0adc0ff243138239ed47505cce7c3f4e.ppt
- Количество слайдов: 71
 cal Disease s Disease s s Diseases Ph y Ps s hyoyogo Pih lh log s Pioi l s e Gen es eas Dis my ato An Novel hips relations sight Deeper in
cal Disease s Disease s s Diseases Ph y Ps s hyoyogo Pih lh log s Pioi l s e Gen es eas Dis my ato An Novel hips relations sight Deeper in
 Integrative Genomics For Understanding Disease Process Anil Jegga Division of Biomedical Informatics, Cincinnati Children’s Hospital Medical Center (CCHMC) Department of Pediatrics, University of Cincinnati, Ohio - 45229 Anil. Jegga@cchmc. org
Integrative Genomics For Understanding Disease Process Anil Jegga Division of Biomedical Informatics, Cincinnati Children’s Hospital Medical Center (CCHMC) Department of Pediatrics, University of Cincinnati, Ohio - 45229 Anil. Jegga@cchmc. org
 Acknowledgement • • • Jing Chen Mrunal Deshmukh Sivakumar Gowrisankar Chandra Gudivada Arvind Muthukrishnan Bruce J Aronow
Acknowledgement • • • Jing Chen Mrunal Deshmukh Sivakumar Gowrisankar Chandra Gudivada Arvind Muthukrishnan Bruce J Aronow
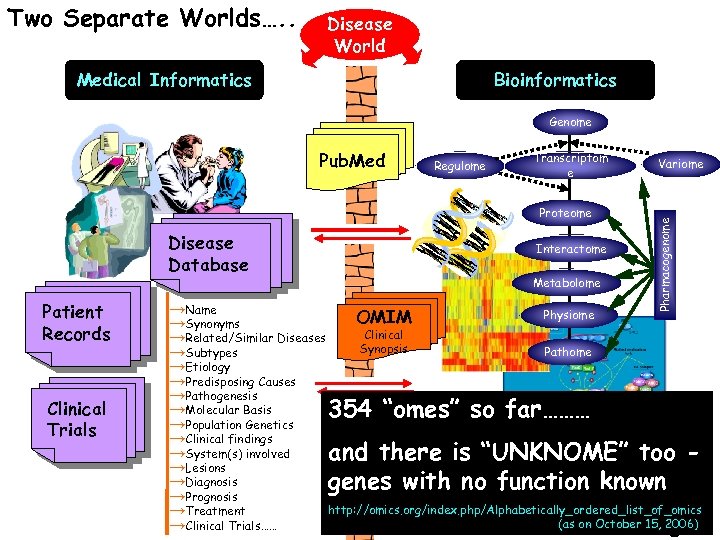 Two Separate Worlds…. . Disease World Medical Informatics Bioinformatics Genome Regulome Transcriptom e Proteome Disease Database Patient Records Clinical Trials Interactome Metabolome Variome Pharmacogenome Pub. Med →Name Physiome OMIM →Synonyms Clinical →Related/Similar Diseases Synopsis Pathome →Subtypes →Etiology →Predisposing Causes →Pathogenesis →Molecular Basis 354 “omes” so far……… →Population Genetics →Clinical findings →System(s) involved and there is “UNKNOME” too →Lesions →Diagnosis genes with no function known →Prognosis http: //omics. org/index. php/Alphabetically_ordered_list_of_omics →Treatment (as October 15, 2006) →Clinical Trials…… With Some Dataon. Exchange…
Two Separate Worlds…. . Disease World Medical Informatics Bioinformatics Genome Regulome Transcriptom e Proteome Disease Database Patient Records Clinical Trials Interactome Metabolome Variome Pharmacogenome Pub. Med →Name Physiome OMIM →Synonyms Clinical →Related/Similar Diseases Synopsis Pathome →Subtypes →Etiology →Predisposing Causes →Pathogenesis →Molecular Basis 354 “omes” so far……… →Population Genetics →Clinical findings →System(s) involved and there is “UNKNOME” too →Lesions →Diagnosis genes with no function known →Prognosis http: //omics. org/index. php/Alphabetically_ordered_list_of_omics →Treatment (as October 15, 2006) →Clinical Trials…… With Some Dataon. Exchange…
 Motivation To correlate diseases with anatomical parts affected, the genes/proteins involved, and the underlying physiological processes (interactions, pathways, processes). In other words, bringing the disciplines of Medical Informatics (MI) and Bio. Informatics (BI) together (Biomedical Informatics - BMI) to support personalized or “tailor-made” medicine. How to integrate multiple types of genome-scale data across experiments and phenotypes in order to find genes associated with diseases
Motivation To correlate diseases with anatomical parts affected, the genes/proteins involved, and the underlying physiological processes (interactions, pathways, processes). In other words, bringing the disciplines of Medical Informatics (MI) and Bio. Informatics (BI) together (Biomedical Informatics - BMI) to support personalized or “tailor-made” medicine. How to integrate multiple types of genome-scale data across experiments and phenotypes in order to find genes associated with diseases
 Model Organism Databases: Common Issues • Heterogeneous Data Sets - Data Integration – From Genotype to Phenotype – Experimental and Consensus Views • Incorporation of Large Datasets – Whole genome annotation pipelines – Large scale mutagenesis/variation projects (db. SNP) • Computational vs. Literature-based Data Collection and Evaluation (Med. Line) • Data Mining – extraction of new knowledge – testable hypotheses (Hypothesis Generation)
Model Organism Databases: Common Issues • Heterogeneous Data Sets - Data Integration – From Genotype to Phenotype – Experimental and Consensus Views • Incorporation of Large Datasets – Whole genome annotation pipelines – Large scale mutagenesis/variation projects (db. SNP) • Computational vs. Literature-based Data Collection and Evaluation (Med. Line) • Data Mining – extraction of new knowledge – testable hypotheses (Hypothesis Generation)
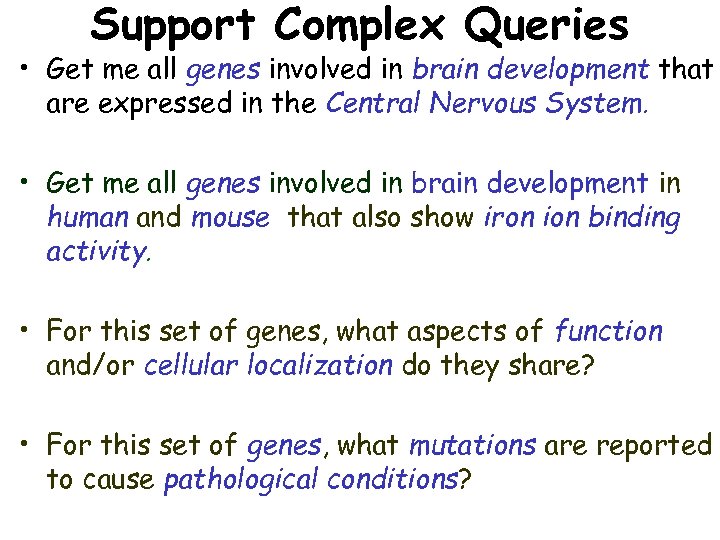 Support Complex Queries • Get me all genes involved in brain development that are expressed in the Central Nervous System. • Get me all genes involved in brain development in human and mouse that also show iron ion binding activity. • For this set of genes, what aspects of function and/or cellular localization do they share? • For this set of genes, what mutations are reported to cause pathological conditions?
Support Complex Queries • Get me all genes involved in brain development that are expressed in the Central Nervous System. • Get me all genes involved in brain development in human and mouse that also show iron ion binding activity. • For this set of genes, what aspects of function and/or cellular localization do they share? • For this set of genes, what mutations are reported to cause pathological conditions?
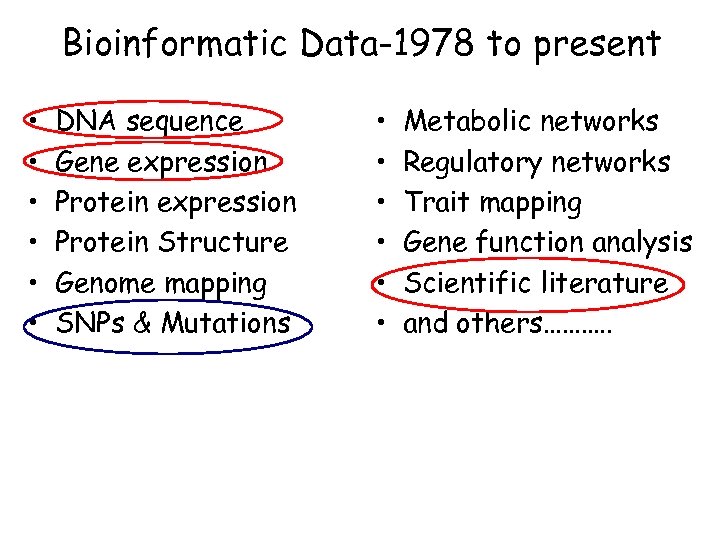 Bioinformatic Data-1978 to present • • • DNA sequence Gene expression Protein Structure Genome mapping SNPs & Mutations • • • Metabolic networks Regulatory networks Trait mapping Gene function analysis Scientific literature and others………. .
