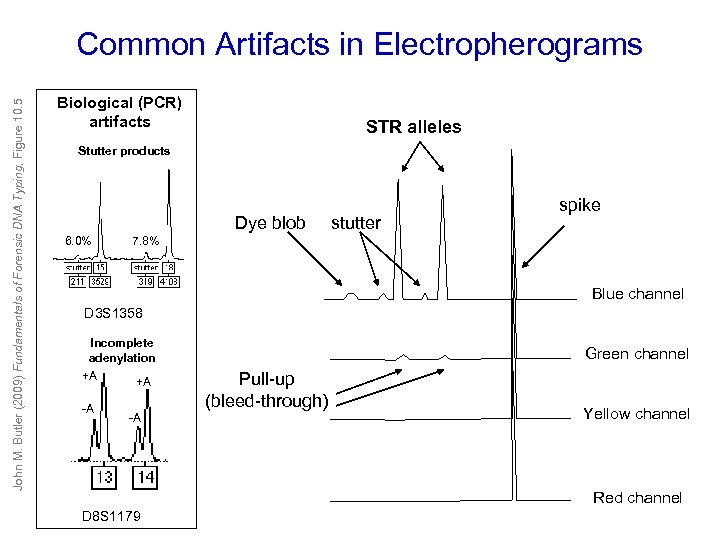 John M. Butler (2009) Fundamentals of Forensic DNA Typing, Figure 10. 5 Common Artifacts in Electropherograms Biological (PCR) artifacts STR alleles Stutter products Dye blob 6. 0% stutter spike 7. 8% Blue channel D 3 S 1358 Incomplete adenylation +A +A -A -A Green channel Pull-up (bleed-through) Yellow channel Red channel D 8 S 1179
John M. Butler (2009) Fundamentals of Forensic DNA Typing, Figure 10. 5 Common Artifacts in Electropherograms Biological (PCR) artifacts STR alleles Stutter products Dye blob 6. 0% stutter spike 7. 8% Blue channel D 3 S 1358 Incomplete adenylation +A +A -A -A Green channel Pull-up (bleed-through) Yellow channel Red channel D 8 S 1179
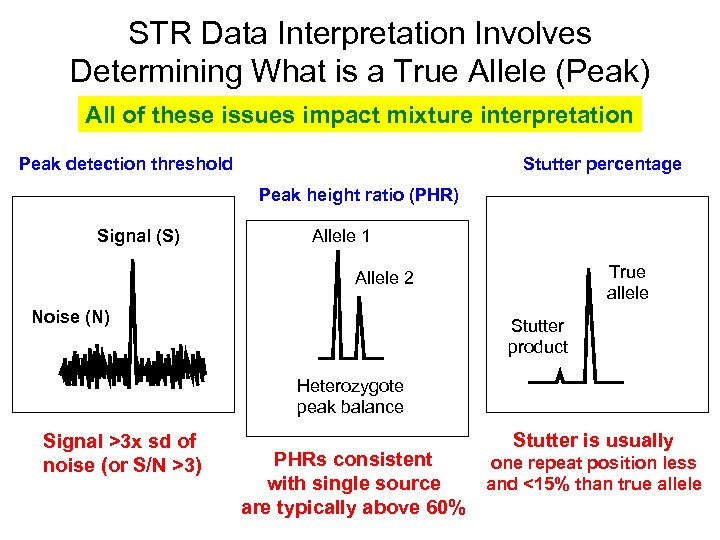 STR Data Interpretation Involves Determining What is a True Allele (Peak) All of these issues impact mixture interpretation Peak detection threshold Stutter percentage Peak height ratio (PHR) Signal (S) Allele 1 True allele Allele 2 Noise (N) Stutter product Heterozygote peak balance Signal >3 x sd of noise (or S/N >3) PHRs consistent with single source are typically above 60% Stutter is usually one repeat position less and <15% than true allele
STR Data Interpretation Involves Determining What is a True Allele (Peak) All of these issues impact mixture interpretation Peak detection threshold Stutter percentage Peak height ratio (PHR) Signal (S) Allele 1 True allele Allele 2 Noise (N) Stutter product Heterozygote peak balance Signal >3 x sd of noise (or S/N >3) PHRs consistent with single source are typically above 60% Stutter is usually one repeat position less and <15% than true allele
 Stutter Products • Peaks that show up primarily one repeat less than the true allele as a result of strand slippage during DNA synthesis • Stutter is less pronounced with larger repeat unit sizes (dinucleotides > tri- > tetra- > penta-) • Longer repeat regions generate more stutter • Each successive stutter product is less intense (allele > repeat-1 > repeat-2) • Stutter peaks make mixture analysis more difficult
Stutter Products • Peaks that show up primarily one repeat less than the true allele as a result of strand slippage during DNA synthesis • Stutter is less pronounced with larger repeat unit sizes (dinucleotides > tri- > tetra- > penta-) • Longer repeat regions generate more stutter • Each successive stutter product is less intense (allele > repeat-1 > repeat-2) • Stutter peaks make mixture analysis more difficult
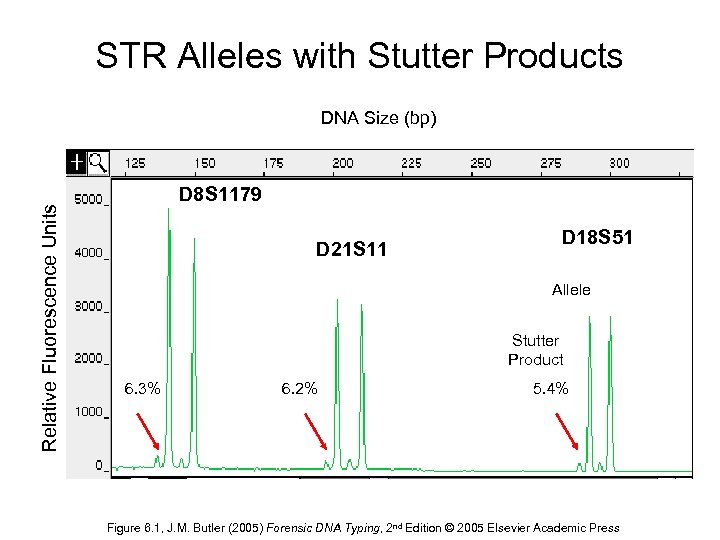 STR Alleles with Stutter Products Relative Fluorescence Units DNA Size (bp) D 8 S 1179 D 21 S 11 D 18 S 51 Allele Stutter Product 6. 3% 6. 2% 5. 4% Figure 6. 1, J. M. Butler (2005) Forensic DNA Typing, 2 nd Edition © 2005 Elsevier Academic Press
STR Alleles with Stutter Products Relative Fluorescence Units DNA Size (bp) D 8 S 1179 D 21 S 11 D 18 S 51 Allele Stutter Product 6. 3% 6. 2% 5. 4% Figure 6. 1, J. M. Butler (2005) Forensic DNA Typing, 2 nd Edition © 2005 Elsevier Academic Press
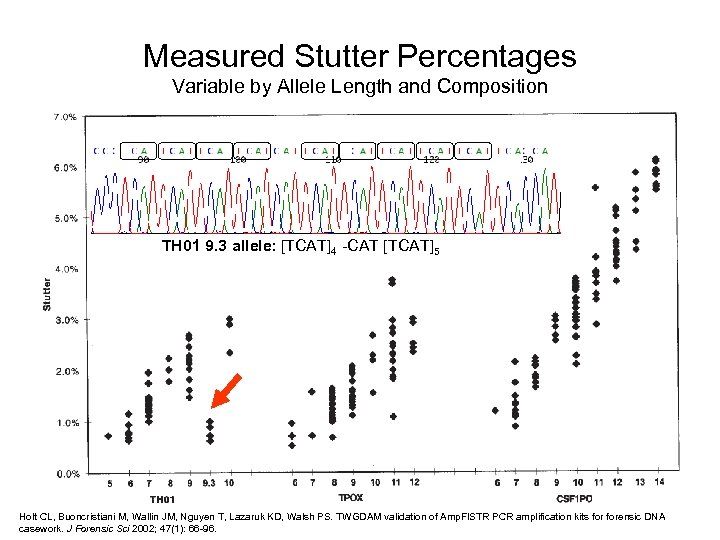 Measured Stutter Percentages Variable by Allele Length and Composition TH 01 9. 3 allele: [TCAT]4 -CAT [TCAT]5 Holt CL, Buoncristiani M, Wallin JM, Nguyen T, Lazaruk KD, Walsh PS. TWGDAM validation of Amp. Fl. STR PCR amplification kits forensic DNA casework. J Forensic Sci 2002; 47(1): 66 -96.
Measured Stutter Percentages Variable by Allele Length and Composition TH 01 9. 3 allele: [TCAT]4 -CAT [TCAT]5 Holt CL, Buoncristiani M, Wallin JM, Nguyen T, Lazaruk KD, Walsh PS. TWGDAM validation of Amp. Fl. STR PCR amplification kits forensic DNA casework. J Forensic Sci 2002; 47(1): 66 -96.
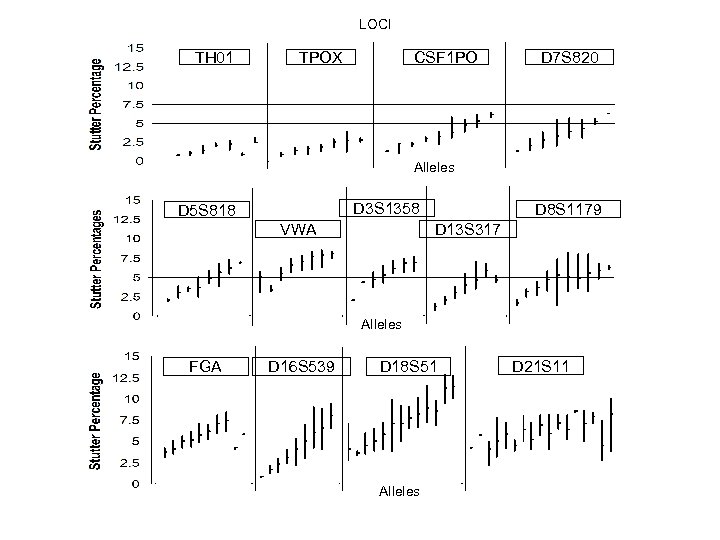 LOCI TH 01 TPOX CSF 1 PO D 7 S 820 Alleles D 3 S 1358 D 5 S 818 VWA D 8 S 1179 D 13 S 317 Alleles FGA D 16 S 539 D 18 S 51 Alleles D 21 S 11
LOCI TH 01 TPOX CSF 1 PO D 7 S 820 Alleles D 3 S 1358 D 5 S 818 VWA D 8 S 1179 D 13 S 317 Alleles FGA D 16 S 539 D 18 S 51 Alleles D 21 S 11
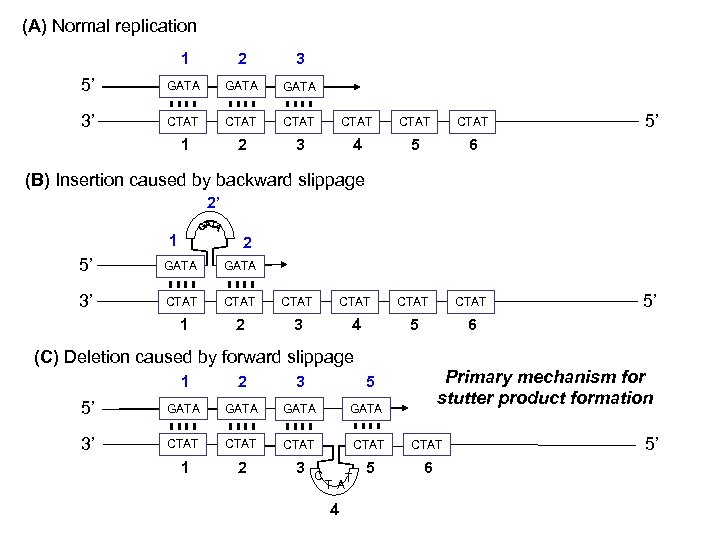 (A) Normal replication 1 2 3 5’ GATA 3’ CTAT CTAT 1 2 3 4 5 6 5’ (B) Insertion caused by backward slippage 2’ 1 2 5’ GATA 3’ CTAT CTAT 1 2 3 4 5 6 (C) Deletion caused by forward slippage Primary mechanism for stutter product formation 1 2 3 5 5’ GATA 3’ CTAT CTAT 1 2 3 5 6 C TA 4 T 5’ 5’
(A) Normal replication 1 2 3 5’ GATA 3’ CTAT CTAT 1 2 3 4 5 6 5’ (B) Insertion caused by backward slippage 2’ 1 2 5’ GATA 3’ CTAT CTAT 1 2 3 4 5 6 (C) Deletion caused by forward slippage Primary mechanism for stutter product formation 1 2 3 5 5’ GATA 3’ CTAT CTAT 1 2 3 5 6 C TA 4 T 5’ 5’
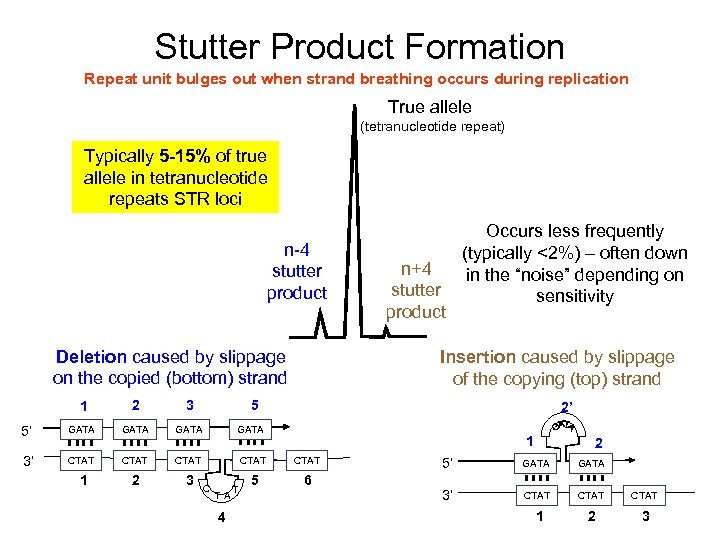 Stutter Product Formation Repeat unit bulges out when strand breathing occurs during replication True allele (tetranucleotide repeat) Typically 5 -15% of true allele in tetranucleotide repeats STR loci n-4 stutter product Deletion caused by slippage on the copied (bottom) strand Occurs less frequently (typically <2%) – often down n+4 in the “noise” depending on stutter sensitivity product Insertion caused by slippage of the copying (top) strand 1 2 3 5 5’ GATA 3’ CTAT CTAT 1 2 3 5 6 C T A 4 T 2’ 1 2 5’ GATA 3’ CTAT 1 2 3
