de6e706d947546202e545a4ae83e0e91.ppt
- Количество слайдов: 43

CEFIC LRI Tools – Ambit 1. 21 Nina Jeliazkova Ideaconsult Ltd. Sofia, Bulgaria 29. 6. 2007 QSAR Awareness Day, JRC, Ispra, Italy
Outline n n Ambit overview Demo : 1. 2. 3. 29. 6. 2007 Finding basic information about a query compound in the database Complex query in the database –retrieve data meeting multiple criteria from Ambit database Import data from EURAS Gold standard Bioconcentration database QSAR Awareness Day, JRC, Ispra, Italy 2
Introduction – why Ambit ? n n Limited free, publicly accessible, methodologically transparent software was identified as one of the roadblocks for broadening use of in-silico methods (ICCA Workshop in Setubal 2002, OECD) Realization that efficient use of existing information on chemicals requires better ways for n Storage n n standardized formats, computer automated verification of structures, capability to store large amounts of data Taking advantage of rapidly evolving field of data mining and extraction of relevant information 29. 6. 2007 QSAR Awareness Day, JRC, Ispra, Italy 3

IT strategy n n Ambit - building blocks for Decision Support System High emphasis on n interoperability for “plug and play” Flexibility modular design Transparency n Open source, relying on open standards. Open source software lowers the user barrier, facilitates the dissemination activities and enables the reproducibility of models and results n n n The cheminformatics functionality relies on the open source Java library – The Chemistry Development Kit http: //cdk. sourceforge. net/ The software is based on My. SQL database (www. mysql. com), which is the most popular open source relational database. Chemical Markup Language (CML) n n n 29. 6. 2007 acknowledged method of encoding chemical data in XML Is being adopted by a large number of chemical organisations, from government, through commercial to academia. The choice of CML for the internal format makes the database independent of the software which is able to access it, in contrast to some proprietary solutions. QSAR Awareness Day, JRC, Ispra, Italy 4

IT strategy n n n Desktop installation: My. SQL database and standalone application (Ambit. Database. Tools) on the same PC Intranet installation: My. SQL database on a server and standalone application (Ambit. Database. Tools) on the user PCs Internet installation – My SQL Database and web server (JSP and Servlets), Web browser as user interface 29. 6. 2007 QSAR Awareness Day, JRC, Ispra, Italy 5
Ambit overview n The AMBIT database: n n n stores chemical structures, their identifiers such as CAS, INCh. I numbers; attributes such as molecular descriptors, experimental data together with test descriptions, and literature references. The database can also store QSAR models. In addition the software can generate a suite of 2 D and 3 D molecular descriptors. can be searched by identifiers, attribute value or range, experimental data value or range, user defined structure and substructure, structural similarity AMBIT database contains over 450 000 chemical compounds with data imported from over a dozen databases [http: //ambit. acad. bg/ambit/stats/]. The number of compounds is growing all the time and one the of system’s great strengths is that any dataset can be imported for comparison and analysis. AMBITDatabase. Tools 1. 21 allows the user to create a local database and to import his own sets of chemical compounds. AMBIT Discovery performs chemical grouping and assesses the applicability domain of a QSAR offering a variety of methods including using different approaches to similarity assessments: statistical that rely on ‘descriptor space’; approaches based on mechanistic understanding; and approaches based on structural similarity. ECB QMRF inventory – a tailored version of Ambit database (under development). Will store information in QMRF. Large effort on standardization 29. 6. 2007 QSAR Awareness Day, JRC, Ispra, Italy 6