Bioinformatic Data-1978 to present • • • DNA sequence Gene expression Protein Structure Genome mapping SNPs & Mutations • • • Metabolic networks Regulatory networks Trait mapping Gene function analysis Scientific literature and others………. .
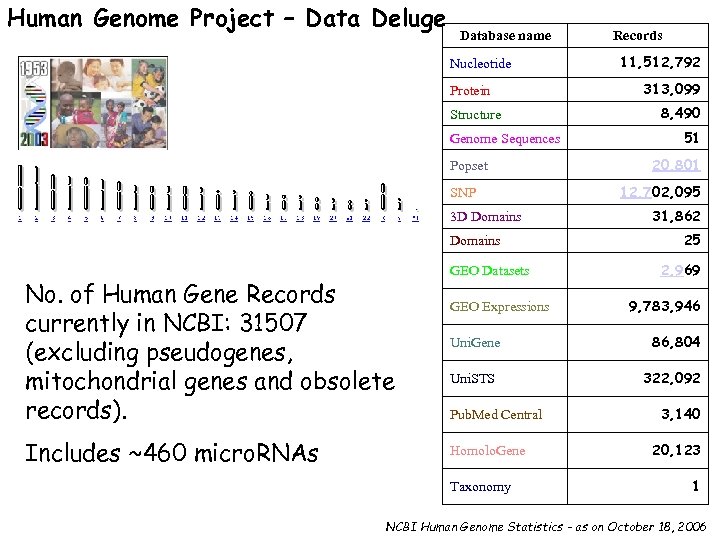 Human Genome Project – Data Deluge Database name Nucleotide Protein Structure Genome Sequences Popset SNP 3 D Domains No. of Human Gene Records currently in NCBI: 31507 (excluding pseudogenes, mitochondrial genes and obsolete records). Includes ~460 micro. RNAs GEO Datasets GEO Expressions Uni. Gene Uni. STS Pub. Med Central Homolo. Gene Taxonomy Records 11, 512, 792 313, 099 8, 490 51 20, 801 12, 702, 095 31, 862 25 2, 969 9, 783, 946 86, 804 322, 092 3, 140 20, 123 1 NCBI Human Genome Statistics – as on October 18, 2006
Human Genome Project – Data Deluge Database name Nucleotide Protein Structure Genome Sequences Popset SNP 3 D Domains No. of Human Gene Records currently in NCBI: 31507 (excluding pseudogenes, mitochondrial genes and obsolete records). Includes ~460 micro. RNAs GEO Datasets GEO Expressions Uni. Gene Uni. STS Pub. Med Central Homolo. Gene Taxonomy Records 11, 512, 792 313, 099 8, 490 51 20, 801 12, 702, 095 31, 862 25 2, 969 9, 783, 946 86, 804 322, 092 3, 140 20, 123 1 NCBI Human Genome Statistics – as on October 18, 2006
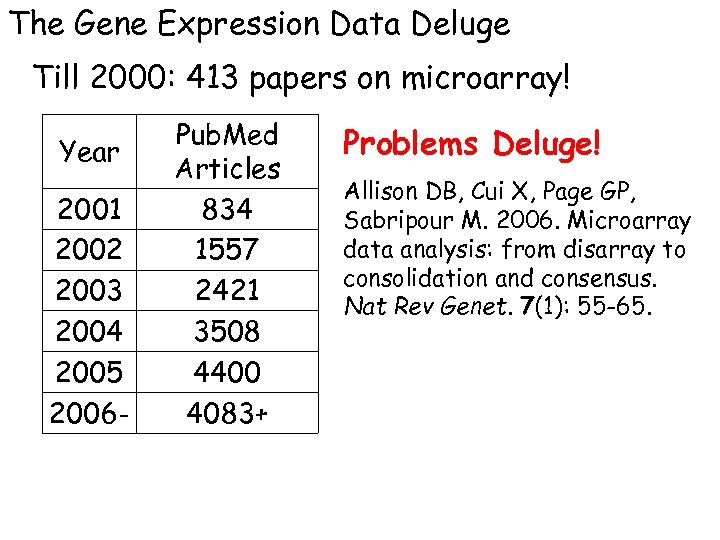 The Gene Expression Data Deluge Till 2000: 413 papers on microarray! Year 2001 2002 2003 2004 2005 2006 - Pub. Med Articles 834 1557 2421 3508 4400 4083+ Problems Deluge! Allison DB, Cui X, Page GP, Sabripour M. 2006. Microarray data analysis: from disarray to consolidation and consensus. Nat Rev Genet. 7(1): 55 -65.
The Gene Expression Data Deluge Till 2000: 413 papers on microarray! Year 2001 2002 2003 2004 2005 2006 - Pub. Med Articles 834 1557 2421 3508 4400 4083+ Problems Deluge! Allison DB, Cui X, Page GP, Sabripour M. 2006. Microarray data analysis: from disarray to consolidation and consensus. Nat Rev Genet. 7(1): 55 -65.
 Information Deluge…. . • 3 scientific journals in 1750 • Now - >120, 000 scientific journals! • >500, 000 medical articles/year • >4, 000 scientific articles/year • >16 million abstracts in Pub. Med derived from >32, 500 journals • >4. 5 billion distinct web pages indexed by Google! Google Search for integrative genomics: ~930, 000 hits “integrative genomics”: ~112, 000 hits A researcher would have to scan 130 different journals and read 27 papers per day to follow a single disease, such as breast cancer (Baasiri et al. , 1999 Oncogene 18: 7958 -7965).
Information Deluge…. . • 3 scientific journals in 1750 • Now - >120, 000 scientific journals! • >500, 000 medical articles/year • >4, 000 scientific articles/year • >16 million abstracts in Pub. Med derived from >32, 500 journals • >4. 5 billion distinct web pages indexed by Google! Google Search for integrative genomics: ~930, 000 hits “integrative genomics”: ~112, 000 hits A researcher would have to scan 130 different journals and read 27 papers per day to follow a single disease, such as breast cancer (Baasiri et al. , 1999 Oncogene 18: 7958 -7965).
