023206320ebbe0633d1208aa83cd6ad5.ppt
- Количество слайдов: 56

Cells: The Living Unit Biology 2121 Chapter 3

Introduction • (1). C ells are the basic units of all living things. • (2). The basic parts of a cell – Plasma membrane – C ytoplasm – Nucleus • (3). Organelles – Specialized cellular components – “little organs”
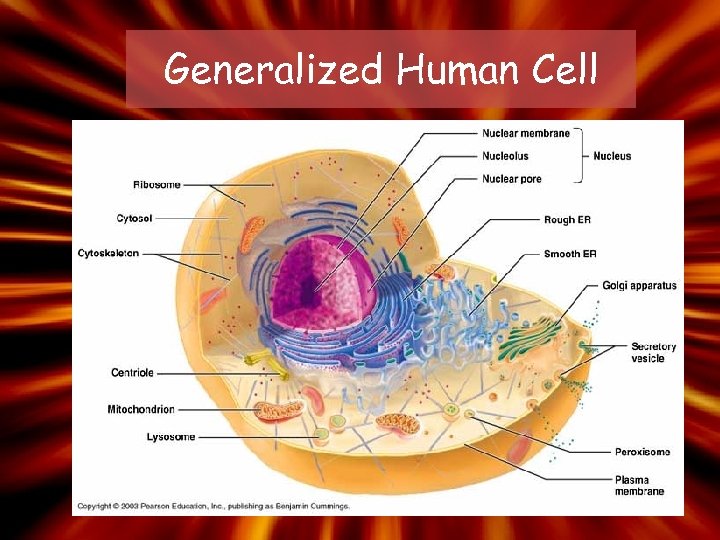
Generalized Human Cell
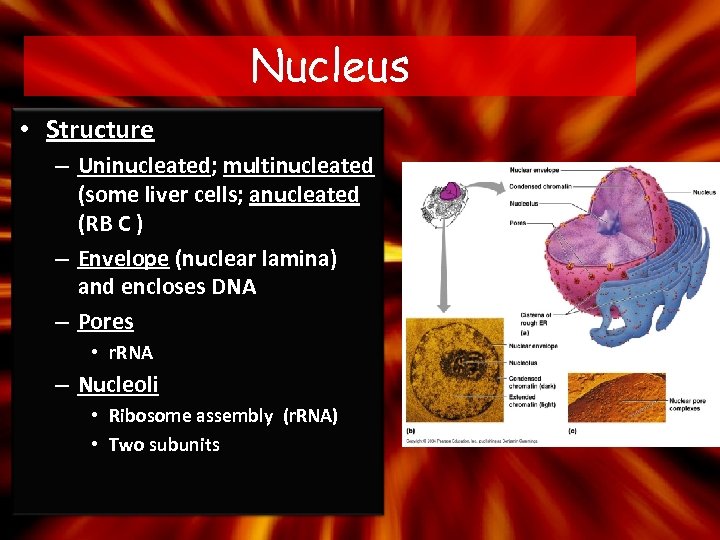
Nucleus • Structure – Uninucleated; multinucleated (some liver cells; anucleated (RB C ) – Envelope (nuclear lamina) and encloses DNA – Pores • r. RNA – Nucleoli • Ribosome assembly (r. RNA) • Two subunits
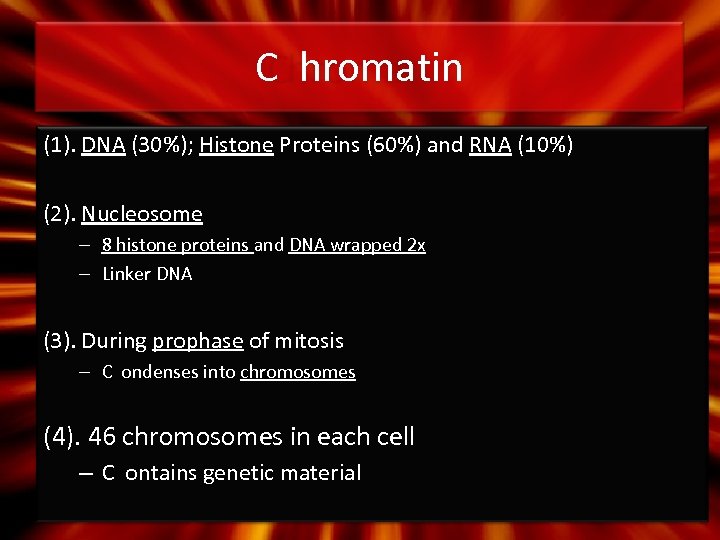
C hromatin (1). DNA (30%); Histone Proteins (60%) and RNA (10%) (2). Nucleosome – 8 histone proteins and DNA wrapped 2 x – Linker DNA (3). During prophase of mitosis – C ondenses into chromosomes (4). 46 chromosomes in each cell – C ontains genetic material
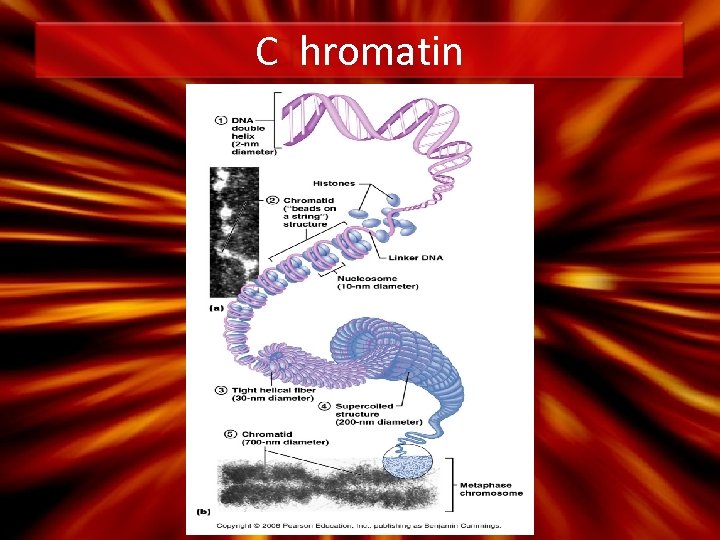
C hromatin
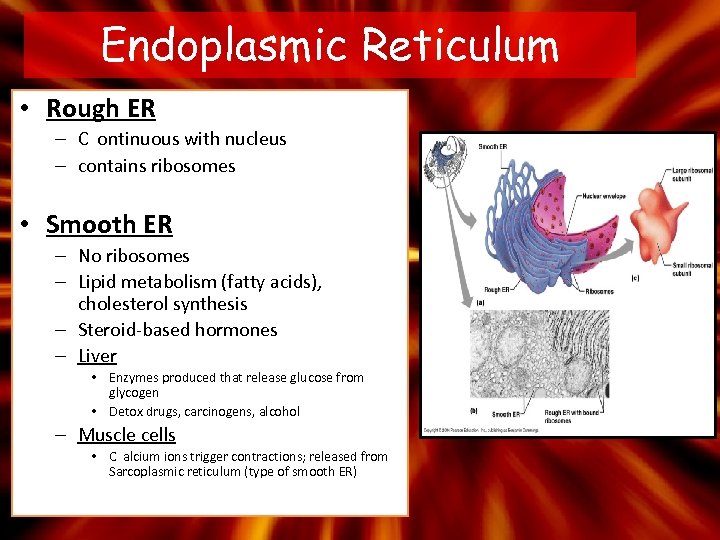
Endoplasmic Reticulum • Rough ER – C ontinuous with nucleus – contains ribosomes • Smooth ER – No ribosomes – Lipid metabolism (fatty acids), cholesterol synthesis – Steroid-based hormones – Liver • Enzymes produced that release glucose from glycogen • Detox drugs, carcinogens, alcohol – Muscle cells • C alcium ions trigger contractions; released from Sarcoplasmic reticulum (type of smooth ER)
Ribosomes • (1). Site of ‘Protein Synthesis’ • (2). Free – Floating in cytoplasm – Produce proteins for cytosol in cytoplasm • (3). Attached – Produce proteins for the cell membrane or ‘export’ • Hormones, integral proteins, enzymes • (4). Structure – r. RNA and proteins (2 subunits)