Stutter Product Formation Repeat unit bulges out when strand breathing occurs during replication True allele (tetranucleotide repeat) Typically 5 -15% of true allele in tetranucleotide repeats STR loci n-4 stutter product Deletion caused by slippage on the copied (bottom) strand Occurs less frequently (typically <2%) – often down n+4 in the “noise” depending on stutter sensitivity product Insertion caused by slippage of the copying (top) strand 1 2 3 5 5’ GATA 3’ CTAT CTAT 1 2 3 5 6 C T A 4 T 2’ 1 2 5’ GATA 3’ CTAT 1 2 3
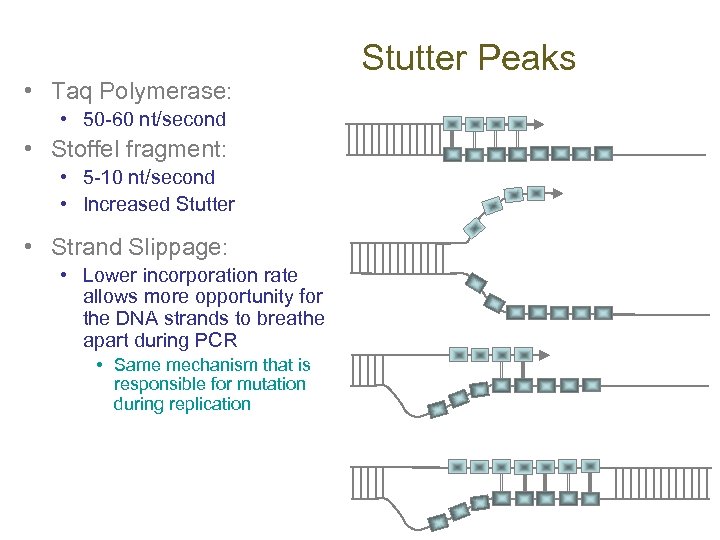 • Taq Polymerase: • 50 -60 nt/second • Stoffel fragment: • 5 -10 nt/second • Increased Stutter • Strand Slippage: • Lower incorporation rate allows more opportunity for the DNA strands to breathe apart during PCR • Same mechanism that is responsible for mutation during replication Stutter Peaks
• Taq Polymerase: • 50 -60 nt/second • Stoffel fragment: • 5 -10 nt/second • Increased Stutter • Strand Slippage: • Lower incorporation rate allows more opportunity for the DNA strands to breathe apart during PCR • Same mechanism that is responsible for mutation during replication Stutter Peaks
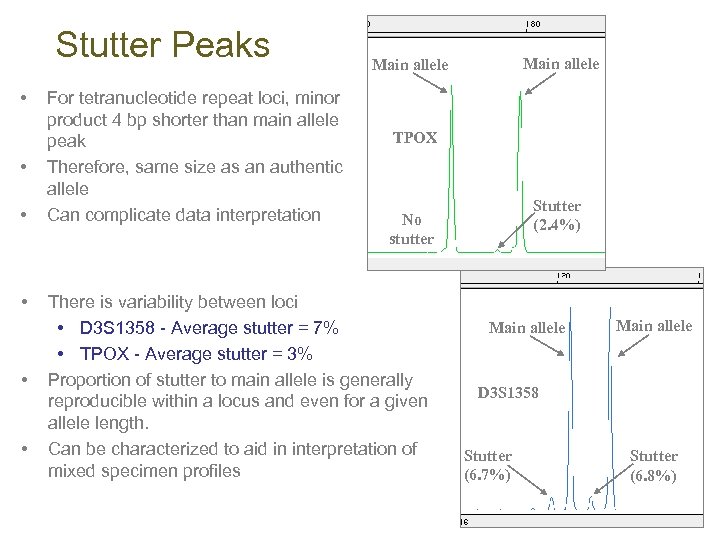 Stutter Peaks • • • For tetranucleotide repeat loci, minor product 4 bp shorter than main allele peak Therefore, same size as an authentic allele Can complicate data interpretation Main allele TPOX Stutter (2. 4%) No stutter There is variability between loci • D 3 S 1358 - Average stutter = 7% • TPOX - Average stutter = 3% Proportion of stutter to main allele is generally reproducible within a locus and even for a given allele length. Can be characterized to aid in interpretation of mixed specimen profiles Main allele D 3 S 1358 Stutter (6. 7%) Stutter (6. 8%)
Stutter Peaks • • • For tetranucleotide repeat loci, minor product 4 bp shorter than main allele peak Therefore, same size as an authentic allele Can complicate data interpretation Main allele TPOX Stutter (2. 4%) No stutter There is variability between loci • D 3 S 1358 - Average stutter = 7% • TPOX - Average stutter = 3% Proportion of stutter to main allele is generally reproducible within a locus and even for a given allele length. Can be characterized to aid in interpretation of mixed specimen profiles Main allele D 3 S 1358 Stutter (6. 7%) Stutter (6. 8%)
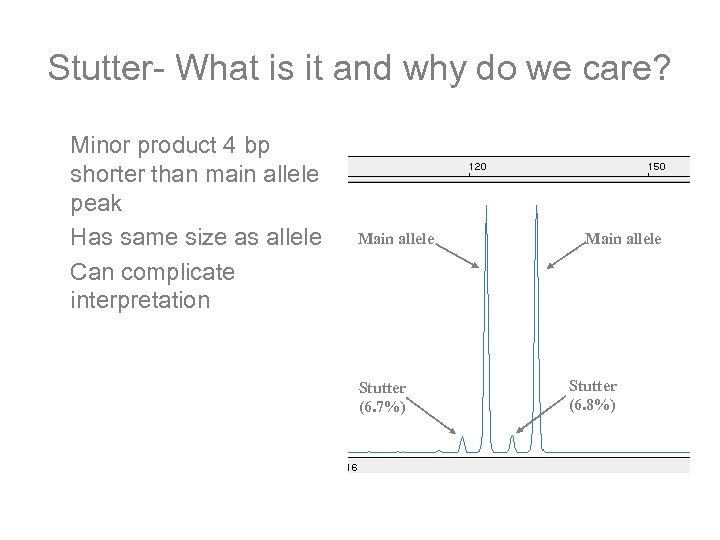 Stutter- What is it and why do we care? Minor product 4 bp shorter than main allele peak Has same size as allele Can complicate interpretation Main allele Stutter (6. 7%) Main allele Stutter (6. 8%)
Stutter- What is it and why do we care? Minor product 4 bp shorter than main allele peak Has same size as allele Can complicate interpretation Main allele Stutter (6. 7%) Main allele Stutter (6. 8%)
 What do we know about stutter peaks? • There is variability between loci – v. WA - Average stutter = 7% – TPOX - Average stutter = 3% • Proportion of stutter to main allele is generally reproducible within a locus and even for a given allele length. • Can be characterized to aid in interpretation of mixed specimen profiles
What do we know about stutter peaks? • There is variability between loci – v. WA - Average stutter = 7% – TPOX - Average stutter = 3% • Proportion of stutter to main allele is generally reproducible within a locus and even for a given allele length. • Can be characterized to aid in interpretation of mixed specimen profiles
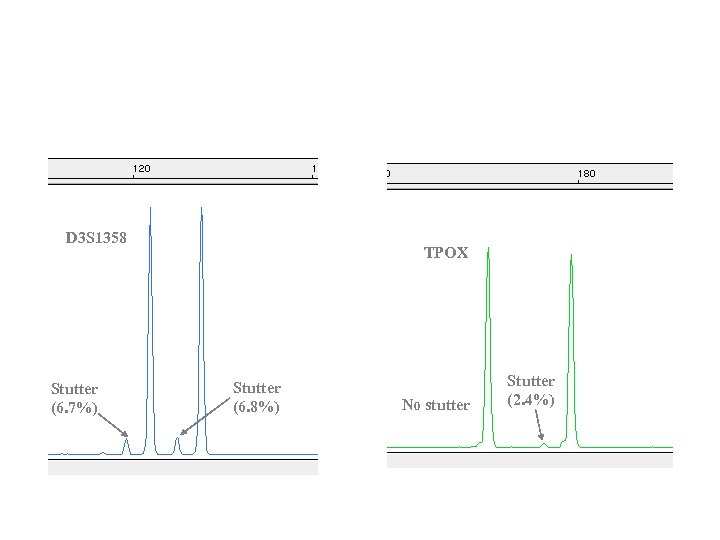 D 3 S 1358 Stutter (6. 7%) TPOX Stutter (6. 8%) No stutter Stutter (2. 4%)
D 3 S 1358 Stutter (6. 7%) TPOX Stutter (6. 8%) No stutter Stutter (2. 4%)
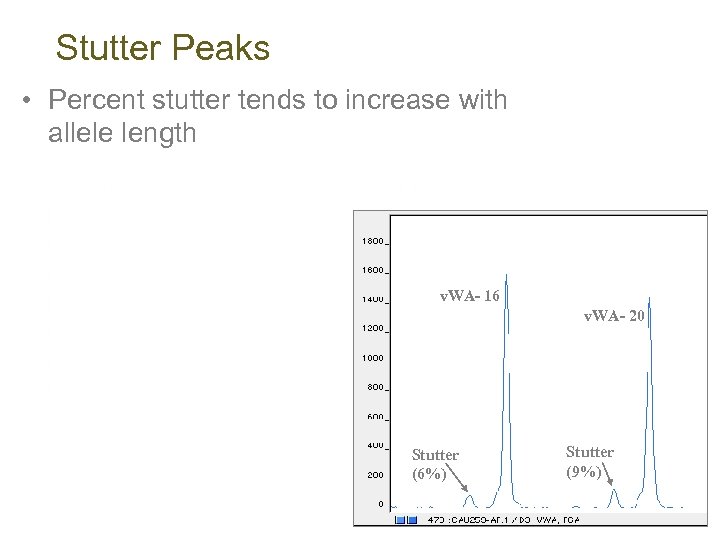 Stutter Peaks • Percent stutter tends to increase with allele length v. WA- 16 v. WA- 20 Stutter (6%) Stutter (9%)
Stutter Peaks • Percent stutter tends to increase with allele length v. WA- 16 v. WA- 20 Stutter (6%) Stutter (9%)
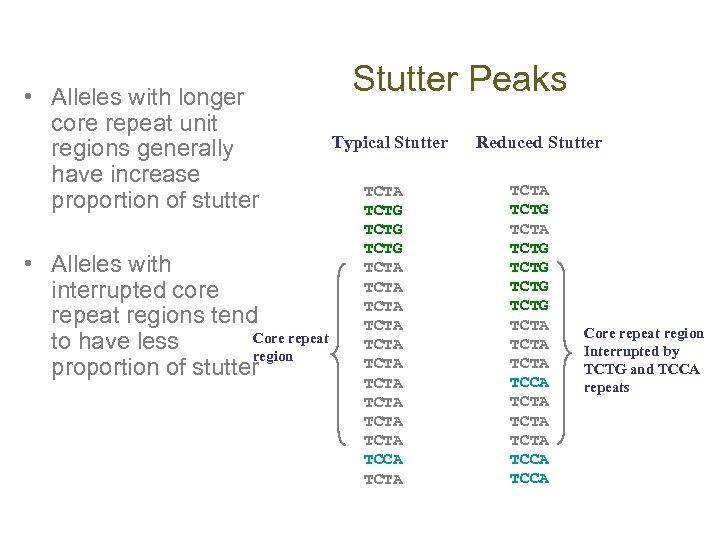 • Alleles with longer core repeat unit regions generally have increase proportion of stutter • Alleles with interrupted core repeat regions tend Core repeat to have less region proportion of stutter Stutter Peaks Typical Stutter TCTA TCTG TCTA TCTA TCTA TCCA TCTA Reduced Stutter TCTA TCTG TCTG TCTA TCTA TCCA Core repeat region Interrupted by TCTG and TCCA repeats
• Alleles with longer core repeat unit regions generally have increase proportion of stutter • Alleles with interrupted core repeat regions tend Core repeat to have less region proportion of stutter Stutter Peaks Typical Stutter TCTA TCTG TCTA TCTA TCTA TCCA TCTA Reduced Stutter TCTA TCTG TCTG TCTA TCTA TCCA Core repeat region Interrupted by TCTG and TCCA repeats