 Data-driven Problems…. . What’s in a name! Rose is a rose! Gene Nomenclature Disease names • Accelerin • Draculin • • Antiquitin • Fidgetin • Bang Senseless • Gleeful • • Bride of Sevenless • Knobhead • • Christmas Factor • Lunatic Fringe • • Cockeye • Mortalin • • Crack • Orphanin • Draculin • Profilactin • Dickie’s small eye • Sonic Hedgehog Mobius Syndrome with Poland’s Anomaly Werner’s syndrome Down’s syndrome Angelman’s syndrome Creutzfeld-Jacob disease 1. Generally, the names refer to some feature of the mutant phenotype 2. Dickie’s small eye (Thieler et al. , 1978, Anat Embryol (Berl), 155: 81 -86) is now Pax 6 3. Gleeful: "This gene encodes a C 2 H 2 zinc finger transcription factor with high sequence similarity to vertebrate Gli proteins, so we have named the gene gleeful (Gfl). " (Furlong et al. , 2001, Science 293: 1632) • How to name or describe proteins, genes, drugs, diseases and conditions consistently and coherently? • How to ascribe and name a function, process or location consistently? • How to describe interactions, partners, reactions and complexes? Some Solutions • Develop/Use controlled or restricted vocabularies (IUPAC-like naming conventions, HGNC, MGI, UMLS, etc. ) • Create/Use thesauruses, central repositories or synonym lists (Me. SH, UMLS, etc. ) • Work towards synoptic reporting and structured abstracting
Data-driven Problems…. . What’s in a name! Rose is a rose! Gene Nomenclature Disease names • Accelerin • Draculin • • Antiquitin • Fidgetin • Bang Senseless • Gleeful • • Bride of Sevenless • Knobhead • • Christmas Factor • Lunatic Fringe • • Cockeye • Mortalin • • Crack • Orphanin • Draculin • Profilactin • Dickie’s small eye • Sonic Hedgehog Mobius Syndrome with Poland’s Anomaly Werner’s syndrome Down’s syndrome Angelman’s syndrome Creutzfeld-Jacob disease 1. Generally, the names refer to some feature of the mutant phenotype 2. Dickie’s small eye (Thieler et al. , 1978, Anat Embryol (Berl), 155: 81 -86) is now Pax 6 3. Gleeful: "This gene encodes a C 2 H 2 zinc finger transcription factor with high sequence similarity to vertebrate Gli proteins, so we have named the gene gleeful (Gfl). " (Furlong et al. , 2001, Science 293: 1632) • How to name or describe proteins, genes, drugs, diseases and conditions consistently and coherently? • How to ascribe and name a function, process or location consistently? • How to describe interactions, partners, reactions and complexes? Some Solutions • Develop/Use controlled or restricted vocabularies (IUPAC-like naming conventions, HGNC, MGI, UMLS, etc. ) • Create/Use thesauruses, central repositories or synonym lists (Me. SH, UMLS, etc. ) • Work towards synoptic reporting and structured abstracting
 Some more ambiguous examples……. . • The yeast homologue of the human gene PMS 1, which codes for a DNA repair protein, is called PMS 2; whereas yeast PMS 1 corresponds to human PMS 2! • Even more confusing, 4, 257 abbreviated names were used to refer to more than one gene. Top of the list was MT 1, used to describe at least 11 members of a cluster of genes encoding small proteins that bind to metal ions (Nature: 411: 631 -632). and there are some weird ones too……. . • AR*E: aryl sulfatase E in all species • f**K: fuculokinase gene in bacteria
Some more ambiguous examples……. . • The yeast homologue of the human gene PMS 1, which codes for a DNA repair protein, is called PMS 2; whereas yeast PMS 1 corresponds to human PMS 2! • Even more confusing, 4, 257 abbreviated names were used to refer to more than one gene. Top of the list was MT 1, used to describe at least 11 members of a cluster of genes encoding small proteins that bind to metal ions (Nature: 411: 631 -632). and there are some weird ones too……. . • AR*E: aryl sulfatase E in all species • f**K: fuculokinase gene in bacteria
 Rose is a rose…. . Not Really! What is a cell? • any small compartment • (biology) the basic structural and functional unit of all organisms; they may exist as independent units of life (as in monads) or may form colonies or tissues as in higher plants and animals • a device that delivers an electric current as a result of chemical reaction • a small unit serving as part of or as the nucleus of a larger political movement • cellular telephone: a hand-held mobile radiotelephone for use in an area divided into small sections, each with its own shortrange transmitter/receiver • small room in which a monk or nun lives • a room where a prisoner is kept Image Sources: Somewhere from the internet…