Golgi Apparatus • Function – Modify, concentrate and package proteins and lipids made in the rough ER • Movement – Proteins move from the ER to the Golgi and to parts of the golgi via transport vesicles • Faces – Enter on the ‘cis’ face and leave via the ‘transface’ • Vesicles Products 1. Proteins and lipids for export 2. Proteins inserted and function in the plasma membrane 3. Enzymes for lysosomes
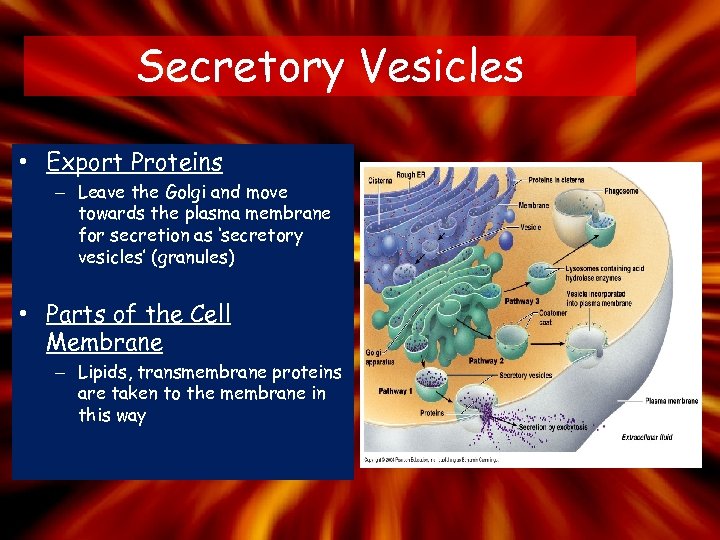
Secretory Vesicles • Export Proteins – Leave the Golgi and move towards the plasma membrane for secretion as ‘secretory vesicles’ (granules) • Parts of the Cell Membrane – Lipids, transmembrane proteins are taken to the membrane in this way
Lysosomes • Function – Dispose of bacteria, old parts of cells, waste Animations- Lysosome • C ontents – Digestive enzymes (hydrolytic) – ‘acidic hydrolases’ function in low p. H environments best • Other Functions – Digest particles that enter the cell via endocytosis – Degrades old organelles, tissues – Breaks down nonuseful tissues (between fingers and toes during development)
Endomembrane System • System of organelles that function together – Produce, store, export molecules – Degradation of substances – Vesicular Transport (animation) • Organelles in the System – 1. Endoplasmic Reticulum (ER) – 2. Golgi Apparatus – 3. Secretory vesicles – 4. Lysosomes – 5. Nuclear membrane
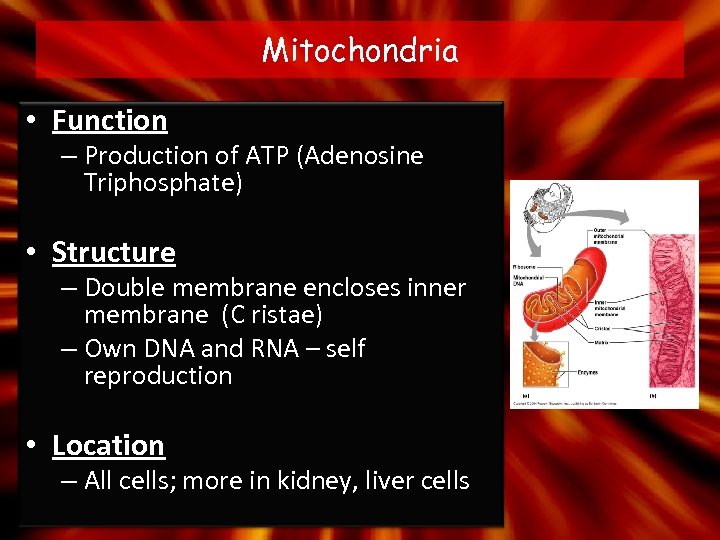
Mitochondria • Function – Production of ATP (Adenosine Triphosphate) • Structure – Double membrane encloses inner membrane (C ristae) – Own DNA and RNA – self reproduction • Location – All cells; more in kidney, liver cells

Peroxisomes C ontains two important enzymes • 1. Oxidase – Detoxes alcohol and formaldehyde. Formaldehyde converted into hydrogen peroxide – Destroys free radicals; converts them into hydrogen peroxide • 2. C atalase – Breaks hydrogen peroxide into water and oxygen • Large amounts in kidney and liver