AMBIT Database Today Not restricted to these datasets! Any dataset can be imported. (e. g. DSSTox, AQUIRE, LLNA …) 29. 6. 2007 QSAR Awareness Day, JRC, Ispra, Italy 7
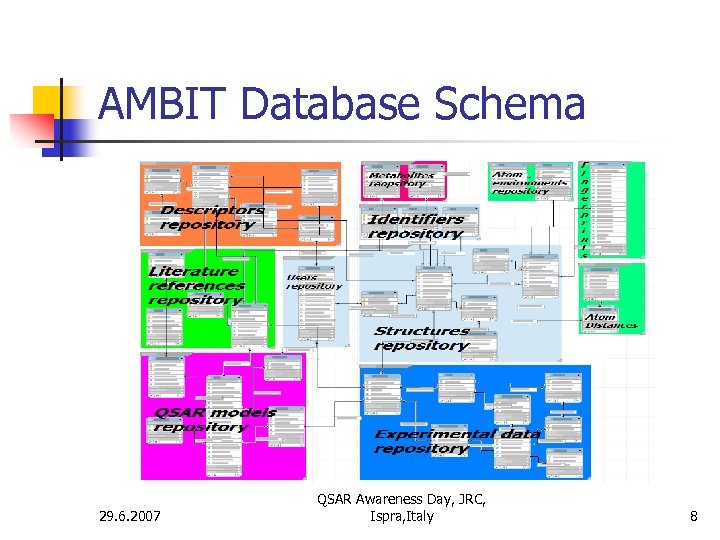
AMBIT Database Schema 29. 6. 2007 QSAR Awareness Day, JRC, Ispra, Italy 8

AMBIT Online: Similarity search 29. 6. 2007 QSAR Awareness Day, JRC, Ispra, Italy 9
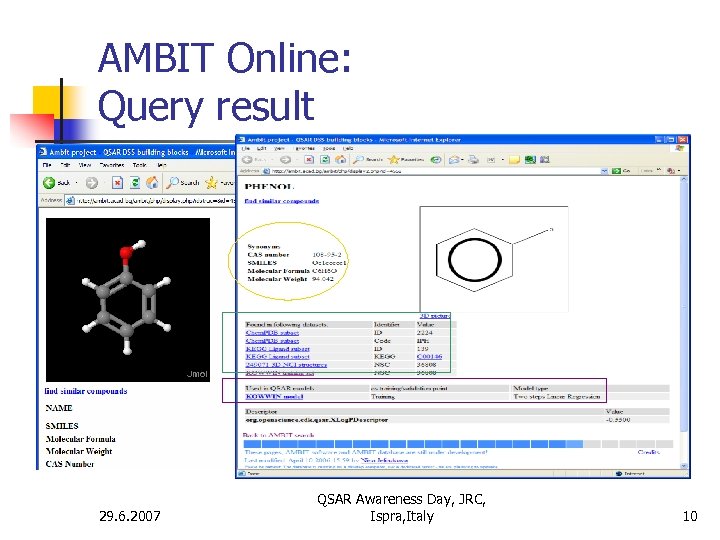
AMBIT Online: Query result 29. 6. 2007 QSAR Awareness Day, JRC, Ispra, Italy 10

Links to other databases: example: KEGG 29. 6. 2007 QSAR Awareness Day, JRC, Ispra, Italy 11

Information about QSAR models 29. 6. 2007 QSAR Awareness Day, JRC, Ispra, Italy 12
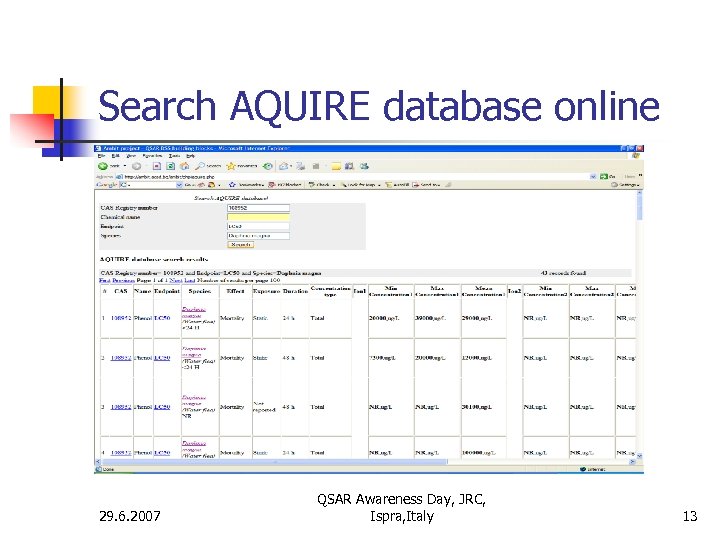
Search AQUIRE database online 29. 6. 2007 QSAR Awareness Day, JRC, Ispra, Italy 13