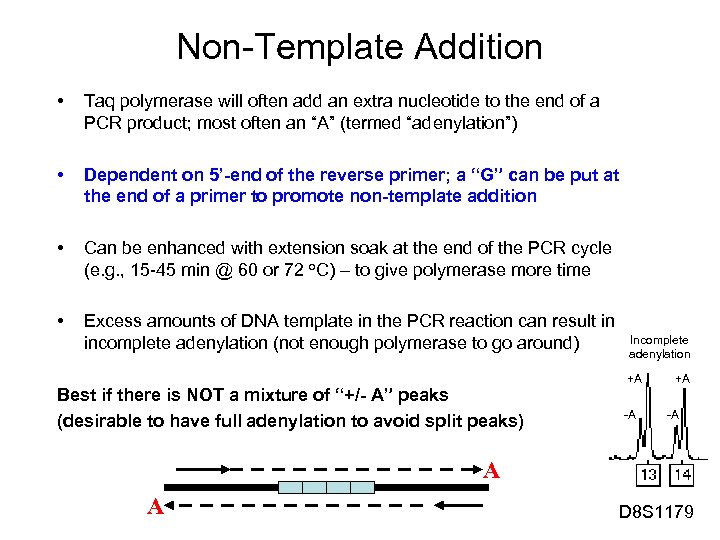 Non-Template Addition • Taq polymerase will often add an extra nucleotide to the end of a PCR product; most often an “A” (termed “adenylation”) • Dependent on 5’-end of the reverse primer; a “G” can be put at the end of a primer to promote non-template addition • Can be enhanced with extension soak at the end of the PCR cycle (e. g. , 15 -45 min @ 60 or 72 o. C) – to give polymerase more time • Excess amounts of DNA template in the PCR reaction can result in incomplete adenylation (not enough polymerase to go around) Best if there is NOT a mixture of “+/- A” peaks (desirable to have full adenylation to avoid split peaks) Incomplete adenylation +A -A A A D 8 S 1179
Non-Template Addition • Taq polymerase will often add an extra nucleotide to the end of a PCR product; most often an “A” (termed “adenylation”) • Dependent on 5’-end of the reverse primer; a “G” can be put at the end of a primer to promote non-template addition • Can be enhanced with extension soak at the end of the PCR cycle (e. g. , 15 -45 min @ 60 or 72 o. C) – to give polymerase more time • Excess amounts of DNA template in the PCR reaction can result in incomplete adenylation (not enough polymerase to go around) Best if there is NOT a mixture of “+/- A” peaks (desirable to have full adenylation to avoid split peaks) Incomplete adenylation +A -A A A D 8 S 1179
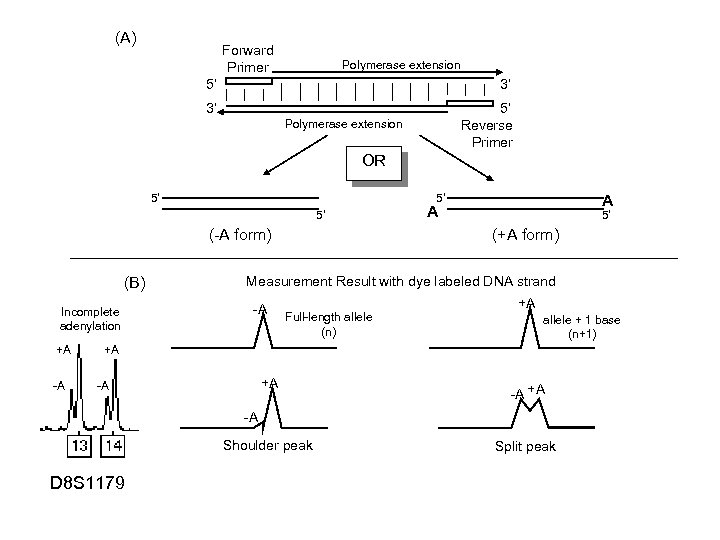 (A) Forward Primer Polymerase extension 5’ 3’ 3’ 5’ Reverse Primer Polymerase extension OR 5’ 5’ 5’ (-A form) (B) Incomplete adenylation +A A A 5’ (+A form) Measurement Result with dye labeled DNA strand -A Full-length allele (n) +A allele + 1 base (n+1) +A +A -A +A Shoulder peak -A Split peak -A -A D 8 S 1179
(A) Forward Primer Polymerase extension 5’ 3’ 3’ 5’ Reverse Primer Polymerase extension OR 5’ 5’ 5’ (-A form) (B) Incomplete adenylation +A A A 5’ (+A form) Measurement Result with dye labeled DNA strand -A Full-length allele (n) +A allele + 1 base (n+1) +A +A -A +A Shoulder peak -A Split peak -A -A D 8 S 1179
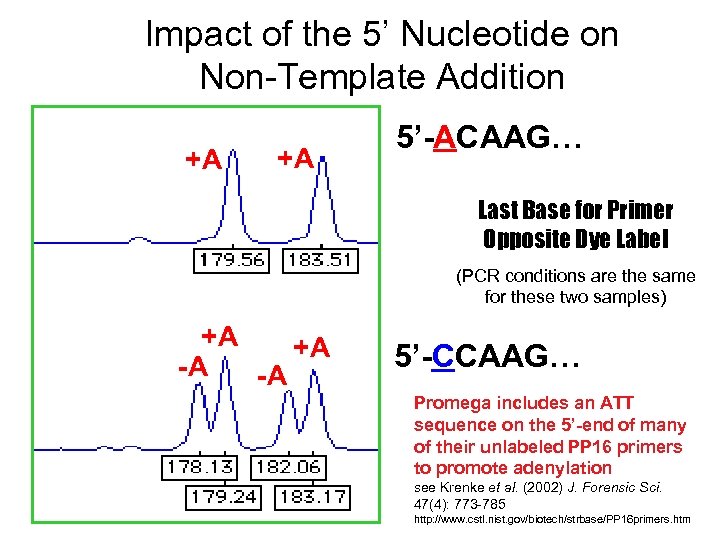 Impact of the 5’ Nucleotide on Non-Template Addition +A +A 5’-ACAAG… Last Base for Primer Opposite Dye Label (PCR conditions are the same for these two samples) +A +A -A -A 5’-CCAAG… Promega includes an ATT sequence on the 5’-end of many of their unlabeled PP 16 primers to promote adenylation see Krenke et al. (2002) J. Forensic Sci. 47(4): 773 -785 http: //www. cstl. nist. gov/biotech/strbase/PP 16 primers. htm
Impact of the 5’ Nucleotide on Non-Template Addition +A +A 5’-ACAAG… Last Base for Primer Opposite Dye Label (PCR conditions are the same for these two samples) +A +A -A -A 5’-CCAAG… Promega includes an ATT sequence on the 5’-end of many of their unlabeled PP 16 primers to promote adenylation see Krenke et al. (2002) J. Forensic Sci. 47(4): 773 -785 http: //www. cstl. nist. gov/biotech/strbase/PP 16 primers. htm
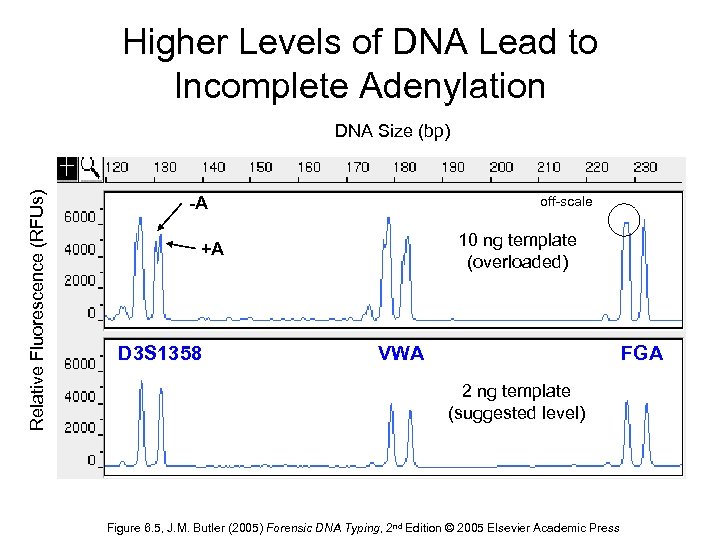 Higher Levels of DNA Lead to Incomplete Adenylation Relative Fluorescence (RFUs) DNA Size (bp) -A off-scale 10 ng template (overloaded) +A D 3 S 1358 VWA FGA 2 ng template (suggested level) Figure 6. 5, J. M. Butler (2005) Forensic DNA Typing, 2 nd Edition © 2005 Elsevier Academic Press
Higher Levels of DNA Lead to Incomplete Adenylation Relative Fluorescence (RFUs) DNA Size (bp) -A off-scale 10 ng template (overloaded) +A D 3 S 1358 VWA FGA 2 ng template (suggested level) Figure 6. 5, J. M. Butler (2005) Forensic DNA Typing, 2 nd Edition © 2005 Elsevier Academic Press
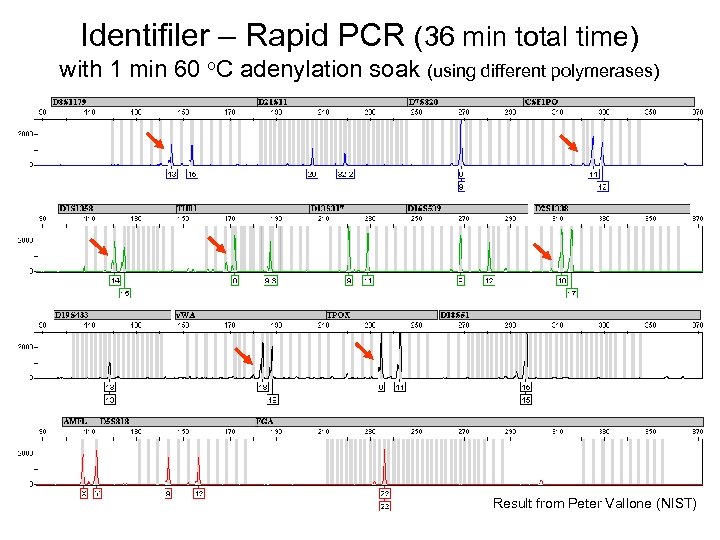 Identifiler – Rapid PCR (36 min total time) with 1 min 60 o. C adenylation soak (using different polymerases) Result from Peter Vallone (NIST)
Identifiler – Rapid PCR (36 min total time) with 1 min 60 o. C adenylation soak (using different polymerases) Result from Peter Vallone (NIST)
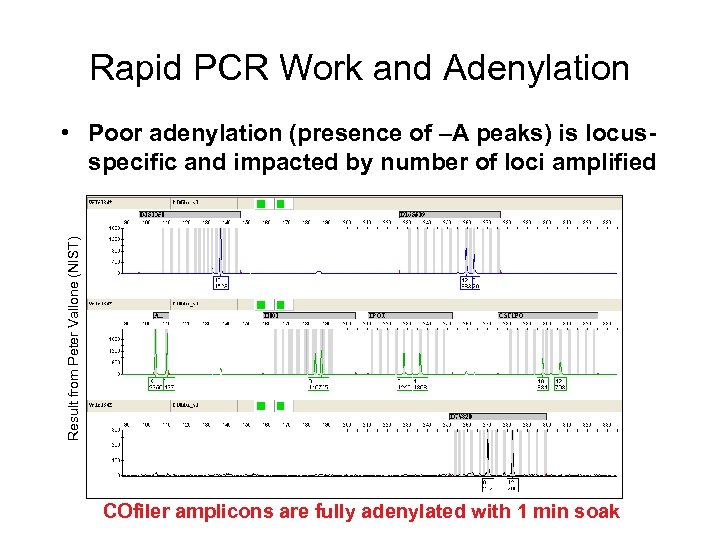 Rapid PCR Work and Adenylation Result from Peter Vallone (NIST) • Poor adenylation (presence of –A peaks) is locusspecific and impacted by number of loci amplified COfiler amplicons are fully adenylated with 1 min soak
Rapid PCR Work and Adenylation Result from Peter Vallone (NIST) • Poor adenylation (presence of –A peaks) is locusspecific and impacted by number of loci amplified COfiler amplicons are fully adenylated with 1 min soak
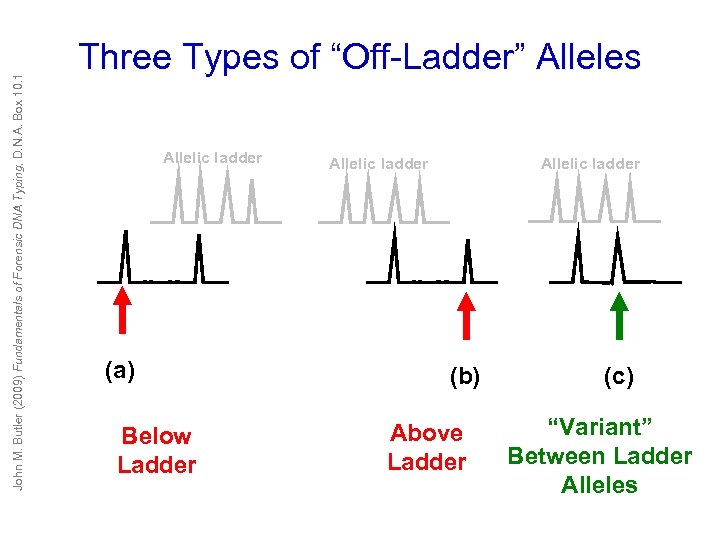 John M. Butler (2009) Fundamentals of Forensic DNA Typing, D. N. A. Box 10. 1 Three Types of “Off-Ladder” Alleles Allelic ladder (a) Below Ladder Allelic ladder (b) Above Ladder (c) “Variant” Between Ladder Alleles
John M. Butler (2009) Fundamentals of Forensic DNA Typing, D. N. A. Box 10. 1 Three Types of “Off-Ladder” Alleles Allelic ladder (a) Below Ladder Allelic ladder (b) Above Ladder (c) “Variant” Between Ladder Alleles
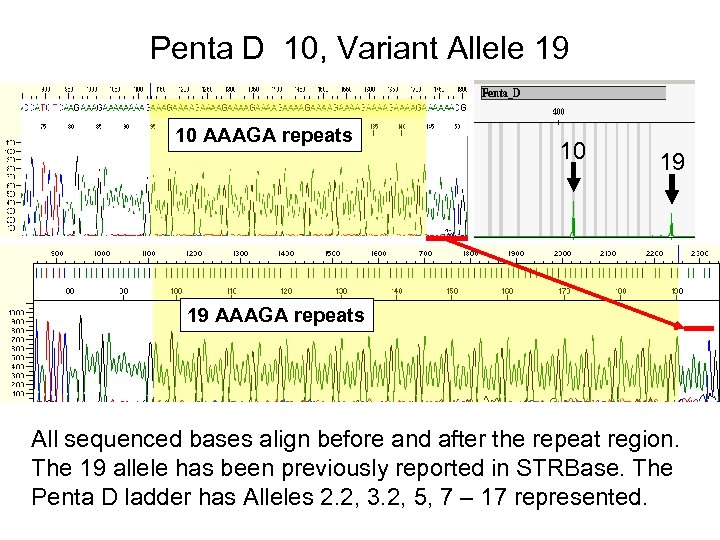 Penta D 10, Variant Allele 19 10 AAAGA repeats 10 19 19 AAAGA repeats All sequenced bases align before and after the repeat region. The 19 allele has been previously reported in STRBase. The Penta D ladder has Alleles 2. 2, 3. 2, 5, 7 – 17 represented.