Other Important Parts 1. C ytoskeleton – Microtubules • Larger; attaches to organelles and pulls them to different locations (motor proteins-ATP) – Microfilaments • Thin; cause cell to change shape due to actin and myosin • C leavage furrow; endo- and exocytosis – Intermediate fibers • Insoluble fibers that attach to desmosomes 2. C entrioles – C ontained within the centrosome – Involved in cell replication or division
The Plasma Membrane 1. Forms a ‘boundry’ – Separates the intracellular and extracellular environments 2. Function – Selectively permeable 3. Structure – Phospholipid bilayer with inserted proteins, cholesterol, glycolipids
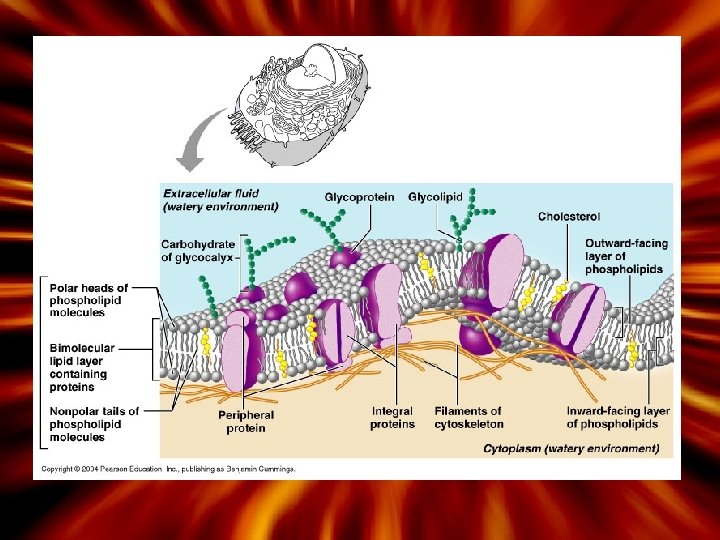