Search EURAS Bioconcentration database online 29. 6. 2007 QSAR Awareness Day, JRC, Ispra, Italy 14
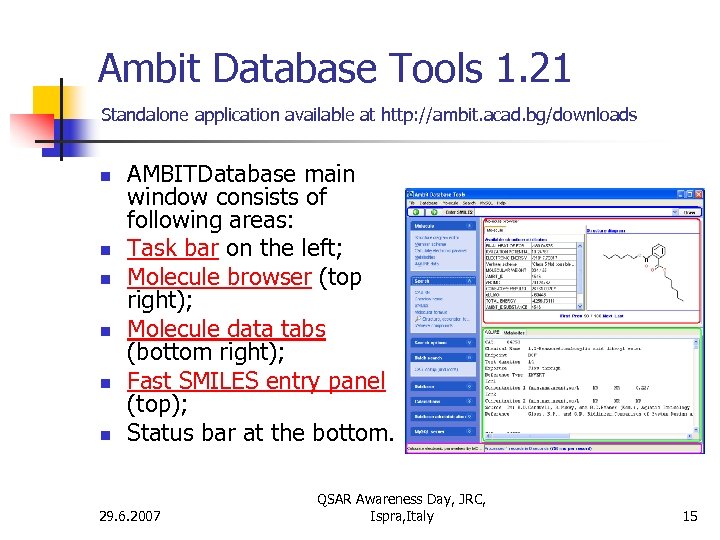
Ambit Database Tools 1. 21 Standalone application available at http: //ambit. acad. bg/downloads n n n AMBITDatabase main window consists of following areas: Task bar on the left; Molecule browser (top right); Molecule data tabs (bottom right); Fast SMILES entry panel (top); Status bar at the bottom. 29. 6. 2007 QSAR Awareness Day, JRC, Ispra, Italy 15

Demo: 1. 2. 3. Finding basic information about a query compound in the database Complex query in the database – retrieve data meeting multiple criteria from Ambit database Import data from EURAS Gold standard Bioconcentration database 29. 6. 2007 QSAR Awareness Day, JRC, Ispra, Italy 16

Exercise 1. Finding basic information about a query compound in the database n Launch Ambit. Database. Tools 1. 20 n Start menu/ All Programs/ CEFICLRI/Ambit 1. 20 Ambit database tools main screen. Various tasks can be started from the menu options at the left panel. This exercise uses Search / CAS RN menu to lookup for compound with specific CAS RN 29. 6. 2007 QSAR Awareness Day, JRC, Ispra, Italy 17
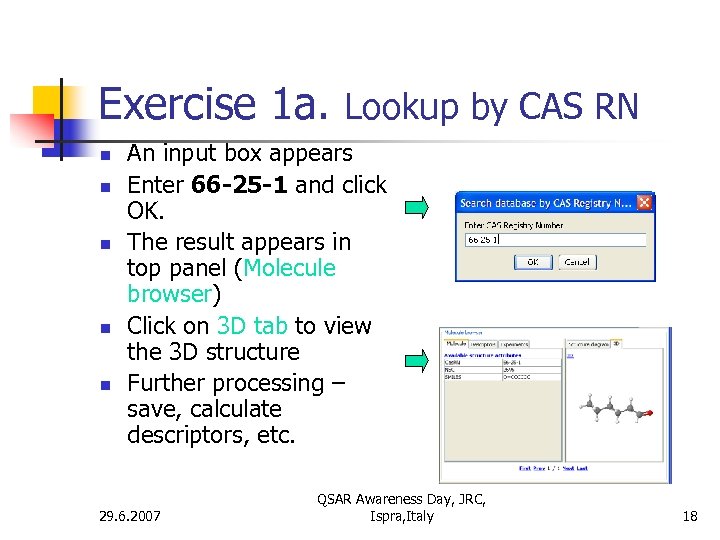
Exercise 1 a. Lookup by CAS RN n n n An input box appears Enter 66 -25 -1 and click OK. The result appears in top panel (Molecule browser) Click on 3 D tab to view the 3 D structure Further processing – save, calculate descriptors, etc. 29. 6. 2007 QSAR Awareness Day, JRC, Ispra, Italy 18