Penta D 10, Variant Allele 19 10 AAAGA repeats 10 19 19 AAAGA repeats All sequenced bases align before and after the repeat region. The 19 allele has been previously reported in STRBase. The Penta D ladder has Alleles 2. 2, 3. 2, 5, 7 – 17 represented.
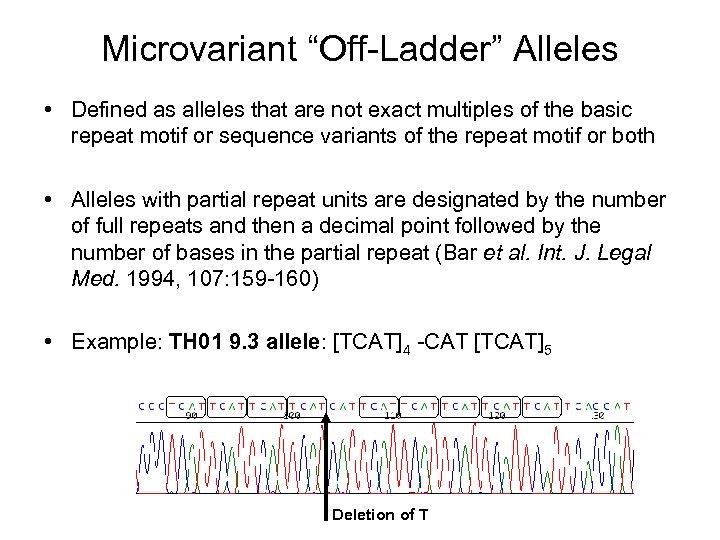 Microvariant “Off-Ladder” Alleles • Defined as alleles that are not exact multiples of the basic repeat motif or sequence variants of the repeat motif or both • Alleles with partial repeat units are designated by the number of full repeats and then a decimal point followed by the number of bases in the partial repeat (Bar et al. Int. J. Legal Med. 1994, 107: 159 -160) • Example: TH 01 9. 3 allele: [TCAT]4 -CAT [TCAT]5 Deletion of T
Microvariant “Off-Ladder” Alleles • Defined as alleles that are not exact multiples of the basic repeat motif or sequence variants of the repeat motif or both • Alleles with partial repeat units are designated by the number of full repeats and then a decimal point followed by the number of bases in the partial repeat (Bar et al. Int. J. Legal Med. 1994, 107: 159 -160) • Example: TH 01 9. 3 allele: [TCAT]4 -CAT [TCAT]5 Deletion of T
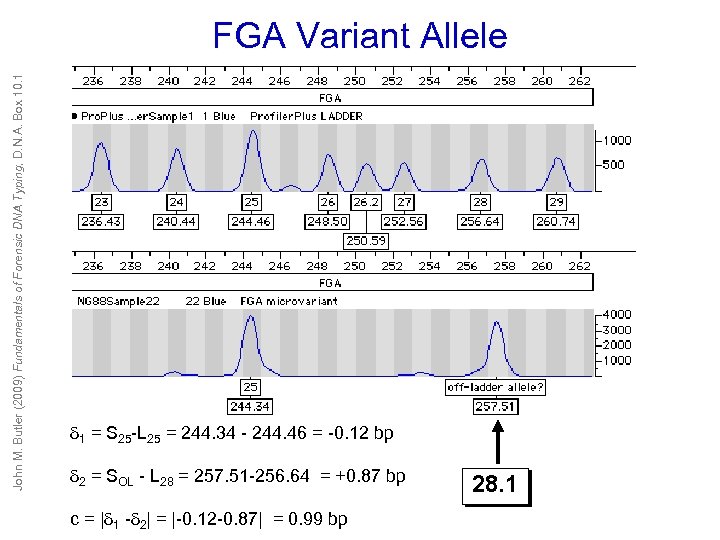 John M. Butler (2009) Fundamentals of Forensic DNA Typing, D. N. A. Box 10. 1 FGA Variant Allele 1 = S 25 -L 25 = 244. 34 - 244. 46 = -0. 12 bp 2 = SOL - L 28 = 257. 51 -256. 64 = +0. 87 bp c = | 1 - 2| = |-0. 12 -0. 87| = 0. 99 bp 28. 1
John M. Butler (2009) Fundamentals of Forensic DNA Typing, D. N. A. Box 10. 1 FGA Variant Allele 1 = S 25 -L 25 = 244. 34 - 244. 46 = -0. 12 bp 2 = SOL - L 28 = 257. 51 -256. 64 = +0. 87 bp c = | 1 - 2| = |-0. 12 -0. 87| = 0. 99 bp 28. 1
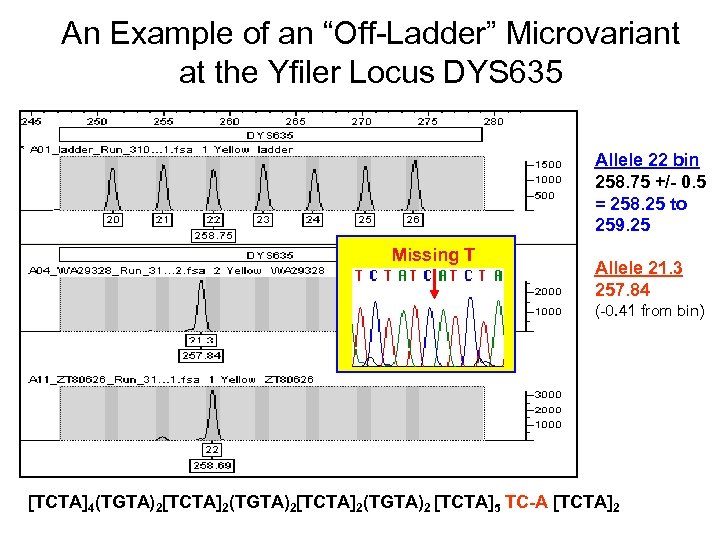 An Example of an “Off-Ladder” Microvariant at the Yfiler Locus DYS 635 Allele 22 bin 258. 75 +/- 0. 5 = 258. 25 to 259. 25 Missing T Allele 21. 3 257. 84 (-0. 41 from bin) [TCTA]4(TGTA)2[TCTA]2(TGTA)2 [TCTA]5 TC-A [TCTA]2
An Example of an “Off-Ladder” Microvariant at the Yfiler Locus DYS 635 Allele 22 bin 258. 75 +/- 0. 5 = 258. 25 to 259. 25 Missing T Allele 21. 3 257. 84 (-0. 41 from bin) [TCTA]4(TGTA)2[TCTA]2(TGTA)2 [TCTA]5 TC-A [TCTA]2
![G C A [TCTA]13 A TCTG [TCTA]11 SNPs within the D 8 S 1179 G C A [TCTA]13 A TCTG [TCTA]11 SNPs within the D 8 S 1179](https://present5.com/presentation/68ca7760cf75b19a8fbd9412490ef507/image-46.jpg) G C A [TCTA]13 A TCTG [TCTA]11 SNPs within the D 8 S 1179 repeat Repeat is TCTA Three NIST samples have genotypes 13, 13. GGA C TCTA TCTG [TCTA]11 TCTA TCTG TGTA [TCTA]10 A CG Analysis by Mass Spec indicates the presence of SNPs (Tom Hall, IBIS) Confirmation of the Mass Spec by sequencing at NIST indicates: [TCTA]2 TCTG [TCTA]10 There are 4 different 13 alleles in these 3 samples.
G C A [TCTA]13 A TCTG [TCTA]11 SNPs within the D 8 S 1179 repeat Repeat is TCTA Three NIST samples have genotypes 13, 13. GGA C TCTA TCTG [TCTA]11 TCTA TCTG TGTA [TCTA]10 A CG Analysis by Mass Spec indicates the presence of SNPs (Tom Hall, IBIS) Confirmation of the Mass Spec by sequencing at NIST indicates: [TCTA]2 TCTG [TCTA]10 There are 4 different 13 alleles in these 3 samples.
 http: //www. cstl. nist. gov/biotech/strbase STRbase has a summary of alleles that have been submitted and sequenced, if the submitting agency agrees to share the information. We require a minimum of 10 ng for the sequencing. We request copies of the electropherograms demonstrating the variant allele. The more information we have up front the better. Please have patience we will get to your samples!
http: //www. cstl. nist. gov/biotech/strbase STRbase has a summary of alleles that have been submitted and sequenced, if the submitting agency agrees to share the information. We require a minimum of 10 ng for the sequencing. We request copies of the electropherograms demonstrating the variant allele. The more information we have up front the better. Please have patience we will get to your samples!