Phospholipid Bilayer • Fluid Mosaic model • Heads are hydrophilic – W ater-loving (polar) • Tails are hydrophobic – W ater-hating (nonpolar) • What is allowed to pass freely through the membrane? – Allow for the passage of small polar molecules such as water, gases, small proteins (amino acids) via facilitate diffusion, lipid soluble – Impermeable to large polar molecules, large proteins, carbohydrates and ions
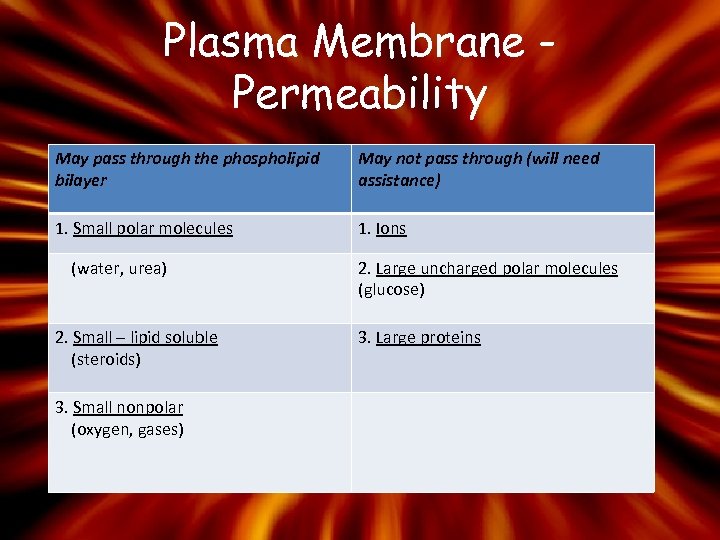
Plasma Membrane Permeability May pass through the phospholipid bilayer May not pass through (will need assistance) 1. Small polar molecules 1. Ions (water, urea) 2. Small – lipid soluble (steroids) 3. Small nonpolar (oxygen, gases) 2. Large uncharged polar molecules (glucose) 3. Large proteins
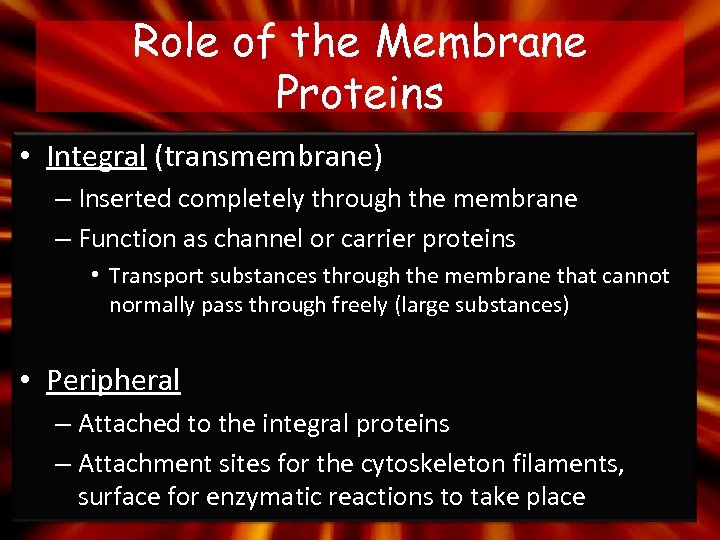
Role of the Membrane Proteins • Integral (transmembrane) – Inserted completely through the membrane – Function as channel or carrier proteins • Transport substances through the membrane that cannot normally pass through freely (large substances) • Peripheral – Attached to the integral proteins – Attachment sites for the cytoskeleton filaments, surface for enzymatic reactions to take place
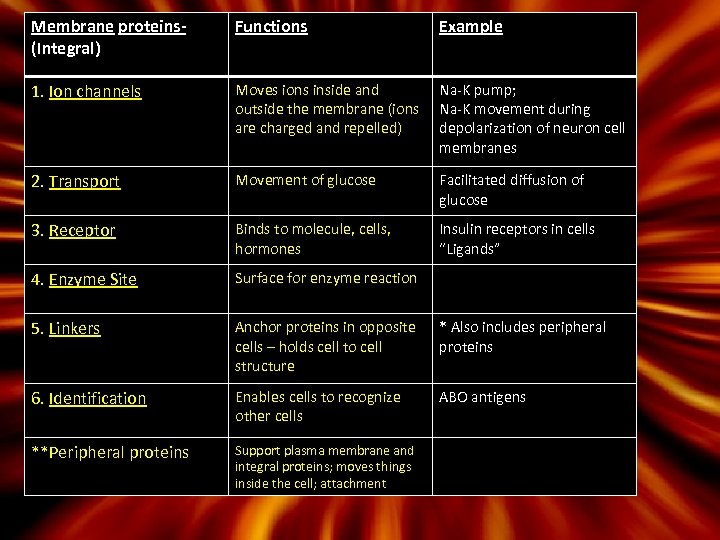
Membrane proteins(Integral) Functions Example 1. Ion channels Moves ions inside and outside the membrane (ions are charged and repelled) Na-K pump; Na-K movement during depolarization of neuron cell membranes 2. Transport Movement of glucose Facilitated diffusion of glucose 3. Receptor Binds to molecule, cells, hormones Insulin receptors in cells “Ligands” 4. Enzyme Site Surface for enzyme reaction 5. Linkers Anchor proteins in opposite cells – holds cell to cell structure * Also includes peripheral proteins 6. Identification Enables cells to recognize other cells ABO antigens **Peripheral proteins Support plasma membrane and integral proteins; moves things inside the cell; attachment
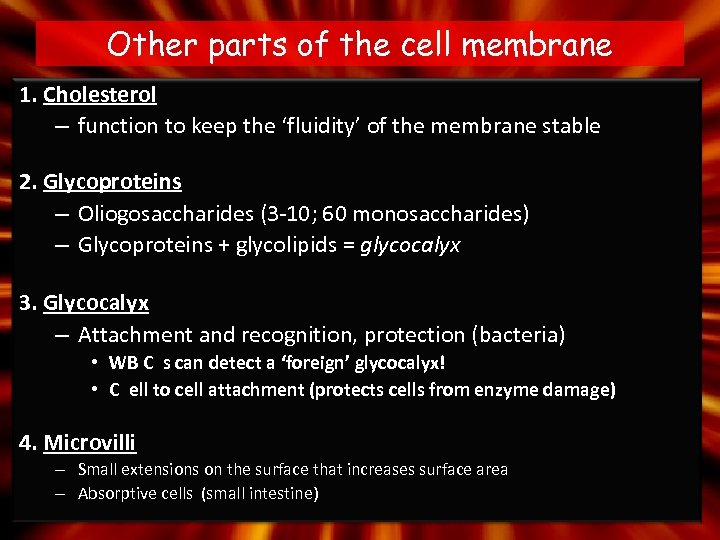
Other parts of the cell membrane 1. Cholesterol – function to keep the ‘fluidity’ of the membrane stable 2. Glycoproteins – Oliogosaccharides (3 -10; 60 monosaccharides) – Glycoproteins + glycolipids = glycocalyx 3. Glycocalyx – Attachment and recognition, protection (bacteria) • WB C s can detect a ‘foreign’ glycocalyx! • C ell to cell attachment (protects cells from enzyme damage) 4. Microvilli – Small extensions on the surface that increases surface area – Absorptive cells (small intestine)
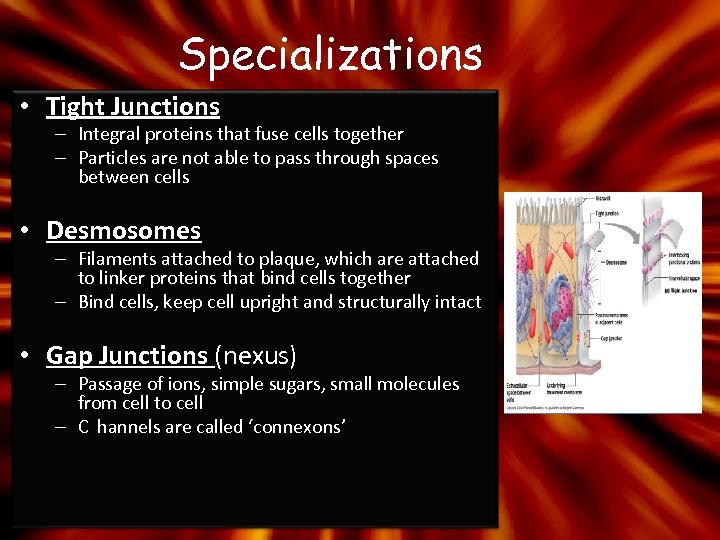
Specializations • Tight Junctions – Integral proteins that fuse cells together – Particles are not able to pass through spaces between cells • Desmosomes – Filaments attached to plaque, which are attached to linker proteins that bind cells together – Bind cells, keep cell upright and structurally intact • Gap Junctions (nexus) – Passage of ions, simple sugars, small molecules from cell to cell – C hannels are called ‘connexons’
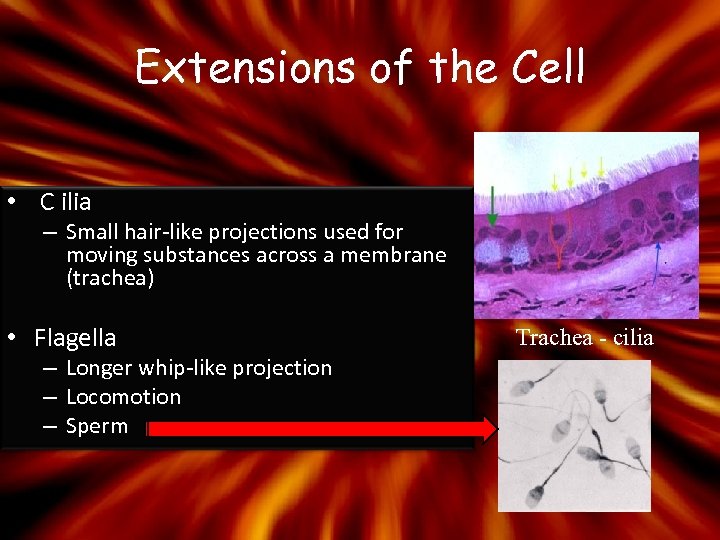
Extensions of the Cell • C ilia – Small hair-like projections used for moving substances across a membrane (trachea) • Flagella – Longer whip-like projection – Locomotion – Sperm Trachea - cilia
Membrane Transport • Passive processes – No energy required, substances move ‘down’ their concentration gradients • Examples are osmosis and diffusion • Active Processes – Require energy in the form of ATP – Substances move ‘against’ their concentration gradients • Examples are the sodium-potassium pump and vesicular transport

Passive Processes • 1. Simple Diffusion – Nonpolar and lipid-soluble substances pass through – Gases, fat-soluble vitamins • 2. Osmosis – Movement of water from areas (gradients) of higher concentration to areas of lower concentration through a semi-permeable membrane • Diffusion: same, but movement of particles other than water. • 3. Facilitated Diffusion – Membrane protein channels or carriers – Example glucose, amino acids, ions