Exercise 1 b. Retrieve descriptors n The objective of this exercise is to retrieve values of several descriptors from the database. The descriptors we are interested are n n n Log. P Crossectional diameter Maximum diameter Molecular weight Use Molecule/Advanced data retrieval menu 29. 6. 2007 QSAR Awareness Day, JRC, Ispra, Italy 19

Exercise 1 b. Retrieve descriptors n n The following window appears Check Read descriptors row The following window appears: Check following descriptors : n n XLog. PDescriptor Weight. Descriptor Crossectional. Diameter. Descriptor Maximum. Diameter. Descriptor 29. 6. 2007 QSAR Awareness Day, JRC, Ispra, Italy 20

Exercise 1 b. Retrieve descriptors n n The results appear in Descriptors tab Further processing – save, etc. 29. 6. 2007 QSAR Awareness Day, JRC, Ispra, Italy 21

Exercise 1 c. Retrieve AQUIRE data n n n Use Molecule/AQUIRE menu to retrieve toxicity data for hexaldehyde The results can be observed in bottom panel, EXPERIMENTAL data tab. Click on each row to view more details. Save to a file using File/Save menu (sdf, csv, xls, txt) 29. 6. 2007 QSAR Awareness Day, JRC, Ispra, Italy 22

SDF file for hexaldehyde n n n n n n n CDK 6/23/07, 13: 23 19 18 0 0. 0021 -0. 0041 0. 0020 C 1. 4167 2. 0553 -0. 0004 C -1. 4333 -0. 5336 0. 0129 C 1. 3963 3. 5622 0. 0079 C ……… 6 18 1 0 0 6 19 1 0 0 M END > <NSC> 2596 0 0 0 0999 0 0 0 0 0 0 0 0 V 2000 0 0 0 -0. 0187 0 0 1. 5258 0. 0104 C 0 0 0 > <Cross. Sectional. Diameter. Descriptor [Angstrom]> 2. 4897 > <XLog. PDescriptor> 1. 7530 > <Maximum. Diameter. Descriptor [Angstrom]> 8. 1759 > <SMILES> O=CCCCCC > <AQUIRE> LC 50=22000, ug/L Pimephales promelas > <Cas. RN> 66 -25 -1 > <Weight. Descriptor> 100. 0888 29. 6. 2007 QSAR Awareness Day, JRC, Ispra, Italy 23
XLS file for hexaldehyde 29. 6. 2007 QSAR Awareness Day, JRC, Ispra, Italy 24

Exercise 2. Complex queries: Use Ambit database to retrieve data that meet multiple criteria n n Use Search options /options menu to configure desired searches Switch to Similarity tab and set 0, 7 for Tanimoto threshold (we will be searching for structures with Tanimoto similarity > 0. 7) 29. 6. 2007 QSAR Awareness Day, JRC, Ispra, Italy 25