Rose is a rose…. . Not Really! What is a cell? • any small compartment • (biology) the basic structural and functional unit of all organisms; they may exist as independent units of life (as in monads) or may form colonies or tissues as in higher plants and animals • a device that delivers an electric current as a result of chemical reaction • a small unit serving as part of or as the nucleus of a larger political movement • cellular telephone: a hand-held mobile radiotelephone for use in an area divided into small sections, each with its own shortrange transmitter/receiver • small room in which a monk or nun lives • a room where a prisoner is kept Image Sources: Somewhere from the internet…
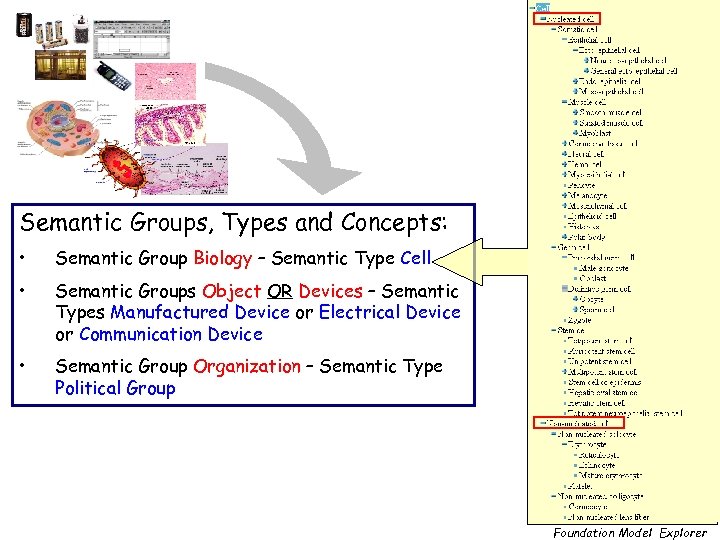 Semantic Groups, Types and Concepts: • Semantic Group Biology – Semantic Type Cell • Semantic Groups Object OR Devices – Semantic Types Manufactured Device or Electrical Device or Communication Device • Semantic Group Organization – Semantic Type Political Group Foundation Model Explorer
Semantic Groups, Types and Concepts: • Semantic Group Biology – Semantic Type Cell • Semantic Groups Object OR Devices – Semantic Types Manufactured Device or Electrical Device or Communication Device • Semantic Group Organization – Semantic Type Political Group Foundation Model Explorer
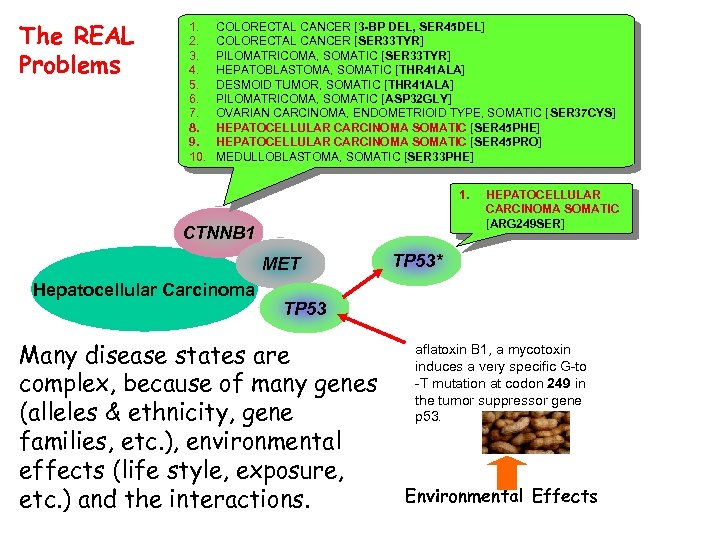 The REAL Problems 1. 2. 3. 4. 5. 6. 7. 8. 9. 10. COLORECTAL CANCER [3 -BP DEL, SER 45 DEL] COLORECTAL CANCER [SER 33 TYR] PILOMATRICOMA, SOMATIC [SER 33 TYR] HEPATOBLASTOMA, SOMATIC [THR 41 ALA] DESMOID TUMOR, SOMATIC [THR 41 ALA] PILOMATRICOMA, SOMATIC [ASP 32 GLY] OVARIAN CARCINOMA, ENDOMETRIOID TYPE, SOMATIC [SER 37 CYS] HEPATOCELLULAR CARCINOMA SOMATIC [SER 45 PHE] HEPATOCELLULAR CARCINOMA SOMATIC [SER 45 PRO] MEDULLOBLASTOMA, SOMATIC [SER 33 PHE] 1. CTNNB 1 MET Hepatocellular Carcinoma HEPATOCELLULAR CARCINOMA SOMATIC [ARG 249 SER] TP 53* TP 53 Many disease states are complex, because of many genes (alleles & ethnicity, gene families, etc. ), environmental effects (life style, exposure, etc. ) and the interactions. aflatoxin B 1, a mycotoxin induces a very specific G-to -T mutation at codon 249 in the tumor suppressor gene p 53. Environmental Effects
The REAL Problems 1. 2. 3. 4. 5. 6. 7. 8. 9. 10. COLORECTAL CANCER [3 -BP DEL, SER 45 DEL] COLORECTAL CANCER [SER 33 TYR] PILOMATRICOMA, SOMATIC [SER 33 TYR] HEPATOBLASTOMA, SOMATIC [THR 41 ALA] DESMOID TUMOR, SOMATIC [THR 41 ALA] PILOMATRICOMA, SOMATIC [ASP 32 GLY] OVARIAN CARCINOMA, ENDOMETRIOID TYPE, SOMATIC [SER 37 CYS] HEPATOCELLULAR CARCINOMA SOMATIC [SER 45 PHE] HEPATOCELLULAR CARCINOMA SOMATIC [SER 45 PRO] MEDULLOBLASTOMA, SOMATIC [SER 33 PHE] 1. CTNNB 1 MET Hepatocellular Carcinoma HEPATOCELLULAR CARCINOMA SOMATIC [ARG 249 SER] TP 53* TP 53 Many disease states are complex, because of many genes (alleles & ethnicity, gene families, etc. ), environmental effects (life style, exposure, etc. ) and the interactions. aflatoxin B 1, a mycotoxin induces a very specific G-to -T mutation at codon 249 in the tumor suppressor gene p 53. Environmental Effects
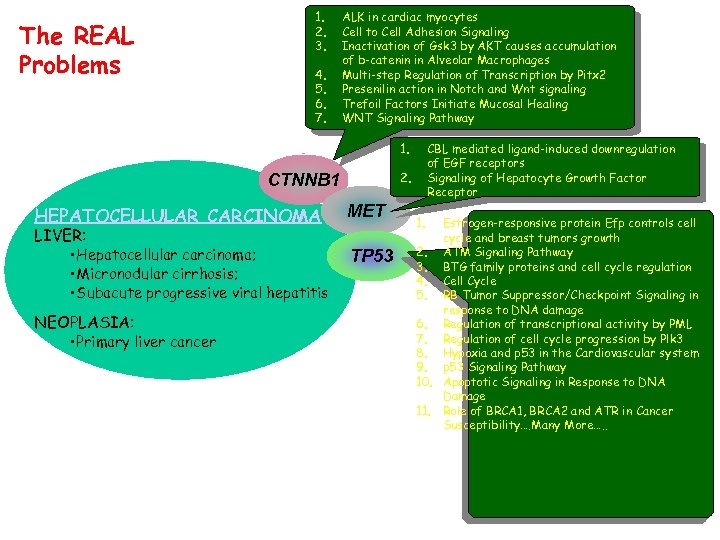 The REAL Problems 1. 2. 3. 4. 5. 6. 7. ALK in cardiac myocytes Cell to Cell Adhesion Signaling Inactivation of Gsk 3 by AKT causes accumulation of b-catenin in Alveolar Macrophages Multi-step Regulation of Transcription by Pitx 2 Presenilin action in Notch and Wnt signaling Trefoil Factors Initiate Mucosal Healing WNT Signaling Pathway 1. 2. CTNNB 1 HEPATOCELLULAR CARCINOMA LIVER: • Hepatocellular carcinoma; • Micronodular cirrhosis; • Subacute progressive viral hepatitis NEOPLASIA: • Primary liver cancer CBL mediated ligand-induced downregulation of EGF receptors Signaling of Hepatocyte Growth Factor Receptor MET TP 53 1. Estrogen-responsive protein Efp controls cell cycle and breast tumors growth 2. ATM Signaling Pathway 3. BTG family proteins and cell cycle regulation 4. Cell Cycle 5. RB Tumor Suppressor/Checkpoint Signaling in response to DNA damage 6. Regulation of transcriptional activity by PML 7. Regulation of cell cycle progression by Plk 3 8. Hypoxia and p 53 in the Cardiovascular system 9. p 53 Signaling Pathway 10. Apoptotic Signaling in Response to DNA Damage 11. Role of BRCA 1, BRCA 2 and ATR in Cancer Susceptibility…. Many More…. .
The REAL Problems 1. 2. 3. 4. 5. 6. 7. ALK in cardiac myocytes Cell to Cell Adhesion Signaling Inactivation of Gsk 3 by AKT causes accumulation of b-catenin in Alveolar Macrophages Multi-step Regulation of Transcription by Pitx 2 Presenilin action in Notch and Wnt signaling Trefoil Factors Initiate Mucosal Healing WNT Signaling Pathway 1. 2. CTNNB 1 HEPATOCELLULAR CARCINOMA LIVER: • Hepatocellular carcinoma; • Micronodular cirrhosis; • Subacute progressive viral hepatitis NEOPLASIA: • Primary liver cancer CBL mediated ligand-induced downregulation of EGF receptors Signaling of Hepatocyte Growth Factor Receptor MET TP 53 1. Estrogen-responsive protein Efp controls cell cycle and breast tumors growth 2. ATM Signaling Pathway 3. BTG family proteins and cell cycle regulation 4. Cell Cycle 5. RB Tumor Suppressor/Checkpoint Signaling in response to DNA damage 6. Regulation of transcriptional activity by PML 7. Regulation of cell cycle progression by Plk 3 8. Hypoxia and p 53 in the Cardiovascular system 9. p 53 Signaling Pathway 10. Apoptotic Signaling in Response to DNA Damage 11. Role of BRCA 1, BRCA 2 and ATR in Cancer Susceptibility…. Many More…. .