Animations • Animation: How Diffusion Works • Animation: How Facilitated Diffusion Works • Animation: How Osmosis Works
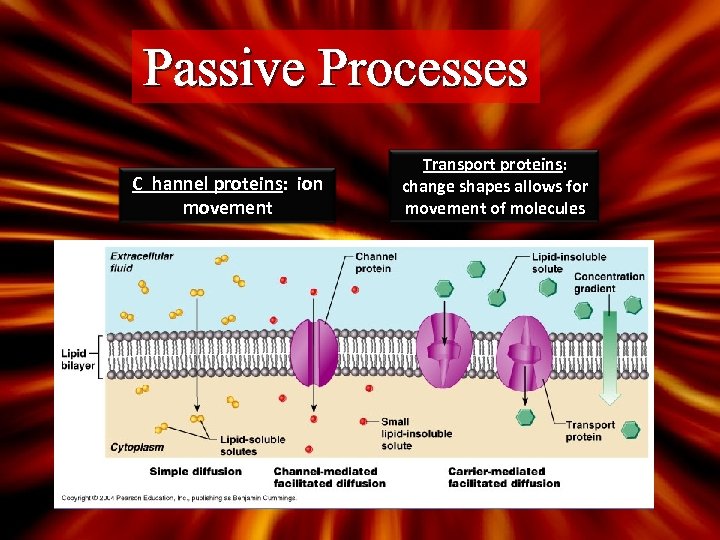
Passive Processes C hannel proteins: ion movement Transport proteins: change shapes allows for movement of molecules
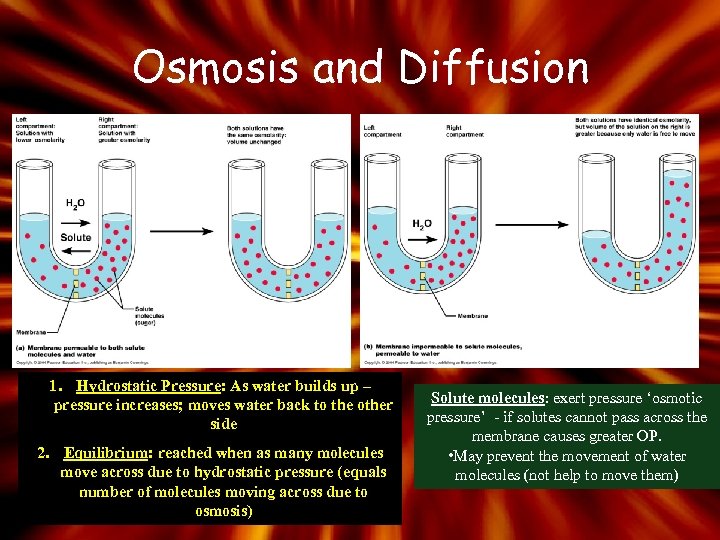
Osmosis and Diffusion 1. Hydrostatic Pressure: As water builds up – pressure increases; moves water back to the other side 2. Equilibrium: reached when as many molecules move across due to hydrostatic pressure (equals number of molecules moving across due to osmosis) Solute molecules: exert pressure ‘osmotic pressure’ - if solutes cannot pass across the membrane causes greater OP. • May prevent the movement of water molecules (not help to move them)
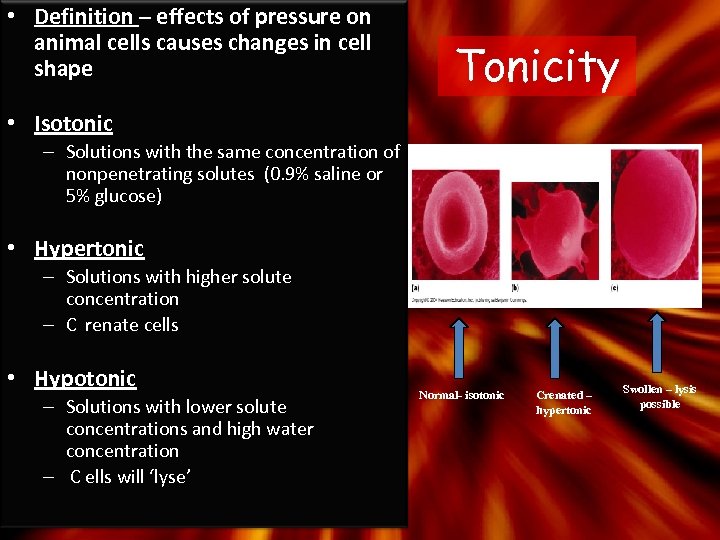
• Definition – effects of pressure on animal cells causes changes in cell shape Tonicity • Isotonic – Solutions with the same concentration of nonpenetrating solutes (0. 9% saline or 5% glucose) • Hypertonic – Solutions with higher solute concentration – C renate cells • Hypotonic – Solutions with lower solute concentrations and high water concentration – C ells will ‘lyse’ Normal- isotonic Crenated – hypertonic Swollen – lysis possible

Review • A beaker is filled with water which is 80% Na C l. A dialysis bag is filled with a solution of water which contains 20% Na C l. The dialysis bag is impermeable to Na C l but permeable to water. • W hich direction will water move?