 Sample Submissions • For those that desire more assurances of confidentiality we can have MOUs signed. • We generally re-type the samples at NIST prior to starting sequencing. • We may run a monoplex assay (single locus). • We return results as Power. Point slides. • We thank all of those agencies that have used this free service (thanks to NIJ)! • Contact Margaret Kline: margaret. kline@nist. gov
Sample Submissions • For those that desire more assurances of confidentiality we can have MOUs signed. • We generally re-type the samples at NIST prior to starting sequencing. • We may run a monoplex assay (single locus). • We return results as Power. Point slides. • We thank all of those agencies that have used this free service (thanks to NIJ)! • Contact Margaret Kline: margaret. kline@nist. gov
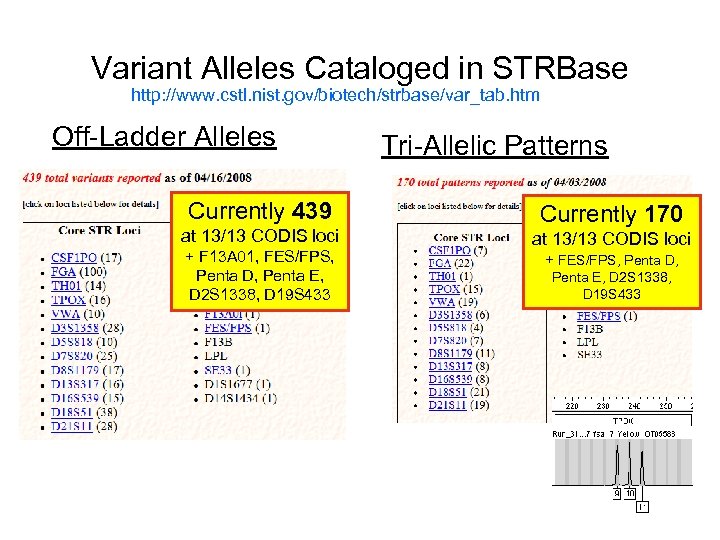 Variant Alleles Cataloged in STRBase http: //www. cstl. nist. gov/biotech/strbase/var_tab. htm Off-Ladder Alleles Currently 439 at 13/13 CODIS loci + F 13 A 01, FES/FPS, Penta D, Penta E, D 2 S 1338, D 19 S 433 Tri-Allelic Patterns Currently 170 at 13/13 CODIS loci + FES/FPS, Penta D, Penta E, D 2 S 1338, D 19 S 433
Variant Alleles Cataloged in STRBase http: //www. cstl. nist. gov/biotech/strbase/var_tab. htm Off-Ladder Alleles Currently 439 at 13/13 CODIS loci + F 13 A 01, FES/FPS, Penta D, Penta E, D 2 S 1338, D 19 S 433 Tri-Allelic Patterns Currently 170 at 13/13 CODIS loci + FES/FPS, Penta D, Penta E, D 2 S 1338, D 19 S 433
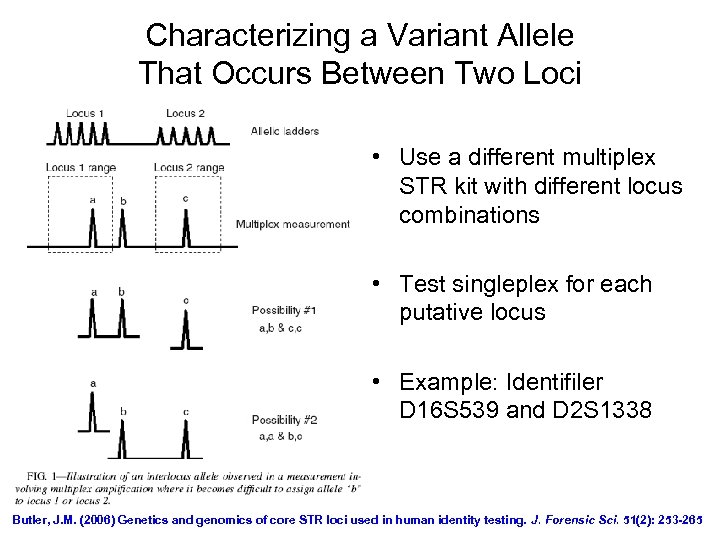 Characterizing a Variant Allele That Occurs Between Two Loci • Use a different multiplex STR kit with different locus combinations • Test singleplex for each putative locus • Example: Identifiler D 16 S 539 and D 2 S 1338 Butler, J. M. (2006) Genetics and genomics of core STR loci used in human identity testing. J. Forensic Sci. 51(2): 253 -265
Characterizing a Variant Allele That Occurs Between Two Loci • Use a different multiplex STR kit with different locus combinations • Test singleplex for each putative locus • Example: Identifiler D 16 S 539 and D 2 S 1338 Butler, J. M. (2006) Genetics and genomics of core STR loci used in human identity testing. J. Forensic Sci. 51(2): 253 -265
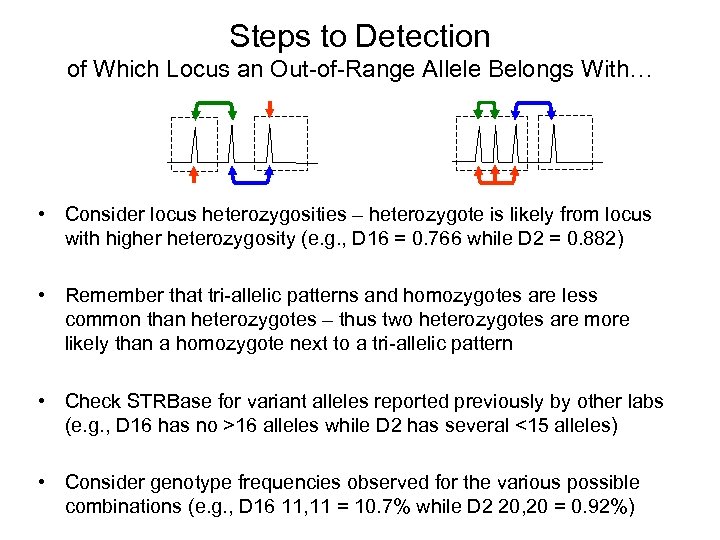 Steps to Detection of Which Locus an Out-of-Range Allele Belongs With… • Consider locus heterozygosities – heterozygote is likely from locus with higher heterozygosity (e. g. , D 16 = 0. 766 while D 2 = 0. 882) • Remember that tri-allelic patterns and homozygotes are less common than heterozygotes – thus two heterozygotes are more likely than a homozygote next to a tri-allelic pattern • Check STRBase for variant alleles reported previously by other labs (e. g. , D 16 has no >16 alleles while D 2 has several <15 alleles) • Consider genotype frequencies observed for the various possible combinations (e. g. , D 16 11, 11 = 10. 7% while D 2 20, 20 = 0. 92%)
Steps to Detection of Which Locus an Out-of-Range Allele Belongs With… • Consider locus heterozygosities – heterozygote is likely from locus with higher heterozygosity (e. g. , D 16 = 0. 766 while D 2 = 0. 882) • Remember that tri-allelic patterns and homozygotes are less common than heterozygotes – thus two heterozygotes are more likely than a homozygote next to a tri-allelic pattern • Check STRBase for variant alleles reported previously by other labs (e. g. , D 16 has no >16 alleles while D 2 has several <15 alleles) • Consider genotype frequencies observed for the various possible combinations (e. g. , D 16 11, 11 = 10. 7% while D 2 20, 20 = 0. 92%)
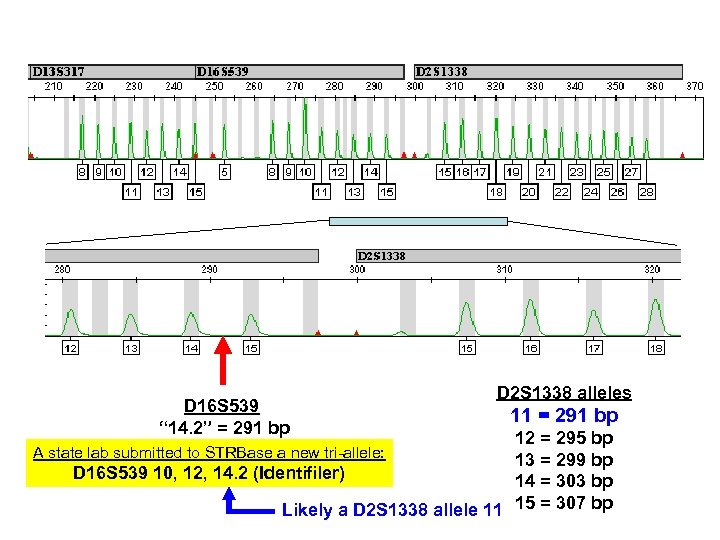 D 16 S 539 “ 14. 2” = 291 bp D 2 S 1338 alleles 11 = 291 bp 12 = 295 bp 13 = 299 bp D 16 S 539 10, 12, 14. 2 (Identifiler) 14 = 303 bp Likely a D 2 S 1338 allele 11 15 = 307 bp A state lab submitted to STRBase a new tri-allele:
D 16 S 539 “ 14. 2” = 291 bp D 2 S 1338 alleles 11 = 291 bp 12 = 295 bp 13 = 299 bp D 16 S 539 10, 12, 14. 2 (Identifiler) 14 = 303 bp Likely a D 2 S 1338 allele 11 15 = 307 bp A state lab submitted to STRBase a new tri-allele:
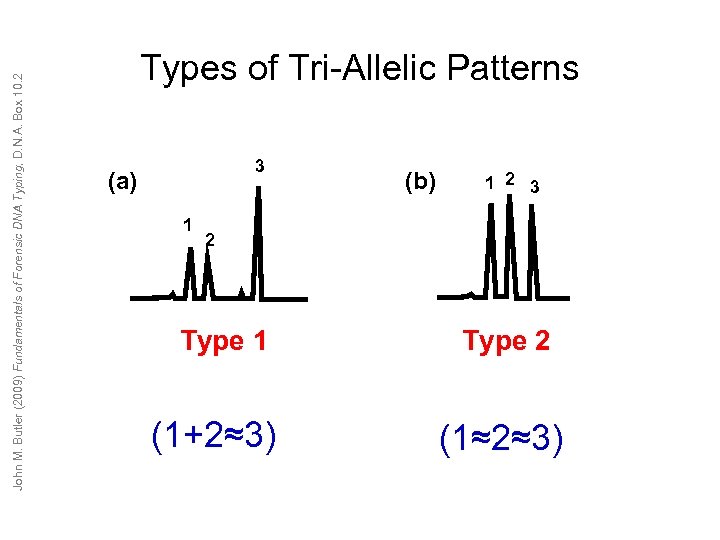 John M. Butler (2009) Fundamentals of Forensic DNA Typing, D. N. A. Box 10. 2 Types of Tri-Allelic Patterns (a) 3 1 Type 1 (1+2≈3) (b) 1 2 3 2 Type 2 (1≈2≈3)
John M. Butler (2009) Fundamentals of Forensic DNA Typing, D. N. A. Box 10. 2 Types of Tri-Allelic Patterns (a) 3 1 Type 1 (1+2≈3) (b) 1 2 3 2 Type 2 (1≈2≈3)
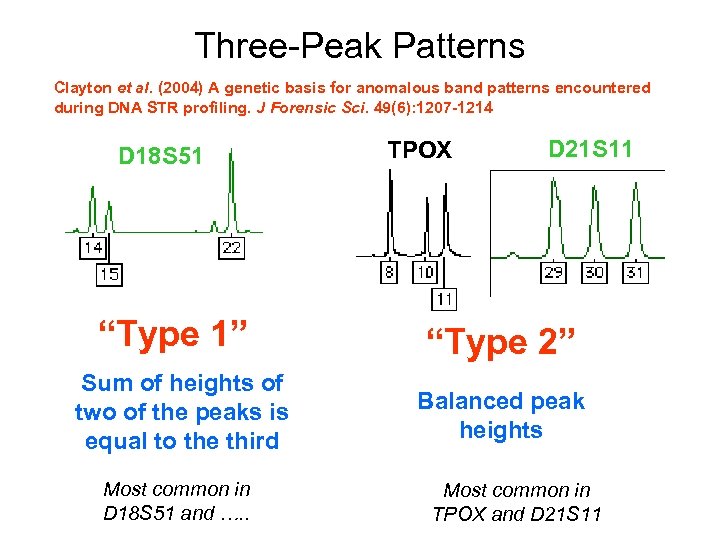 Three-Peak Patterns Clayton et al. (2004) A genetic basis for anomalous band patterns encountered during DNA STR profiling. J Forensic Sci. 49(6): 1207 -1214 D 18 S 51 “Type 1” Sum of heights of two of the peaks is equal to the third Most common in D 18 S 51 and …. . TPOX D 21 S 11 “Type 2” Balanced peak heights Most common in TPOX and D 21 S 11
Three-Peak Patterns Clayton et al. (2004) A genetic basis for anomalous band patterns encountered during DNA STR profiling. J Forensic Sci. 49(6): 1207 -1214 D 18 S 51 “Type 1” Sum of heights of two of the peaks is equal to the third Most common in D 18 S 51 and …. . TPOX D 21 S 11 “Type 2” Balanced peak heights Most common in TPOX and D 21 S 11
![Three Banded Patterns: FGA 20, 25, 26 Alleles [TTTC]3 TTTT TTCT [CTTT]12 CTCC [TTCC]2 Three Banded Patterns: FGA 20, 25, 26 Alleles [TTTC]3 TTTT TTCT [CTTT]12 CTCC [TTCC]2](https://present5.com/presentation/68ca7760cf75b19a8fbd9412490ef507/image-55.jpg) Three Banded Patterns: FGA 20, 25, 26 Alleles [TTTC]3 TTTT TTCT [CTTT]12 CTCC [TTCC]2 20 repeats [TTTC]3 TTTT TTCT [CTTT]17 CTCC [TTCC]2 25 repeats [TTTC]3 TTTT TTCT [CTTT]18 CTCC [TTCC]2 26 repeats This particular tri-allelic pattern has not been reported in STRBase
Three Banded Patterns: FGA 20, 25, 26 Alleles [TTTC]3 TTTT TTCT [CTTT]12 CTCC [TTCC]2 20 repeats [TTTC]3 TTTT TTCT [CTTT]17 CTCC [TTCC]2 25 repeats [TTTC]3 TTTT TTCT [CTTT]18 CTCC [TTCC]2 26 repeats This particular tri-allelic pattern has not been reported in STRBase
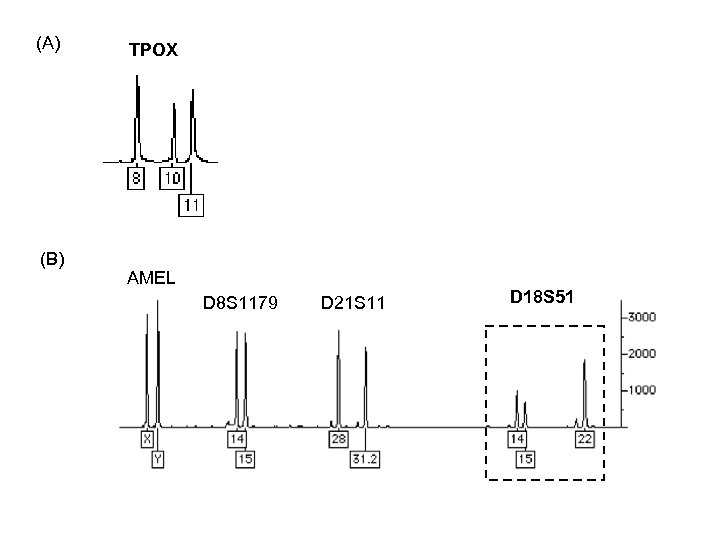 (A) (B) TPOX AMEL D 8 S 1179 D 21 S 11 D 18 S 51
(A) (B) TPOX AMEL D 8 S 1179 D 21 S 11 D 18 S 51
 TPOX Tri-Allelic Patterns FSI Genetics 2008; 2(2): 134– 137 Approximately 2. 4% of indigenous South Africans have three rather than two TPOX alleles. Data collected during routine paternity testing revealed that the extra allele is almost always allele 10 and that it segregates independently of those at the main TPOX locus. Approximately twice as many females as males have tri-allelic genotypes which suggested that the extra allele is on an X chromosome.