 Integrative Genomics - what is it? Another buzzword or a meaningful concept useful for biomedical research? Acquisition, Integration, Curation, and Analysis of biological data Hypothesis Integrative Genomics: the study of complex interactions between genes, organism and environment, the triple helix of biology. Gene <–> Organism <-> Environment It is definitely beyond the buzzword stage - Universities now have programs named 'Integrated Genomics. ' Information is not knowledge - Albert Einstein
Integrative Genomics - what is it? Another buzzword or a meaningful concept useful for biomedical research? Acquisition, Integration, Curation, and Analysis of biological data Hypothesis Integrative Genomics: the study of complex interactions between genes, organism and environment, the triple helix of biology. Gene <–> Organism <-> Environment It is definitely beyond the buzzword stage - Universities now have programs named 'Integrated Genomics. ' Information is not knowledge - Albert Einstein
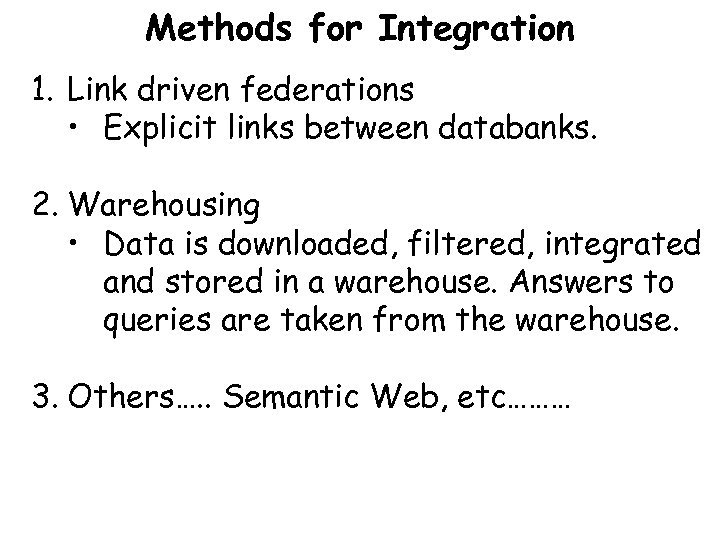 Methods for Integration 1. Link driven federations • Explicit links between databanks. 2. Warehousing • Data is downloaded, filtered, integrated and stored in a warehouse. Answers to queries are taken from the warehouse. 3. Others…. . Semantic Web, etc………
Methods for Integration 1. Link driven federations • Explicit links between databanks. 2. Warehousing • Data is downloaded, filtered, integrated and stored in a warehouse. Answers to queries are taken from the warehouse. 3. Others…. . Semantic Web, etc………
 Link-driven Federations 1. Creates explicit links between databanks 2. query: get interesting results and use web links to reach related data in other databanks Examples: NCBI-Entrez, SRS
Link-driven Federations 1. Creates explicit links between databanks 2. query: get interesting results and use web links to reach related data in other databanks Examples: NCBI-Entrez, SRS
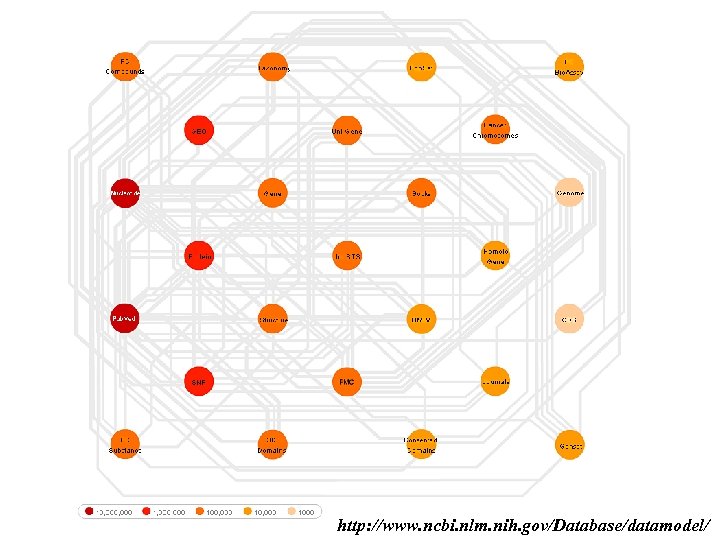 http: //www. ncbi. nlm. nih. gov/Database/datamodel/
http: //www. ncbi. nlm. nih. gov/Database/datamodel/
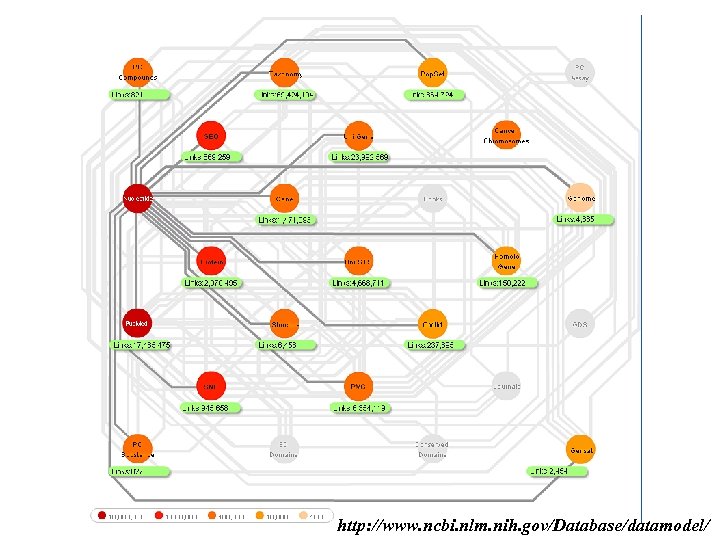 http: //www. ncbi. nlm. nih. gov/Database/datamodel/
http: //www. ncbi. nlm. nih. gov/Database/datamodel/
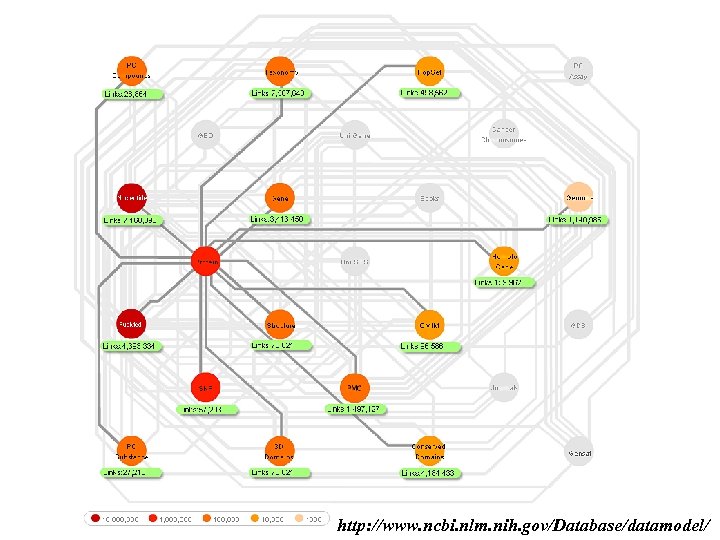 http: //www. ncbi. nlm. nih. gov/Database/datamodel/
http: //www. ncbi. nlm. nih. gov/Database/datamodel/
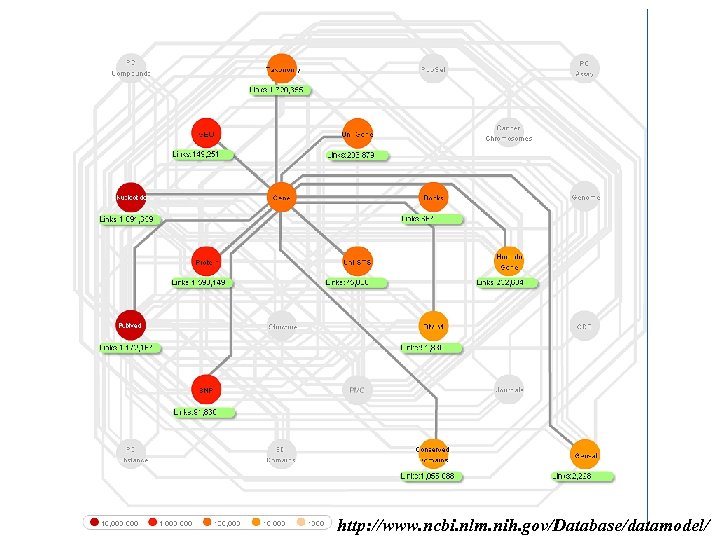 http: //www. ncbi. nlm. nih. gov/Database/datamodel/
http: //www. ncbi. nlm. nih. gov/Database/datamodel/
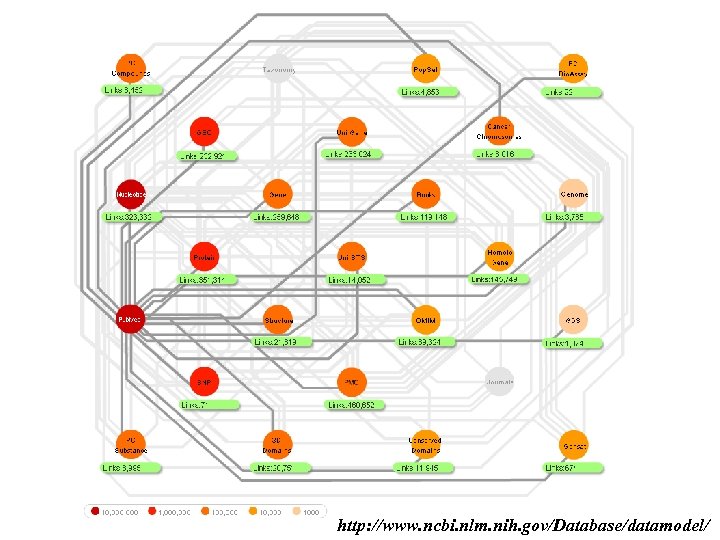 http: //www. ncbi. nlm. nih. gov/Database/datamodel/
http: //www. ncbi. nlm. nih. gov/Database/datamodel/
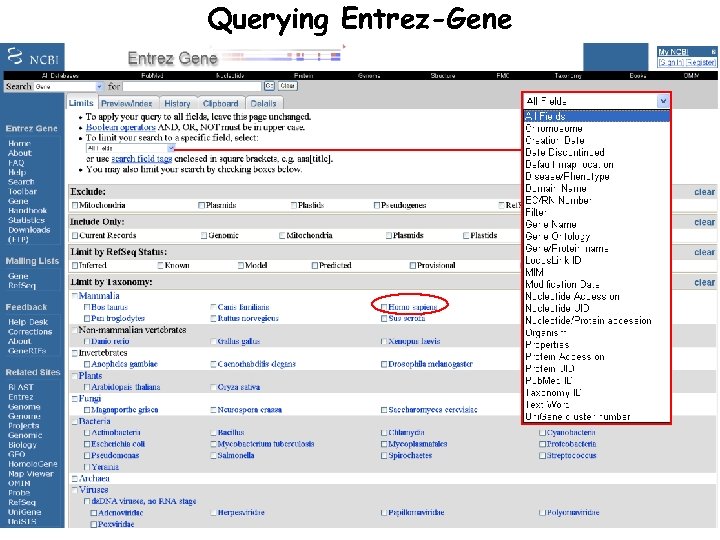 Querying Entrez-Gene
Querying Entrez-Gene
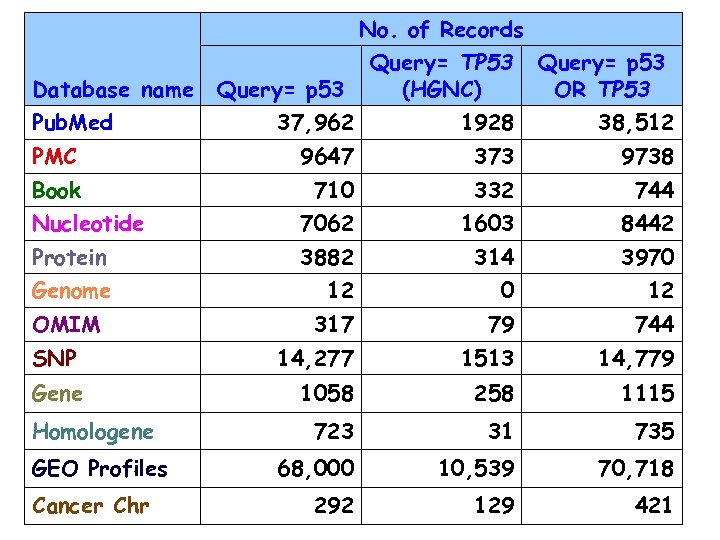 No. of Records Database name Pub. Med Query= p 53 Query= TP 53 (HGNC) Query= p 53 OR TP 53 37, 962 1928 38, 512 PMC 9647 373 9738 Book 710 332 744 Nucleotide 7062 1603 8442 Protein 3882 314 3970 Genome 12 0 12 317 79 744 SNP 14, 277 1513 14, 779 Gene 1058 258 1115 723 31 735 68, 000 10, 539 70, 718 292 129 421 OMIM Homologene GEO Profiles Cancer Chr
No. of Records Database name Pub. Med Query= p 53 Query= TP 53 (HGNC) Query= p 53 OR TP 53 37, 962 1928 38, 512 PMC 9647 373 9738 Book 710 332 744 Nucleotide 7062 1603 8442 Protein 3882 314 3970 Genome 12 0 12 317 79 744 SNP 14, 277 1513 14, 779 Gene 1058 258 1115 723 31 735 68, 000 10, 539 70, 718 292 129 421 OMIM Homologene GEO Profiles Cancer Chr
 Link-driven Federations 1. Advantages • • complex queries Fast • • • require good knowledge syntax based terminology problem not solved 2. Disadvantages
Link-driven Federations 1. Advantages • • complex queries Fast • • • require good knowledge syntax based terminology problem not solved 2. Disadvantages
 Data Warehousing Data is downloaded, filtered, integrated and stored in a warehouse. Answers to queries are taken from the warehouse. Advantages Disadvantages 1. Good for very-specific, task-based queries and studies. 1. Can become quickly outdated – needs constant updates. 2. Since it is custom-built and usually expertcurated, relatively less error-prone. 2. Limited functionality – For e. g. , one diseasebased or one systembased.