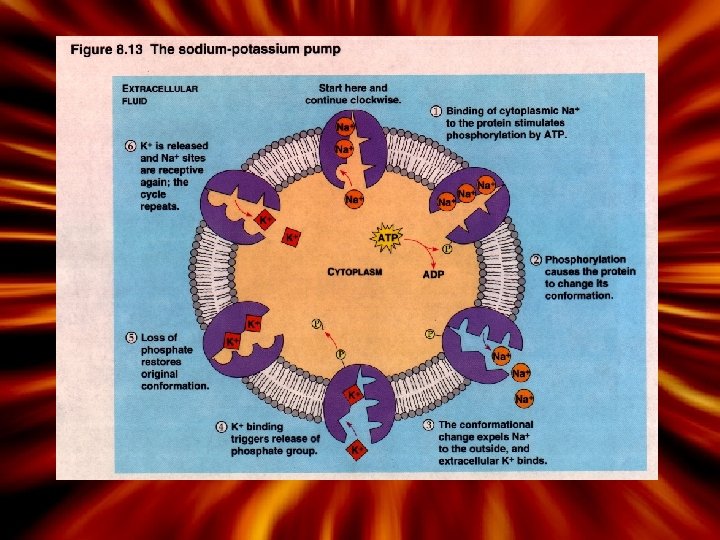
Active Processes • Active Transport – Requires ‘energy’ (ATP) and a protein carrier – Called ‘solute pumps’ • Types of Active Transport – 1. Primary – ‘Sodium potassium pump’ • Uses energy from the ‘hydrolysis of ATP’ • ATP causes the protein to change conformation – 2. Secondary- Sodium and glucose (cotransport) • Uses energy stored in in an ionic concentration gradient • Movement of ions or particles ‘down’ a concentration gradient • 1. As Na+ moves across the cell membrane, drives the transport of another ion. – Same for hydrogen, calcium • For example – Movement of Na drives glucose; movement of Na drives amino acid transport
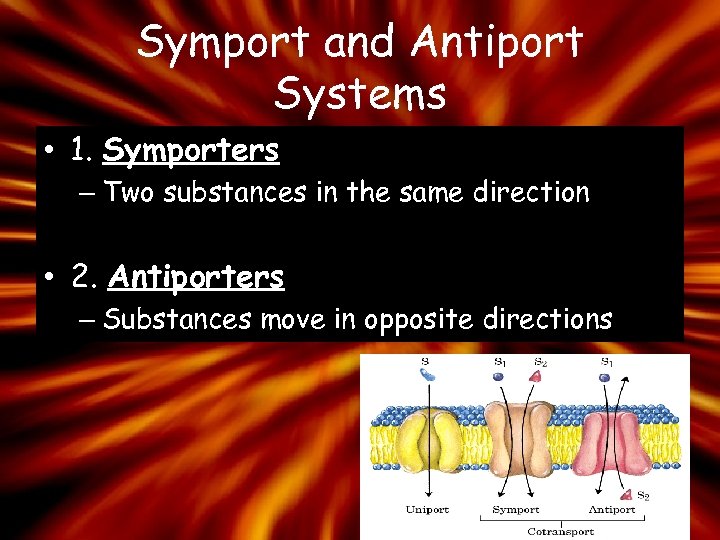
Symport and Antiport Systems • 1. Symporters – Two substances in the same direction • 2. Antiporters – Substances move in opposite directions

Animations • Animation: How the Sodium Potassium Pump Works • Animation: Sodium-Potassium Exchange Pump (Quiz 1)
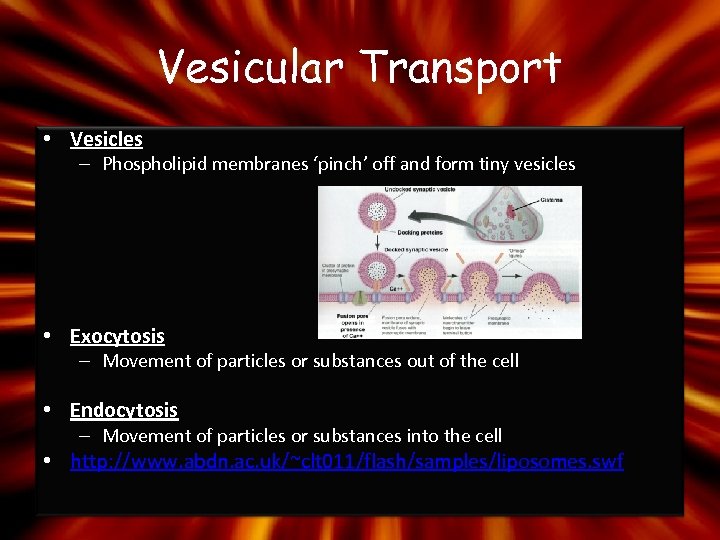
Vesicular Transport • Vesicles – Phospholipid membranes ‘pinch’ off and form tiny vesicles • Exocytosis – Movement of particles or substances out of the cell • Endocytosis – Movement of particles or substances into the cell • http: //www. abdn. ac. uk/~clt 011/flash/samples/liposomes. swf
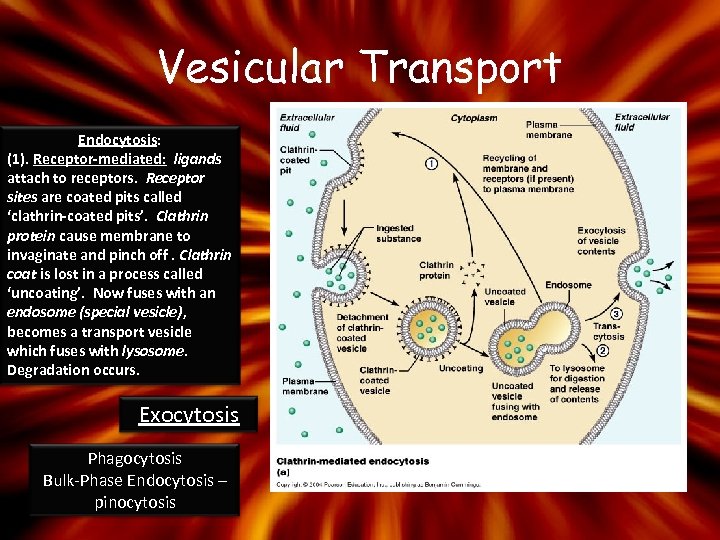
Vesicular Transport Endocytosis: (1). Receptor-mediated: ligands attach to receptors. Receptor sites are coated pits called ‘clathrin-coated pits’. Clathrin protein cause membrane to invaginate and pinch off. Clathrin coat is lost in a process called ‘uncoating’. Now fuses with an endosome (special vesicle), becomes a transport vesicle which fuses with lysosome. Degradation occurs. Exocytosis Phagocytosis Bulk-Phase Endocytosis – pinocytosis
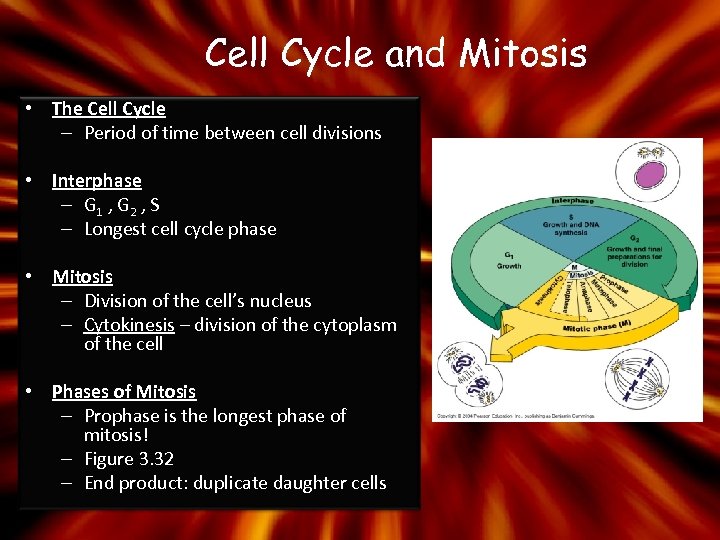
Cell Cycle and Mitosis • The Cell Cycle – Period of time between cell divisions • Interphase – G 1 , G 2 , S – Longest cell cycle phase • Mitosis – Division of the cell’s nucleus – Cytokinesis – division of the cytoplasm of the cell • Phases of Mitosis – Prophase is the longest phase of mitosis! – Figure 3. 32 – End product: duplicate daughter cells