TPOX Tri-Allelic Patterns FSI Genetics 2008; 2(2): 134– 137 Approximately 2. 4% of indigenous South Africans have three rather than two TPOX alleles. Data collected during routine paternity testing revealed that the extra allele is almost always allele 10 and that it segregates independently of those at the main TPOX locus. Approximately twice as many females as males have tri-allelic genotypes which suggested that the extra allele is on an X chromosome.
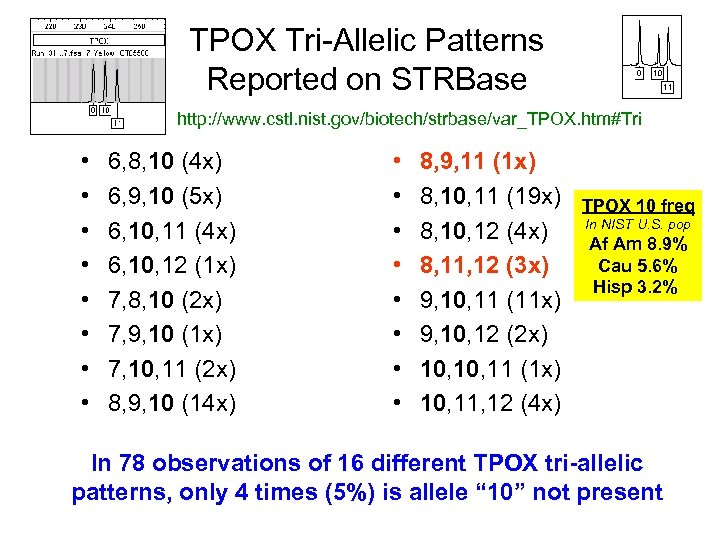 TPOX Tri-Allelic Patterns Reported on STRBase http: //www. cstl. nist. gov/biotech/strbase/var_TPOX. htm#Tri • • 6, 8, 10 (4 x) 6, 9, 10 (5 x) 6, 10, 11 (4 x) 6, 10, 12 (1 x) 7, 8, 10 (2 x) 7, 9, 10 (1 x) 7, 10, 11 (2 x) 8, 9, 10 (14 x) • • 8, 9, 11 (1 x) 8, 10, 11 (19 x) 8, 10, 12 (4 x) 8, 11, 12 (3 x) 9, 10, 11 (11 x) 9, 10, 12 (2 x) 10, 11 (1 x) 10, 11, 12 (4 x) TPOX 10 freq In NIST U. S. pop Af Am 8. 9% Cau 5. 6% Hisp 3. 2% In 78 observations of 16 different TPOX tri-allelic patterns, only 4 times (5%) is allele “ 10” not present
TPOX Tri-Allelic Patterns Reported on STRBase http: //www. cstl. nist. gov/biotech/strbase/var_TPOX. htm#Tri • • 6, 8, 10 (4 x) 6, 9, 10 (5 x) 6, 10, 11 (4 x) 6, 10, 12 (1 x) 7, 8, 10 (2 x) 7, 9, 10 (1 x) 7, 10, 11 (2 x) 8, 9, 10 (14 x) • • 8, 9, 11 (1 x) 8, 10, 11 (19 x) 8, 10, 12 (4 x) 8, 11, 12 (3 x) 9, 10, 11 (11 x) 9, 10, 12 (2 x) 10, 11 (1 x) 10, 11, 12 (4 x) TPOX 10 freq In NIST U. S. pop Af Am 8. 9% Cau 5. 6% Hisp 3. 2% In 78 observations of 16 different TPOX tri-allelic patterns, only 4 times (5%) is allele “ 10” not present
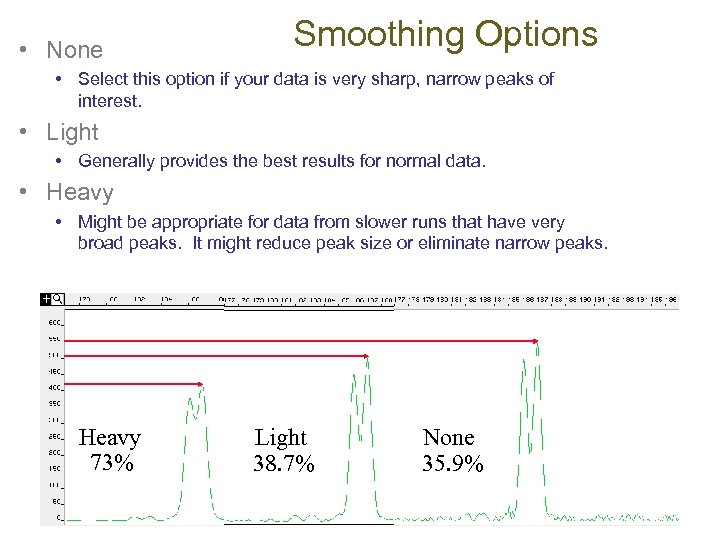 • None Smoothing Options • Select this option if your data is very sharp, narrow peaks of interest. • Light • Generally provides the best results for normal data. • Heavy • Might be appropriate for data from slower runs that have very broad peaks. It might reduce peak size or eliminate narrow peaks. Heavy 73% Light 38. 7% None 35. 9%
• None Smoothing Options • Select this option if your data is very sharp, narrow peaks of interest. • Light • Generally provides the best results for normal data. • Heavy • Might be appropriate for data from slower runs that have very broad peaks. It might reduce peak size or eliminate narrow peaks. Heavy 73% Light 38. 7% None 35. 9%
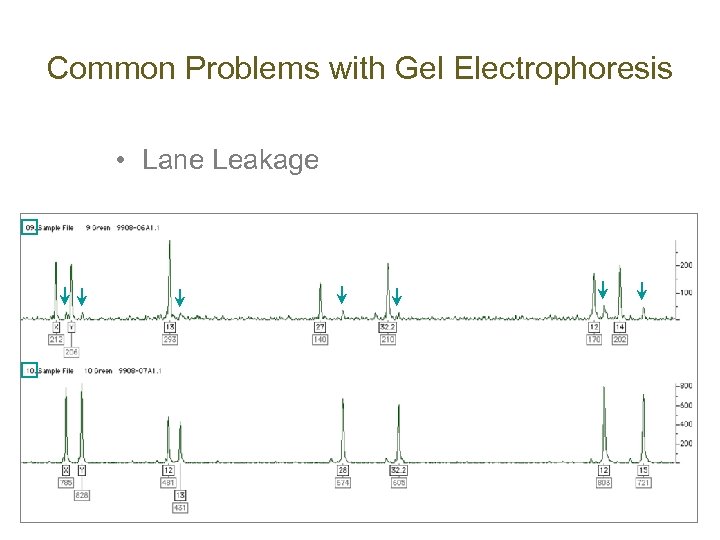 Common Problems with Gel Electrophoresis • Lane Leakage
Common Problems with Gel Electrophoresis • Lane Leakage
 Null Alleles • Allele is present in the DNA sample but fails to be amplified due to a nucleotide change in a primer binding site • Allele dropout is a problem because a heterozygous sample appears falsely as a homozygote • Two PCR primer sets can yield different results on samples originating from the same source • This phenomenon impacts DNA databases • Large concordance studies are typically performed prior to use of new STR kits For more information, see J. M. Butler (2005) Forensic DNA Typing, 2 nd Edition, pp. 133 -138
Null Alleles • Allele is present in the DNA sample but fails to be amplified due to a nucleotide change in a primer binding site • Allele dropout is a problem because a heterozygous sample appears falsely as a homozygote • Two PCR primer sets can yield different results on samples originating from the same source • This phenomenon impacts DNA databases • Large concordance studies are typically performed prior to use of new STR kits For more information, see J. M. Butler (2005) Forensic DNA Typing, 2 nd Edition, pp. 133 -138
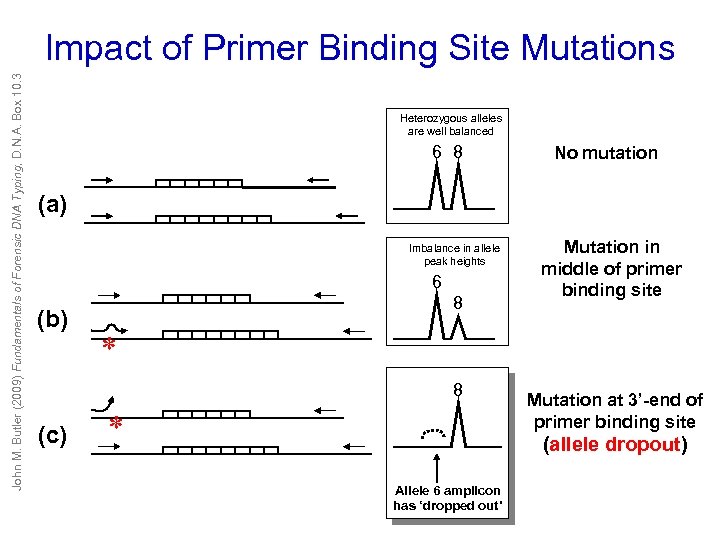 John M. Butler (2009) Fundamentals of Forensic DNA Typing, D. N. A. Box 10. 3 Impact of Primer Binding Site Mutations Heterozygous alleles are well balanced 6 8 No mutation (a) Imbalance in allele peak heights 6 (b) 8 * 8 (c) Mutation in middle of primer binding site * Mutation at 3’-end of primer binding site (allele dropout) Allele 6 amplicon has ‘dropped out’
John M. Butler (2009) Fundamentals of Forensic DNA Typing, D. N. A. Box 10. 3 Impact of Primer Binding Site Mutations Heterozygous alleles are well balanced 6 8 No mutation (a) Imbalance in allele peak heights 6 (b) 8 * 8 (c) Mutation in middle of primer binding site * Mutation at 3’-end of primer binding site (allele dropout) Allele 6 amplicon has ‘dropped out’
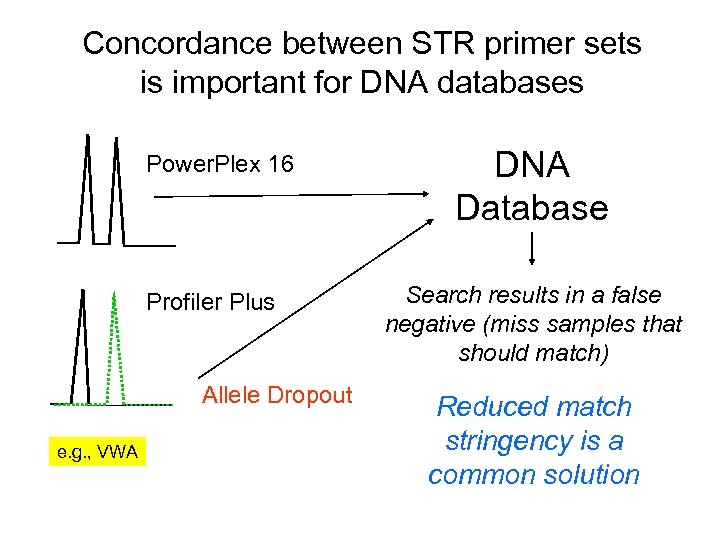 Concordance between STR primer sets is important for DNA databases Power. Plex 16 Profiler Plus Allele Dropout e. g. , VWA DNA Database Search results in a false negative (miss samples that should match) Reduced match stringency is a common solution
Concordance between STR primer sets is important for DNA databases Power. Plex 16 Profiler Plus Allele Dropout e. g. , VWA DNA Database Search results in a false negative (miss samples that should match) Reduced match stringency is a common solution
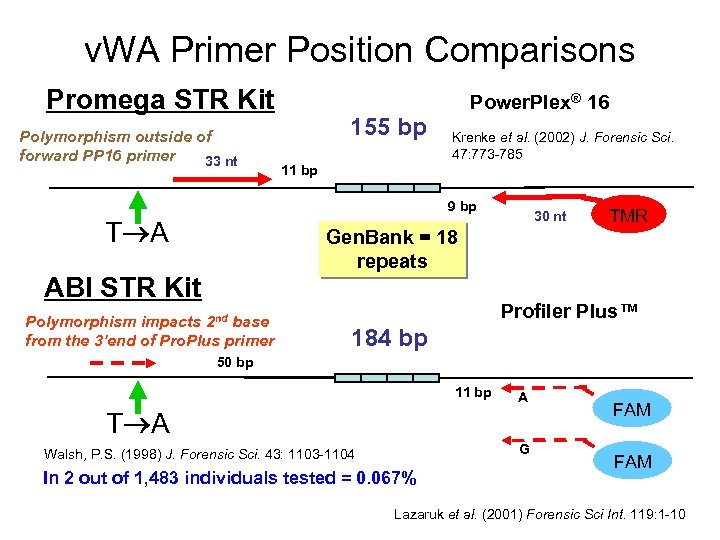 v. WA Primer Position Comparisons Promega STR Kit Polymorphism outside of forward PP 16 primer 33 nt 155 bp Power. Plex® 16 Krenke et al. (2002) J. Forensic Sci. 47: 773 -785 11 bp 9 bp T A 30 nt TMR Gen. Bank = 18 repeats ABI STR Kit Polymorphism impacts 2 nd base from the 3’end of Pro. Plus primer Profiler Plus™ 184 bp 50 bp 11 bp A T A G Walsh, P. S. (1998) J. Forensic Sci. 43: 1103 -1104 In 2 out of 1, 483 individuals tested = 0. 067% FAM Lazaruk et al. (2001) Forensic Sci Int. 119: 1 -10
v. WA Primer Position Comparisons Promega STR Kit Polymorphism outside of forward PP 16 primer 33 nt 155 bp Power. Plex® 16 Krenke et al. (2002) J. Forensic Sci. 47: 773 -785 11 bp 9 bp T A 30 nt TMR Gen. Bank = 18 repeats ABI STR Kit Polymorphism impacts 2 nd base from the 3’end of Pro. Plus primer Profiler Plus™ 184 bp 50 bp 11 bp A T A G Walsh, P. S. (1998) J. Forensic Sci. 43: 1103 -1104 In 2 out of 1, 483 individuals tested = 0. 067% FAM Lazaruk et al. (2001) Forensic Sci Int. 119: 1 -10