Data Warehousing Data is downloaded, filtered, integrated and stored in a warehouse. Answers to queries are taken from the warehouse. Advantages Disadvantages 1. Good for very-specific, task-based queries and studies. 1. Can become quickly outdated – needs constant updates. 2. Since it is custom-built and usually expertcurated, relatively less error-prone. 2. Limited functionality – For e. g. , one diseasebased or one systembased.
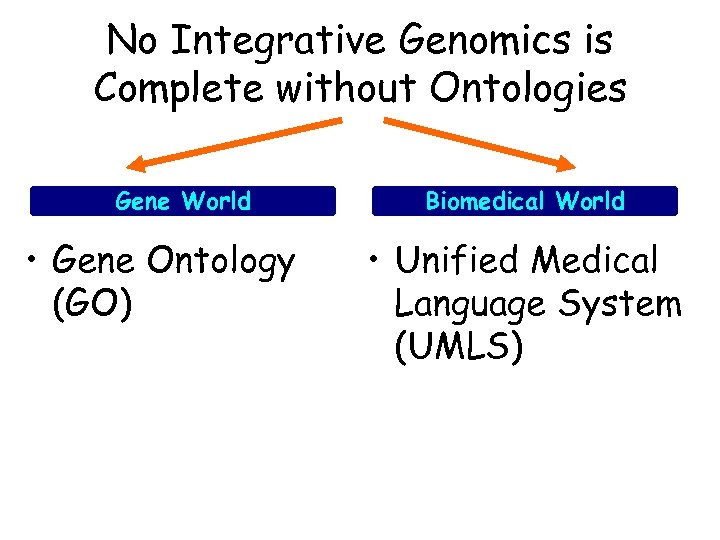 No Integrative Genomics is Complete without Ontologies Gene World • Gene Ontology (GO) Biomedical World • Unified Medical Language System (UMLS)
No Integrative Genomics is Complete without Ontologies Gene World • Gene Ontology (GO) Biomedical World • Unified Medical Language System (UMLS)
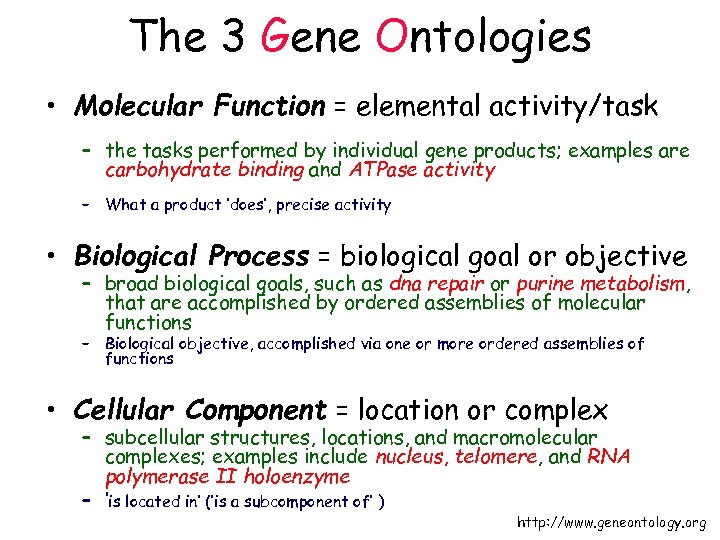 The 3 Gene Ontologies • Molecular Function = elemental activity/task – the tasks performed by individual gene products; examples are carbohydrate binding and ATPase activity – What a product ‘does’, precise activity • Biological Process = biological goal or objective – broad biological goals, such as dna repair or purine metabolism, that are accomplished by ordered assemblies of molecular functions – Biological objective, accomplished via one or more ordered assemblies of functions • Cellular Component = location or complex – subcellular structures, locations, and macromolecular complexes; examples include nucleus, telomere, and RNA polymerase II holoenzyme – ‘is located in’ (‘is a subcomponent of’ ) http: //www. geneontology. org
The 3 Gene Ontologies • Molecular Function = elemental activity/task – the tasks performed by individual gene products; examples are carbohydrate binding and ATPase activity – What a product ‘does’, precise activity • Biological Process = biological goal or objective – broad biological goals, such as dna repair or purine metabolism, that are accomplished by ordered assemblies of molecular functions – Biological objective, accomplished via one or more ordered assemblies of functions • Cellular Component = location or complex – subcellular structures, locations, and macromolecular complexes; examples include nucleus, telomere, and RNA polymerase II holoenzyme – ‘is located in’ (‘is a subcomponent of’ ) http: //www. geneontology. org
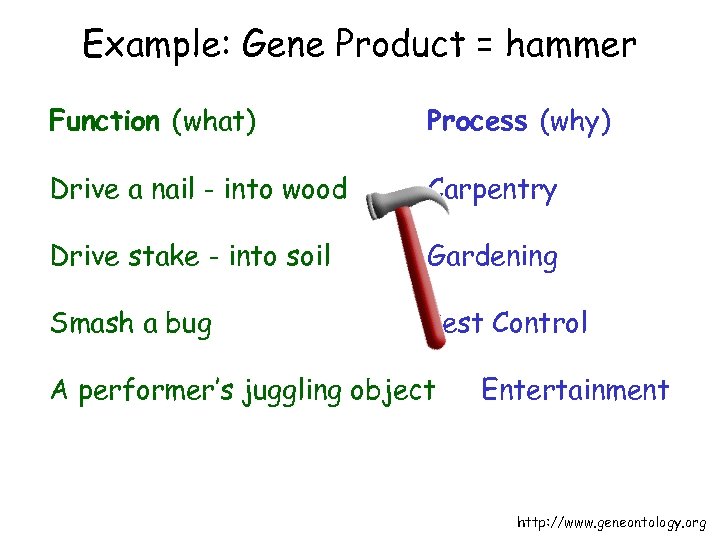 Example: Gene Product = hammer Function (what) Process (why) Drive a nail - into wood Carpentry Drive stake - into soil Gardening Smash a bug Pest Control A performer’s juggling object Entertainment http: //www. geneontology. org
Example: Gene Product = hammer Function (what) Process (why) Drive a nail - into wood Carpentry Drive stake - into soil Gardening Smash a bug Pest Control A performer’s juggling object Entertainment http: //www. geneontology. org
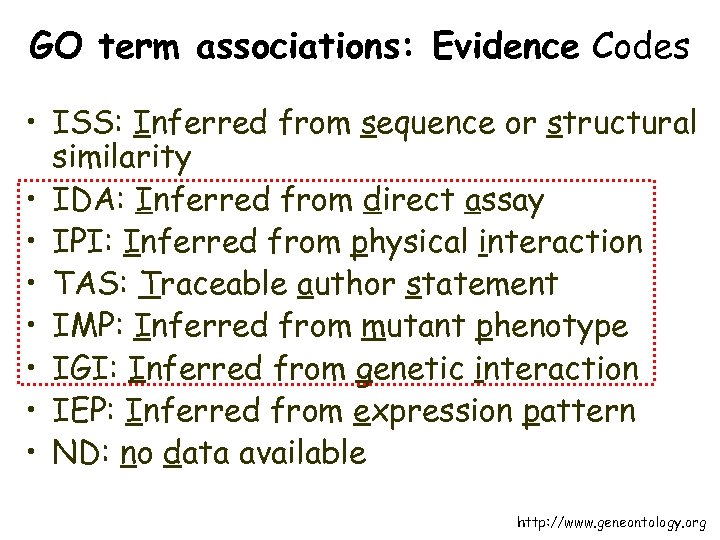 GO term associations: Evidence Codes • ISS: Inferred from sequence or structural similarity • IDA: Inferred from direct assay • IPI: Inferred from physical interaction • TAS: Traceable author statement • IMP: Inferred from mutant phenotype • IGI: Inferred from genetic interaction • IEP: Inferred from expression pattern • ND: no data available http: //www. geneontology. org