Exercise 2 a. Similarity search n n n Use Search/Structure search menu to invoke advanced query window Draw dimetylphtalate as shown at the figure Click Similarity button n Browse the 7 compounds found (in Molecule Browser) Go to Search/options and lower threshold to 0. 6 Use Search/Structure search/Similarity again with the same compound 29. 6. 2007 QSAR Awareness Day, JRC, Ispra, Italy 26

Exercise 2 a. Similarity search n n Now there are 156 compounds with Tanimoto similarity > 0. 6 We will be using Molecule/Save as dataset menu to store the query results into the database n Hint: you can store query results directly into database, without loading into Molecule Browser, by setting Search Options/Result destination – DATABASE and then performing the query 29. 6. 2007 QSAR Awareness Day, JRC, Ispra, Italy 27
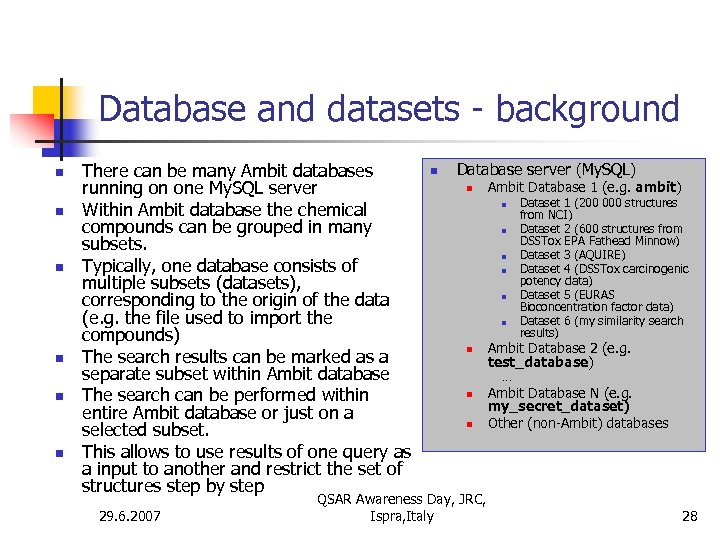
Database and datasets - background n n n There can be many Ambit databases running on one My. SQL server Within Ambit database the chemical compounds can be grouped in many subsets. Typically, one database consists of multiple subsets (datasets), corresponding to the origin of the data (e. g. the file used to import the compounds) The search results can be marked as a separate subset within Ambit database The search can be performed within entire Ambit database or just on a selected subset. This allows to use results of one query as a input to another and restrict the set of structures step by step 29. 6. 2007 n Database server (My. SQL) n Ambit Database 1 (e. g. ambit) n n n n Dataset 1 (200 000 structures from NCI) Dataset 2 (600 structures from DSSTox EPA Fathead Minnow) Dataset 3 (AQUIRE) Dataset 4 (DSSTox carcinogenic potency data) Dataset 5 (EURAS Bioconcentration factor data) Dataset 6 (my similarity search results) Ambit Database 2 (e. g. test_database) … n n QSAR Awareness Day, JRC, Ispra, Italy Ambit Database N (e. g. my_secret_dataset) Other (non-Ambit) databases 28
Exercise 2 a. Similarity search n n n Use Molecule/Save as dataset menu to store the query results into the database In the dialog box (as at right), add “+” button to add a new entry for the dataset. Type in the name for the dataset (e. g. “Similarity search Tanimoto > 0. 6”) n Click OK 29. 6. 2007 QSAR Awareness Day, JRC, Ispra, Italy 29

Exercise 2 a. Similarity search n n Now the new dataset is available in the datasets list and can be used to restrict subsequent queries Use Search options/Dataset menu to select which dataset to be searched, select “Similarity search Tanimoto > 0. 6” and click OK Note: this will not load any structures into Molecule browser! 29. 6. 2007 QSAR Awareness Day, JRC, Ispra, Italy 30