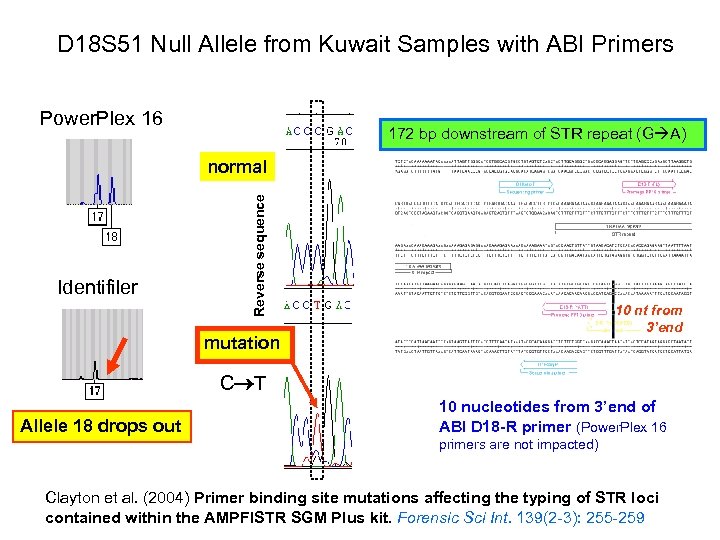 D 18 S 51 Null Allele from Kuwait Samples with ABI Primers Power. Plex 16 172 bp downstream of STR repeat (G A) Identifiler Reverse sequence normal 10 nt from 3’end mutation C T Allele 18 drops out 10 nucleotides from 3’end of ABI D 18 -R primer (Power. Plex 16 primers are not impacted) Clayton et al. (2004) Primer binding site mutations affecting the typing of STR loci contained within the AMPFl. STR SGM Plus kit. Forensic Sci Int. 139(2 -3): 255 -259
D 18 S 51 Null Allele from Kuwait Samples with ABI Primers Power. Plex 16 172 bp downstream of STR repeat (G A) Identifiler Reverse sequence normal 10 nt from 3’end mutation C T Allele 18 drops out 10 nucleotides from 3’end of ABI D 18 -R primer (Power. Plex 16 primers are not impacted) Clayton et al. (2004) Primer binding site mutations affecting the typing of STR loci contained within the AMPFl. STR SGM Plus kit. Forensic Sci Int. 139(2 -3): 255 -259
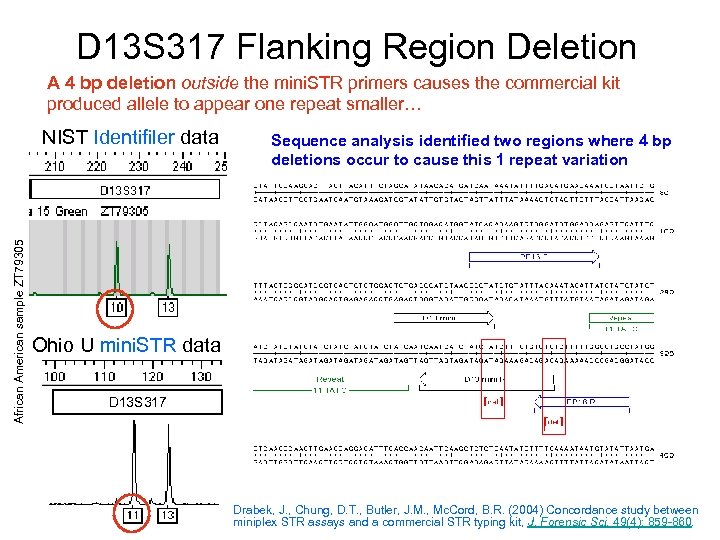 D 13 S 317 Flanking Region Deletion A 4 bp deletion outside the mini. STR primers causes the commercial kit produced allele to appear one repeat smaller… African American sample ZT 79305 NIST Identifiler data Sequence analysis identified two regions where 4 bp deletions occur to cause this 1 repeat variation Ohio U mini. STR data D 13 S 317 Drabek, J. , Chung, D. T. , Butler, J. M. , Mc. Cord, B. R. (2004) Concordance study between miniplex STR assays and a commercial STR typing kit, J. Forensic Sci. 49(4): 859 -860.
D 13 S 317 Flanking Region Deletion A 4 bp deletion outside the mini. STR primers causes the commercial kit produced allele to appear one repeat smaller… African American sample ZT 79305 NIST Identifiler data Sequence analysis identified two regions where 4 bp deletions occur to cause this 1 repeat variation Ohio U mini. STR data D 13 S 317 Drabek, J. , Chung, D. T. , Butler, J. M. , Mc. Cord, B. R. (2004) Concordance study between miniplex STR assays and a commercial STR typing kit, J. Forensic Sci. 49(4): 859 -860.
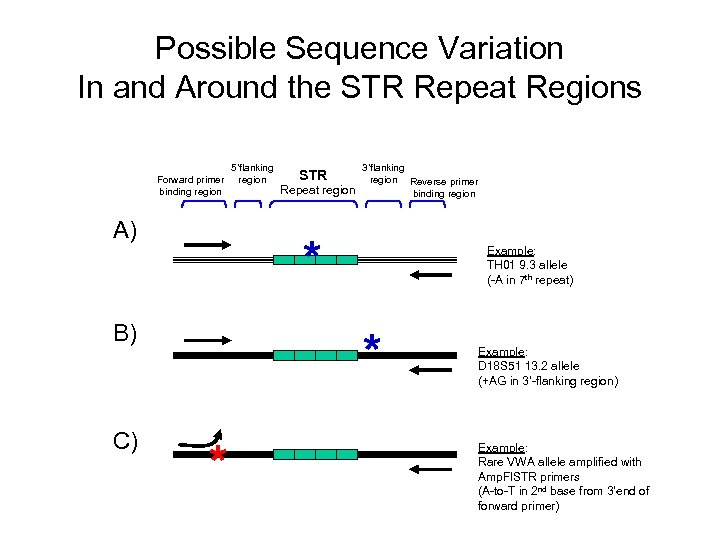 Possible Sequence Variation In and Around the STR Repeat Regions Forward primer binding region A) STR Repeat region 3’flanking region Reverse primer binding region * B) C) 5’flanking region Example: TH 01 9. 3 allele (-A in 7 th repeat) * * Example: D 18 S 51 13. 2 allele (+AG in 3’-flanking region) Example: Rare VWA allele amplified with Amp. Fl. STR primers (A-to-T in 2 nd base from 3’end of forward primer)
Possible Sequence Variation In and Around the STR Repeat Regions Forward primer binding region A) STR Repeat region 3’flanking region Reverse primer binding region * B) C) 5’flanking region Example: TH 01 9. 3 allele (-A in 7 th repeat) * * Example: D 18 S 51 13. 2 allele (+AG in 3’-flanking region) Example: Rare VWA allele amplified with Amp. Fl. STR primers (A-to-T in 2 nd base from 3’end of forward primer)
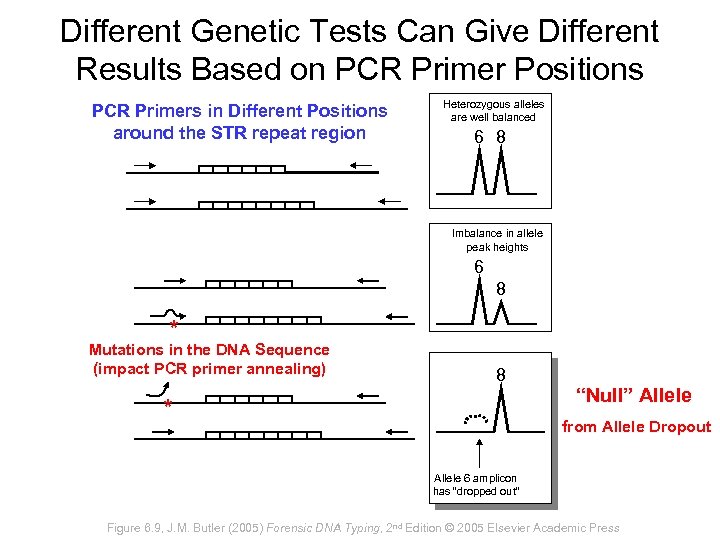 Different Genetic Tests Can Give Different Results Based on PCR Primer Positions PCR Primers in Different Positions around the STR repeat region Heterozygous alleles are well balanced 6 8 Imbalance in allele peak heights 6 8 * Mutations in the DNA Sequence (impact PCR primer annealing) 8 * “Null” Allele from Allele Dropout Allele 6 amplicon has “dropped out” Figure 6. 9, J. M. Butler (2005) Forensic DNA Typing, 2 nd Edition © 2005 Elsevier Academic Press
Different Genetic Tests Can Give Different Results Based on PCR Primer Positions PCR Primers in Different Positions around the STR repeat region Heterozygous alleles are well balanced 6 8 Imbalance in allele peak heights 6 8 * Mutations in the DNA Sequence (impact PCR primer annealing) 8 * “Null” Allele from Allele Dropout Allele 6 amplicon has “dropped out” Figure 6. 9, J. M. Butler (2005) Forensic DNA Typing, 2 nd Edition © 2005 Elsevier Academic Press
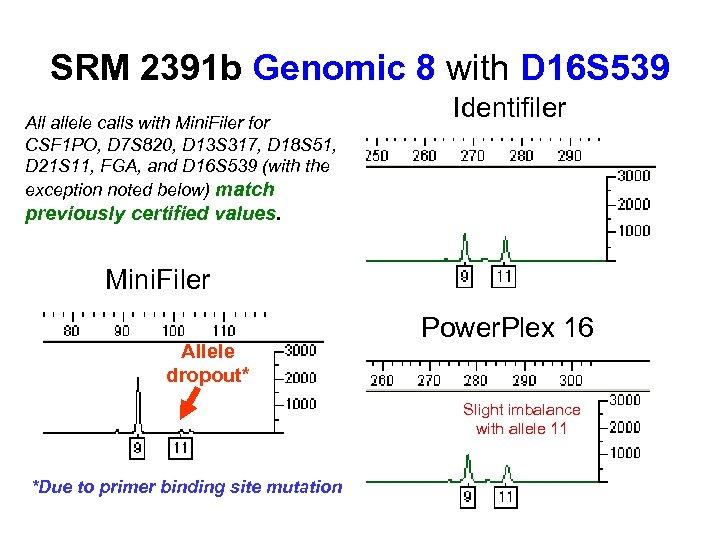 SRM 2391 b Genomic 8 with D 16 S 539 All allele calls with Mini. Filer for CSF 1 PO, D 7 S 820, D 13 S 317, D 18 S 51, D 21 S 11, FGA, and D 16 S 539 (with the exception noted below) match Identifiler previously certified values. Mini. Filer Allele dropout* Power. Plex 16 Slight imbalance with allele 11 *Due to primer binding site mutation
SRM 2391 b Genomic 8 with D 16 S 539 All allele calls with Mini. Filer for CSF 1 PO, D 7 S 820, D 13 S 317, D 18 S 51, D 21 S 11, FGA, and D 16 S 539 (with the exception noted below) match Identifiler previously certified values. Mini. Filer Allele dropout* Power. Plex 16 Slight imbalance with allele 11 *Due to primer binding site mutation
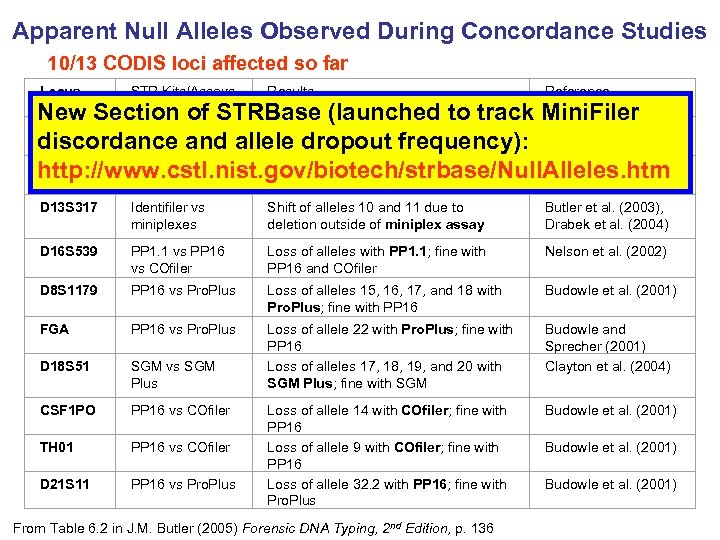 Apparent Null Alleles Observed During Concordance Studies 10/13 CODIS loci affected so far Locus STR Kits/Assays Compared Results Reference D 13 S 317 Identifiler vs miniplexes Shift of alleles 10 and 11 due to deletion outside of miniplex assay Butler et al. (2003), Drabek et al. (2004) D 16 S 539 PP 1. 1 vs PP 16 vs COfiler Loss of alleles with PP 1. 1; fine with PP 16 and COfiler Nelson et al. (2002) D 8 S 1179 PP 16 vs Pro. Plus Loss of alleles 15, 16, 17, and 18 with Pro. Plus; fine with PP 16 Budowle et al. (2001) FGA PP 16 vs Pro. Plus D 18 S 51 SGM vs SGM Plus Loss of allele 22 with Pro. Plus; fine with PP 16 Loss of alleles 17, 18, 19, and 20 with SGM Plus; fine with SGM Budowle and Sprecher (2001) Clayton et al. (2004) CSF 1 PO PP 16 vs COfiler Budowle et al. (2001) TH 01 PP 16 vs COfiler D 21 S 11 PP 16 vs Pro. Plus Loss of allele 14 with COfiler; fine with PP 16 Loss of allele 9 with COfiler; fine with PP 16 Loss of allele 32. 2 with PP 16; fine with Pro. Plus New Section of STRBase (launched to track Mini. Filer VWA PP 1. 1 vs Loss of allele 19 with Pro. Plus; fine with Kline et al. (1998) discordance and allele dropout frequency): Pro. Plus PP 1. 1 D 5 S 818 PP 16 vs Pro. Plus Loss of alleles 10 and 11 with PP 16; Alves et al. (2003) http: //www. cstl. nist. gov/biotech/strbase/Null. Alleles. htm fine with Pro. Plus From Table 6. 2 in J. M. Butler (2005) Forensic DNA Typing, 2 nd Edition, p. 136 Budowle et al. (2001)