GO term associations: Evidence Codes • ISS: Inferred from sequence or structural similarity • IDA: Inferred from direct assay • IPI: Inferred from physical interaction • TAS: Traceable author statement • IMP: Inferred from mutant phenotype • IGI: Inferred from genetic interaction • IEP: Inferred from expression pattern • ND: no data available http: //www. geneontology. org
 What can researchers do with GO? • Access gene product functional information • Find how much of a proteome is involved in a process/ function/ component in the cell • Map GO terms and incorporate manual annotations into own databases • Provide a link between biological knowledge and • gene expression profiles • proteomics data And how? • Getting the GO and GO_Association Files • Data Mining – My Favorite Gene – By GO – By Sequence • Analysis of Data – Clustering by function/process • Other Tools
What can researchers do with GO? • Access gene product functional information • Find how much of a proteome is involved in a process/ function/ component in the cell • Map GO terms and incorporate manual annotations into own databases • Provide a link between biological knowledge and • gene expression profiles • proteomics data And how? • Getting the GO and GO_Association Files • Data Mining – My Favorite Gene – By GO – By Sequence • Analysis of Data – Clustering by function/process • Other Tools
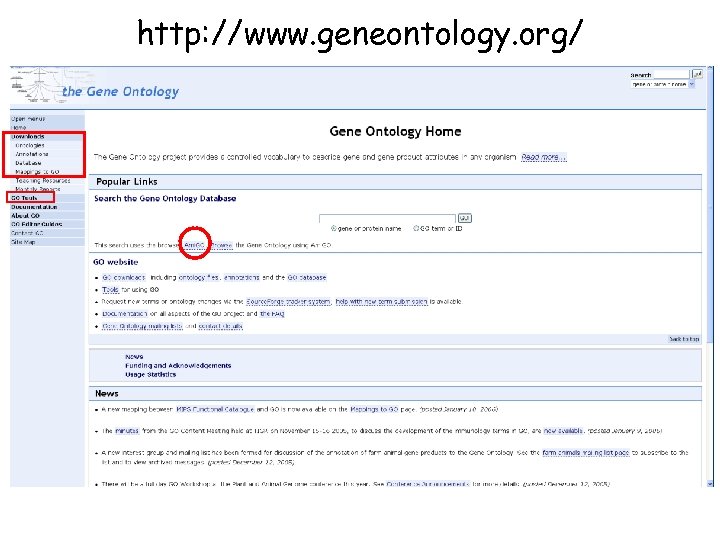 http: //www. geneontology. org/
http: //www. geneontology. org/
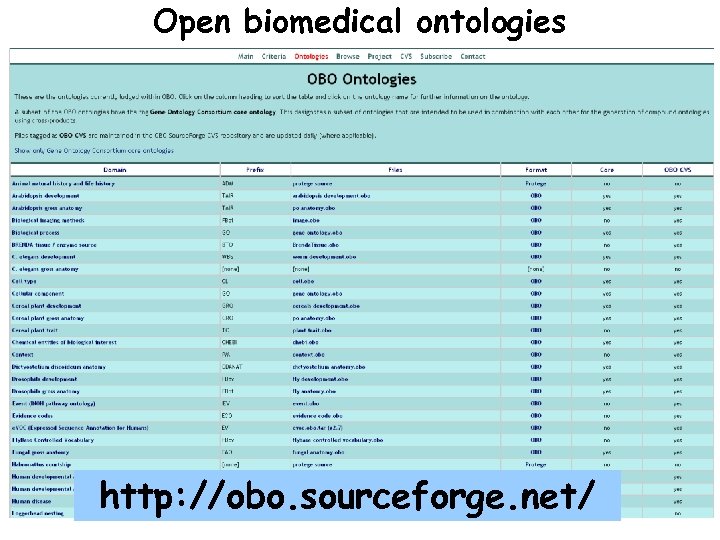 Open biomedical ontologies http: //obo. sourceforge. net/
Open biomedical ontologies http: //obo. sourceforge. net/
 Unified Medical Language System Knowledge Server– UMLSKS http: //umlsks. nlm. nih. gov/kss/ • The UMLS Metathesaurus contains information about biomedical concepts and terms from many controlled vocabularies and classifications used in patient records, administrative health data, bibliographic and full-text databases, and expert systems. • The Semantic Network, through its semantic types, provides a consistent categorization of all concepts represented in the UMLS Metathesaurus. The links between the semantic types provide the structure for the Network and represent important relationships in the biomedical domain. • The SPECIALIST Lexicon is an English language lexicon with many biomedical terms, containing syntactic, morphological, and orthographic information for each term or word.
Unified Medical Language System Knowledge Server– UMLSKS http: //umlsks. nlm. nih. gov/kss/ • The UMLS Metathesaurus contains information about biomedical concepts and terms from many controlled vocabularies and classifications used in patient records, administrative health data, bibliographic and full-text databases, and expert systems. • The Semantic Network, through its semantic types, provides a consistent categorization of all concepts represented in the UMLS Metathesaurus. The links between the semantic types provide the structure for the Network and represent important relationships in the biomedical domain. • The SPECIALIST Lexicon is an English language lexicon with many biomedical terms, containing syntactic, morphological, and orthographic information for each term or word.