Phases of Mitosis Prophase 1. Longest phase of ‘mitosis’! 2. Chromatin condenses into ‘chromosomes’ 3. Spindle fibers appear from the area around the ‘centrioles’. Centrioles migrate to the opposite poles of the parent cell. Spindle fibers come from asters (protein microtubules). 4. Nuclear membrane breaks down and the chromosomes are free to move (manipulated by the spindle fiber proteins) Metaphase 1. Spindle fibers move the chromosomes to the middle or center area of the cell or ‘equator’ (metaphase plate) Anaphase 1. Spindle fibers separate the two sister ‘chromatids’; each chromatid is moved to the opposite pole. Telophase 1. Begins as chromosome movement stops! 2. A ‘reversal’ of prophase 3. ‘Binucleate’ for a short time 4. Cytokinesis follows.

Animations • 1. Cell Cycle • 2. Mitosis and Cytokinesis • 3. Mitosis (Alt)

How is Cell Division Controlled? • 1. Before cell division, cell volume increases (64 fold) more than cell surface area enlargement (16 fold). – Daughter cells produced have appropriate volume to surface area ratios • 2. C hemical Signals (notes) – Growth factors, hormones released by other cells • 3. C ontact Inhibition – When cells come into contact with one another or touching, growth may stop • Proteins Involved (notes) – C yclins – C yclin-dependent kinases (Cdks)