Apparent Null Alleles Observed During Concordance Studies 10/13 CODIS loci affected so far Locus STR Kits/Assays Compared Results Reference D 13 S 317 Identifiler vs miniplexes Shift of alleles 10 and 11 due to deletion outside of miniplex assay Butler et al. (2003), Drabek et al. (2004) D 16 S 539 PP 1. 1 vs PP 16 vs COfiler Loss of alleles with PP 1. 1; fine with PP 16 and COfiler Nelson et al. (2002) D 8 S 1179 PP 16 vs Pro. Plus Loss of alleles 15, 16, 17, and 18 with Pro. Plus; fine with PP 16 Budowle et al. (2001) FGA PP 16 vs Pro. Plus D 18 S 51 SGM vs SGM Plus Loss of allele 22 with Pro. Plus; fine with PP 16 Loss of alleles 17, 18, 19, and 20 with SGM Plus; fine with SGM Budowle and Sprecher (2001) Clayton et al. (2004) CSF 1 PO PP 16 vs COfiler Budowle et al. (2001) TH 01 PP 16 vs COfiler D 21 S 11 PP 16 vs Pro. Plus Loss of allele 14 with COfiler; fine with PP 16 Loss of allele 9 with COfiler; fine with PP 16 Loss of allele 32. 2 with PP 16; fine with Pro. Plus New Section of STRBase (launched to track Mini. Filer VWA PP 1. 1 vs Loss of allele 19 with Pro. Plus; fine with Kline et al. (1998) discordance and allele dropout frequency): Pro. Plus PP 1. 1 D 5 S 818 PP 16 vs Pro. Plus Loss of alleles 10 and 11 with PP 16; Alves et al. (2003) http: //www. cstl. nist. gov/biotech/strbase/Null. Alleles. htm fine with Pro. Plus From Table 6. 2 in J. M. Butler (2005) Forensic DNA Typing, 2 nd Edition, p. 136 Budowle et al. (2001)
 STR Typing Measurement Issues • STR genotypes are generated using PCR amplification and electrophoretic sizing that involves an internal size standard with each sample. • The forensic DNA community almost exclusively uses STR typing kits to obtain results (there are different kits available that examine the same common markers). • PCR amplification is expected to generate consistent genotypes as long as primer positions are not changed between kits. Primer changes can result in allele dropout due to primer site mutations. • Occasionally new commercial kits are created with additional loci. • General STR repeat nomenclature rules have been established but do have some subjectivity in them permitting possible differences in how STR alleles are named.
STR Typing Measurement Issues • STR genotypes are generated using PCR amplification and electrophoretic sizing that involves an internal size standard with each sample. • The forensic DNA community almost exclusively uses STR typing kits to obtain results (there are different kits available that examine the same common markers). • PCR amplification is expected to generate consistent genotypes as long as primer positions are not changed between kits. Primer changes can result in allele dropout due to primer site mutations. • Occasionally new commercial kits are created with additional loci. • General STR repeat nomenclature rules have been established but do have some subjectivity in them permitting possible differences in how STR alleles are named.
 Two Different Independent Methods Used • Size Analysis/Genotyping – Electrophoretic separation and sizing of PCR product compared to an internal size standard followed by comparison to the sizes of one or more sequenced alleles (could be commercially available allelic ladder) run in-house with the same conditions, instrument, and internal size standard • DNA Sequence Analysis – Isolation of each individual allele – DNA sequence analysis followed by direct counting of the number of repeats (and correlation to size variation observed during STR typing)
Two Different Independent Methods Used • Size Analysis/Genotyping – Electrophoretic separation and sizing of PCR product compared to an internal size standard followed by comparison to the sizes of one or more sequenced alleles (could be commercially available allelic ladder) run in-house with the same conditions, instrument, and internal size standard • DNA Sequence Analysis – Isolation of each individual allele – DNA sequence analysis followed by direct counting of the number of repeats (and correlation to size variation observed during STR typing)
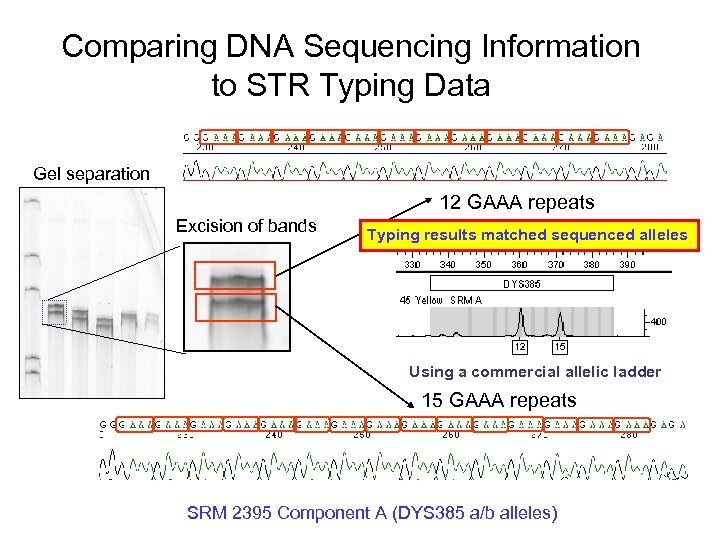 Comparing DNA Sequencing Information to STR Typing Data Gel separation 12 GAAA repeats Excision of bands Typing results matched sequenced alleles Using a commercial allelic ladder 15 GAAA repeats SRM 2395 Component A (DYS 385 a/b alleles)
Comparing DNA Sequencing Information to STR Typing Data Gel separation 12 GAAA repeats Excision of bands Typing results matched sequenced alleles Using a commercial allelic ladder 15 GAAA repeats SRM 2395 Component A (DYS 385 a/b alleles)
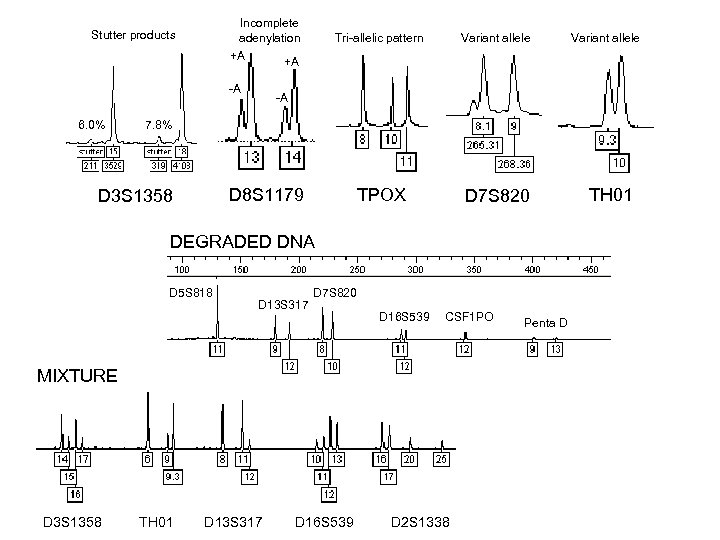 Incomplete adenylation +A +A Stutter products -A 6. 0% Tri-allelic pattern Variant allele TPOX D 7 S 820 -A 7. 8% D 8 S 1179 D 3 S 1358 DEGRADED DNA D 5 S 818 D 13 S 317 D 7 S 820 D 16 S 539 CSF 1 PO MIXTURE D 3 S 1358 Variant allele TH 01 D 13 S 317 D 16 S 539 D 2 S 1338 Penta D TH 01
Incomplete adenylation +A +A Stutter products -A 6. 0% Tri-allelic pattern Variant allele TPOX D 7 S 820 -A 7. 8% D 8 S 1179 D 3 S 1358 DEGRADED DNA D 5 S 818 D 13 S 317 D 7 S 820 D 16 S 539 CSF 1 PO MIXTURE D 3 S 1358 Variant allele TH 01 D 13 S 317 D 16 S 539 D 2 S 1338 Penta D TH 01
 Chapter 10 – Points for Discussion • Is it better to prevent or promote non-template addition? Explain your reasons. • How is a degenerate primer used in STR typing assays? Discuss some advantages and disadvantages to using a degenerate primer. • What observations indicate that a peak in an electropherogram is a spike as opposed to DNA? • What steps are typically taken to confirm the presence of an offladder allele? • Why are mutations and mutation rates a concern with parentage and kinship analysis but not with forensic DNA testing?
Chapter 10 – Points for Discussion • Is it better to prevent or promote non-template addition? Explain your reasons. • How is a degenerate primer used in STR typing assays? Discuss some advantages and disadvantages to using a degenerate primer. • What observations indicate that a peak in an electropherogram is a spike as opposed to DNA? • What steps are typically taken to confirm the presence of an offladder allele? • Why are mutations and mutation rates a concern with parentage and kinship analysis but not with forensic DNA testing?