Exercise 2 b. Pre-set physicochemical profile n n The objective is to extract compounds that have physicochemical properties, relevant for bioaccumulation from the set of structurally similar compounds found by previous query. The recommended descriptors and ranges are: n n Log. P < 4. 5 Molecular weight < 1100 Cross sectional diameter < 17. 4 Å Maximum diameter < 43 Å 29. 6. 2007 QSAR Awareness Day, JRC, Ispra, Italy 31

Exercise 2 b. Pre-set physicochemical profile n n n Use Search/Structure search menu The window with options for structure, descriptors and experimental data queries appears. Click on Descriptors icon to obtain a list of descriptors available in the database 29. 6. 2007 QSAR Awareness Day, JRC, Ispra, Italy 32

Exercise 2 b. Pre-set physicochemical profile n n Select XLog. P descriptor (click on first column Click on Condition column and select “<” sign. Double click on the next column and enter 4. 5 Repeat with descriptors: n n Weight. Descriptor (Molecular weight) < 1100 Crosssectional. Diameter. Descriptor (crossectional diameter) < 17. 4 Maximum. Diameter. Descriptor (maximum diameter or maximum length) < 43 Click the Search button 29. 6. 2007 QSAR Awareness Day, JRC, Ispra, Italy 33

Exercise 2 b. Pre-set physicochemical profile n n 123 out of the 156 structurally similar compounds have the predefined profile. The descriptor values can be inspected in the Descriptors tab 29. 6. 2007 QSAR Awareness Day, JRC, Ispra, Italy 34
Exercise 2 c. Retrieve available toxicity data n n n Use Search Options/Options menu to select he endpoint Select AQUIRE tab Select LC 50 (Lethal concentration to 50% of test compounds) from the first list box 29. 6. 2007 QSAR Awareness Day, JRC, Ispra, Italy 35

Exercise 2 c. Retrieve available toxicity data n n n The next step is to tell the software we want to retrieve the data for all retrieved compounds (not only for the current structure). To do this: Select Molecule processing tab Select Molecule Browser: Current set of structures from the first list box 29. 6. 2007 QSAR Awareness Day, JRC, Ispra, Italy 36

Exercise 2 c. Retrieve available toxicity data n Use Molecule/AQUIRE n menu to retrieve LC 50 data for the current set of compounds Click Start button. 29. 6. 2007 QSAR Awareness Day, JRC, Ispra, Italy 37

Exercise 2 c. Retrieve available toxicity data n n Browse the compounds to view AQUIRE data at the bottom panel Repeat the same procedure to retrieve BCF data from AQUIRE 29. 6. 2007 QSAR Awareness Day, JRC, Ispra, Italy 38

Exercise 2 d. Retrieve available toxicity data (ER Binding) n Structure/Search menu Click experiments n Select DSSTox- n n n ERBinding Select Endpoint=“ER RBA” Click Search 29. 6. 2007 QSAR Awareness Day, JRC, Ispra, Italy 39

Exercise 2 d. Retrieve available toxicity data (ER Binding) n Browse ER Binding data, save results into file 29. 6. 2007 QSAR Awareness Day, JRC, Ispra, Italy 40

More exercises n n n Batch search Import structures into database Import descriptors and experimental data (e. g. bioconcentration factor dataset) Import QSAR models Database processing n n n Descriptor calculation Atom environments, Fingerprint, SMILES generation Create new (empty) database. n n Create users for the new database Import compounds 29. 6. 2007 QSAR Awareness Day, JRC, Ispra, Italy 41
Ambit - Summary n n n AMBIT software is a set of libraries and tools, providing various chemoinformatics functionalities for data management. The AMBIT system consists of a database and functional modules allowing a variety of flexible searches and mining of the data stored in the database. The unique feature of AMBIT is the ability to store multifaceted information about chemical structures and provide a searchable interface linking these diverse components. 29. 6. 2007 QSAR Awareness Day, JRC, Ispra, Italy 42

Thank you! Questions? 29. 6. 2007 QSAR Awareness Day, JRC, Ispra, Italy
de6e706d947546202e545a4ae83e0e91.ppt