Animation • 1. Control of the Cell Cycle
S-Phase and DNA Replication • Steps are listed on page 100 – 1. helicase – 2. RNA primer is laid down (primase) – 3. Formation of ‘complementary strands’ – 4. DNA polymerase III attaches the correct base pairs to the old strand – 5. DNA ligase ‘glues’ together the ‘lagging strand’ • Semi-C onservative Replication
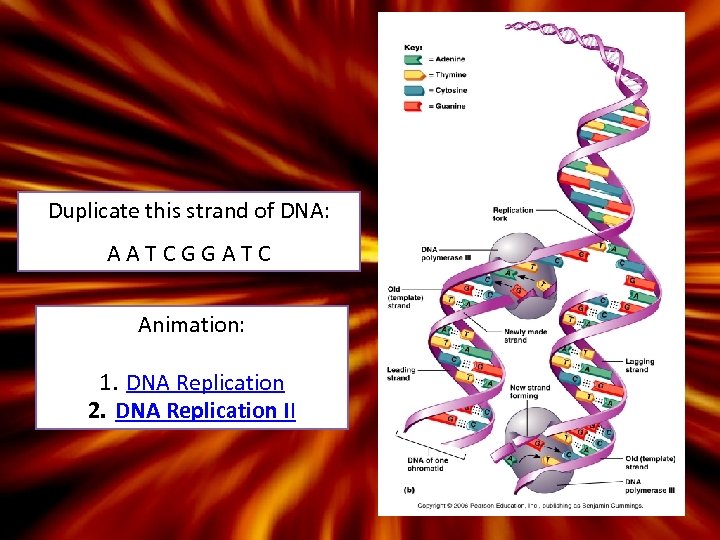
Duplicate this strand of DNA: AATCGGATC Animation: 1. DNA Replication 2. DNA Replication II
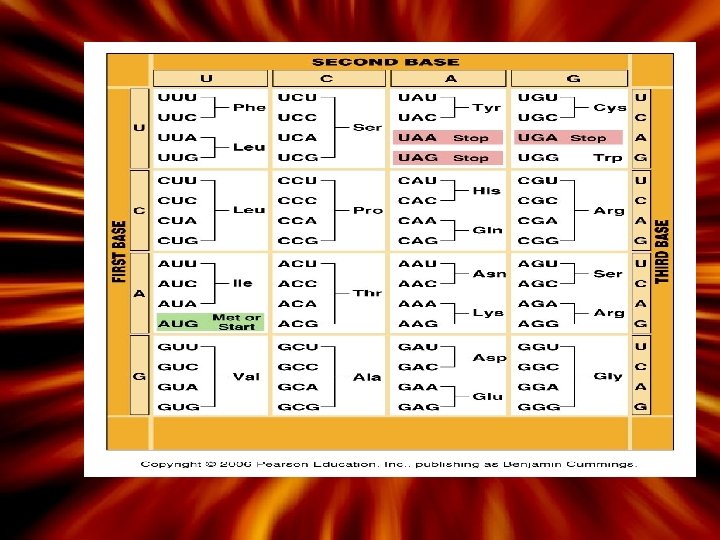
Protein Synthesis • Genes are sequences of DNA that code for one polypeptide sequence – A ‘triplet’ sequence of bases - one amino acid – Example: AAA codes for the amino acid Lysine • See the genetic code (figure 3. 35) • Genes contain exons (coding sequences) and introns (non-coding sequences) – Introns will be cut-out before the making of the polypeptide
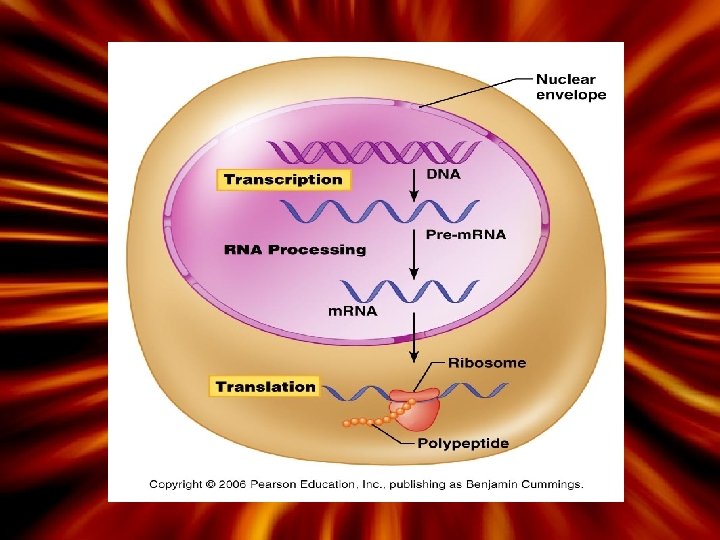
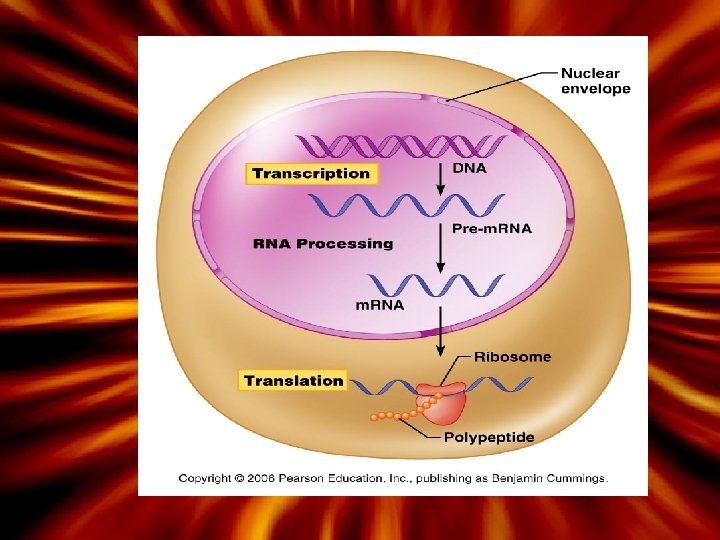
How is the information in DNA copied? Transcription is a process by which DNA is copied into m. RNA (messenger RNA) – Takes place inside the nucleus (DNA does not leave the nucleus) • After transcription is complete, the m. RNA will leave the nucleus through the nuclear pores – Each 3 -base sequence in m. RNA is called a ‘codon’ which codes for one amino acid • Editing: introns are lost and exons are left on the final m. RNA strand (leaves the nucleus)
More About Transcription After DNA is unzipped, the process begins at the place where a DNA promoter binds to the start point of the gene. RNA synthesis RNA nucleotides are laid down, using DNA as the template RNA polymerase C odons Three nucleotides 64 possible codons made (43); 3 stop codons and one start Redundancy (3. 35 - Genetic code) Animation 1. Transcription
Transcribe this sequence of DNA • Base change in m. RNA: U (uracil) replaces T (thymine) • READ FROM THE m. RNA! • DNA: A C C • m. RNA: AG C
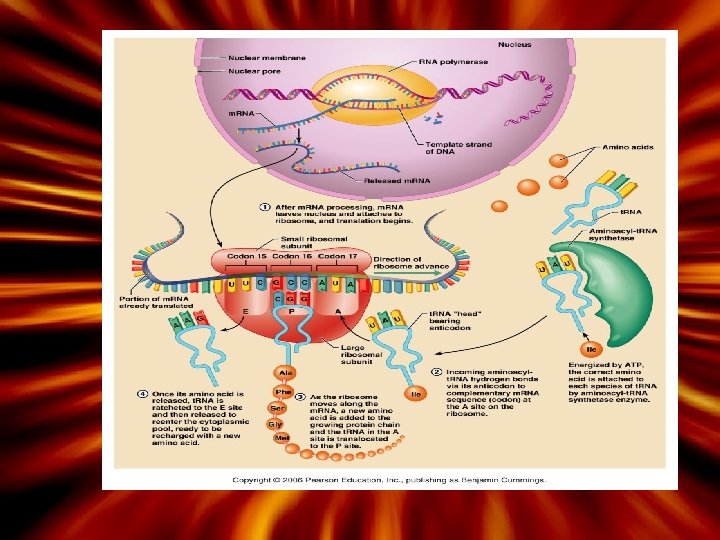
How is a polypeptide chain made? • Translation is the process of converting the language of DNA into amino acid language. – Occurs in the cytoplasm • 1. m. RNA reaches the ribosomes on the surface of the ER (page 108) – m. RNA will base pair to the r. RNA (ribosomes) • 2. t. RNA (transfer RNA) brings the appropriate amino acid to the surface of the m. RNA – Anticodon on the t. RNA will base pair with the m. RNA codon – The first codon is always a ‘start’ codon (AUG)

How is a polypeptide chain made? • 3. The first codon base pairs with the first anti-codon in the P-site – The second codon and anticodon base pair in the A-site • The P and A site amino acids then form a peptide bond. – The P-t. RNA then is released to the E site and goes to pick up another amino acid – The A-site t. RNA now holding the chain moves to the Psite • Another t. RNA carrying an amino acid then moves into the A site • Sequence ends with a ‘stop’ codon (UGA)
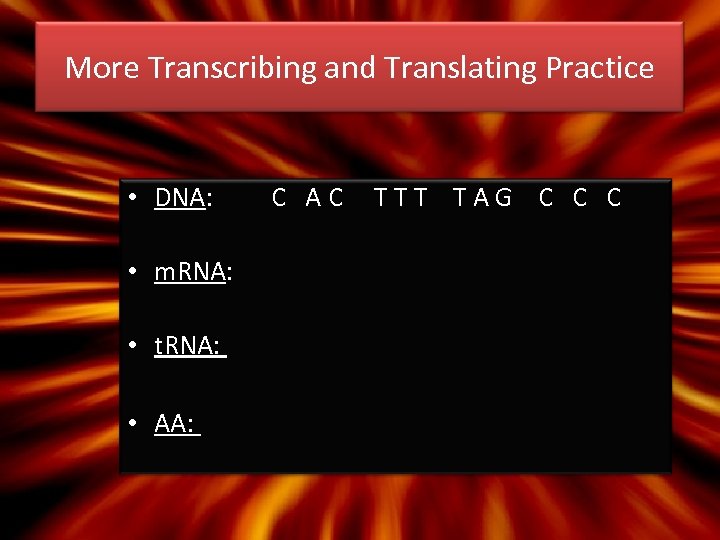
More Transcribing and Translating Practice • DNA: • m. RNA: • t. RNA: • AA: C AC TTT TAG C C C
Animations • 1. Translation
023206320ebbe0633d1208aa83cd6ad5.ppt