 • • • Unified Medical Language System Metathesaurus about over 1 million biomedical concepts About 5 million concept names from more than 100 controlled vocabularies and classifications (some in multiple languages) used in patient records, administrative health data, bibliographic and full-text databases and expert systems. The Metathesaurus is organized by concept or meaning. Alternate names for the same concept (synonyms, lexical variants, and translations) are linked together. Each Metathesaurus concept has attributes that help to define its meaning, e. g. , the semantic type(s) or categories to which it belongs, its position in the hierarchical contexts from various source vocabularies, and, for many concepts, a definition. Customizable: Users can exclude vocabularies that are not relevant for specific purposes or not licensed for use in their institutions. Metamorpho. Sys, the multi-platform Java install and customization program distributed with the UMLS resources, helps users to generate pre-defined or custom subsets of the Metathesaurus. • Uses: – linking between different clinical or biomedical vocabularies – information retrieval from databases with human assigned subject index terms and from free-text information sources – linking patient records to related information in bibliographic, full-text, or factual databases – natural language processing and automated indexing research
• • • Unified Medical Language System Metathesaurus about over 1 million biomedical concepts About 5 million concept names from more than 100 controlled vocabularies and classifications (some in multiple languages) used in patient records, administrative health data, bibliographic and full-text databases and expert systems. The Metathesaurus is organized by concept or meaning. Alternate names for the same concept (synonyms, lexical variants, and translations) are linked together. Each Metathesaurus concept has attributes that help to define its meaning, e. g. , the semantic type(s) or categories to which it belongs, its position in the hierarchical contexts from various source vocabularies, and, for many concepts, a definition. Customizable: Users can exclude vocabularies that are not relevant for specific purposes or not licensed for use in their institutions. Metamorpho. Sys, the multi-platform Java install and customization program distributed with the UMLS resources, helps users to generate pre-defined or custom subsets of the Metathesaurus. • Uses: – linking between different clinical or biomedical vocabularies – information retrieval from databases with human assigned subject index terms and from free-text information sources – linking patient records to related information in bibliographic, full-text, or factual databases – natural language processing and automated indexing research
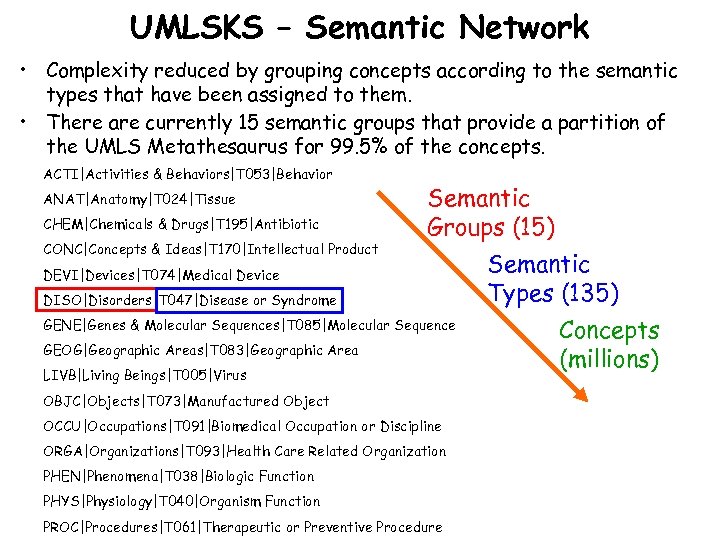 UMLSKS – Semantic Network • Complexity reduced by grouping concepts according to the semantic types that have been assigned to them. • There are currently 15 semantic groups that provide a partition of the UMLS Metathesaurus for 99. 5% of the concepts. ACTI|Activities & Behaviors|T 053|Behavior ANAT|Anatomy|T 024|Tissue CHEM|Chemicals & Drugs|T 195|Antibiotic CONC|Concepts & Ideas|T 170|Intellectual Product Semantic Groups (15) DEVI|Devices|T 074|Medical Device DISO|Disorders|T 047|Disease or Syndrome GENE|Genes & Molecular Sequences|T 085|Molecular Sequence GEOG|Geographic Areas|T 083|Geographic Area LIVB|Living Beings|T 005|Virus OBJC|Objects|T 073|Manufactured Object OCCU|Occupations|T 091|Biomedical Occupation or Discipline ORGA|Organizations|T 093|Health Care Related Organization PHEN|Phenomena|T 038|Biologic Function PHYS|Physiology|T 040|Organism Function PROC|Procedures|T 061|Therapeutic or Preventive Procedure Semantic Types (135) Concepts (millions)
UMLSKS – Semantic Network • Complexity reduced by grouping concepts according to the semantic types that have been assigned to them. • There are currently 15 semantic groups that provide a partition of the UMLS Metathesaurus for 99. 5% of the concepts. ACTI|Activities & Behaviors|T 053|Behavior ANAT|Anatomy|T 024|Tissue CHEM|Chemicals & Drugs|T 195|Antibiotic CONC|Concepts & Ideas|T 170|Intellectual Product Semantic Groups (15) DEVI|Devices|T 074|Medical Device DISO|Disorders|T 047|Disease or Syndrome GENE|Genes & Molecular Sequences|T 085|Molecular Sequence GEOG|Geographic Areas|T 083|Geographic Area LIVB|Living Beings|T 005|Virus OBJC|Objects|T 073|Manufactured Object OCCU|Occupations|T 091|Biomedical Occupation or Discipline ORGA|Organizations|T 093|Health Care Related Organization PHEN|Phenomena|T 038|Biologic Function PHYS|Physiology|T 040|Organism Function PROC|Procedures|T 061|Therapeutic or Preventive Procedure Semantic Types (135) Concepts (millions)
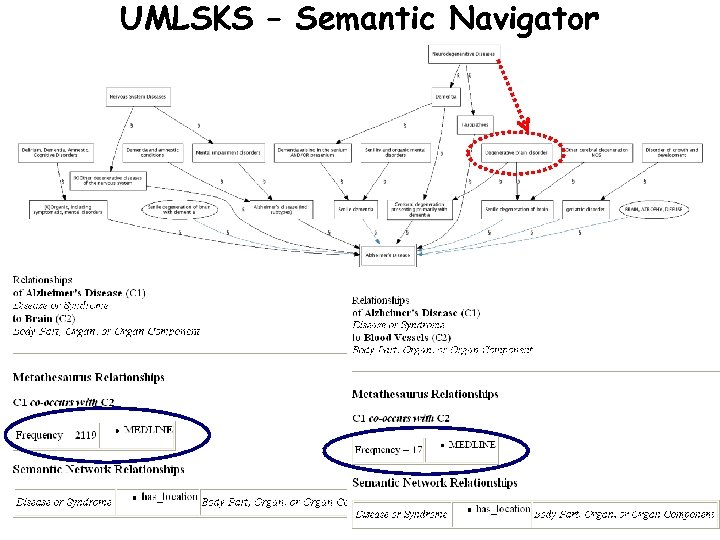 UMLSKS – Semantic Navigator
UMLSKS – Semantic Navigator
 Alzheimer’s Disease – Alarming Statistics • The number of patients with AD in any community depends on the proportion of older people in the group. Traditionally, the developed countries had large proportions of elderly people, and so they had very many cases of Alzheimer’s disease in the community at one time. • 4. 5 million AD patients in the United States today. • Expected to increase to 11 to 16 million by 2050. • In 2000, health care costs for AD patients in the United States totaled approximately $31. 9 billion, which is expected to reach $49. 3 billion by 2010 (http: //www. alz. org) • World-wide: ~18 million (projected to nearly double by 2025 to 34 million). • Demographic transition - Developing countries: • • 1991 India Census: 70 million people were over 60 years. • 2001 India Census: 77 million, or 7. 6% of the population. • • Increased life expectancy (current life expectancy in India is >60 years). By 2025, we will have 177 million elderly people. Currently, more than 50% of people with Alzheimer’s disease live in developing countries and by 2025, this will be over 70%. Source: WHO & NIA
Alzheimer’s Disease – Alarming Statistics • The number of patients with AD in any community depends on the proportion of older people in the group. Traditionally, the developed countries had large proportions of elderly people, and so they had very many cases of Alzheimer’s disease in the community at one time. • 4. 5 million AD patients in the United States today. • Expected to increase to 11 to 16 million by 2050. • In 2000, health care costs for AD patients in the United States totaled approximately $31. 9 billion, which is expected to reach $49. 3 billion by 2010 (http: //www. alz. org) • World-wide: ~18 million (projected to nearly double by 2025 to 34 million). • Demographic transition - Developing countries: • • 1991 India Census: 70 million people were over 60 years. • 2001 India Census: 77 million, or 7. 6% of the population. • • Increased life expectancy (current life expectancy in India is >60 years). By 2025, we will have 177 million elderly people. Currently, more than 50% of people with Alzheimer’s disease live in developing countries and by 2025, this will be over 70%. Source: WHO & NIA
 Alzheimer’s Disease – Why Computational Approaches? • The goal of applying computational data-mining approaches is to extract useful information from large amounts of data by employing mathematical methods that should be as automated as possible. • Computational data-mining approaches are particularly appropriate in areas with much data but few explanations, such as gerontology. If researchers can find/derive patterns in data to perceive information, then information may enhance our knowledge over aging. • The complexity and broad range of cellular and biochemical events make researchers believe that there must be a sophisticated network of AD signal transduction, gene regulation, and protein-protein interaction events. • Therefore, deciphering AD-related molecular network “circuitry” can help researchers understand AD disease better, model details, and propose treatment ideas.
Alzheimer’s Disease – Why Computational Approaches? • The goal of applying computational data-mining approaches is to extract useful information from large amounts of data by employing mathematical methods that should be as automated as possible. • Computational data-mining approaches are particularly appropriate in areas with much data but few explanations, such as gerontology. If researchers can find/derive patterns in data to perceive information, then information may enhance our knowledge over aging. • The complexity and broad range of cellular and biochemical events make researchers believe that there must be a sophisticated network of AD signal transduction, gene regulation, and protein-protein interaction events. • Therefore, deciphering AD-related molecular network “circuitry” can help researchers understand AD disease better, model details, and propose treatment ideas.
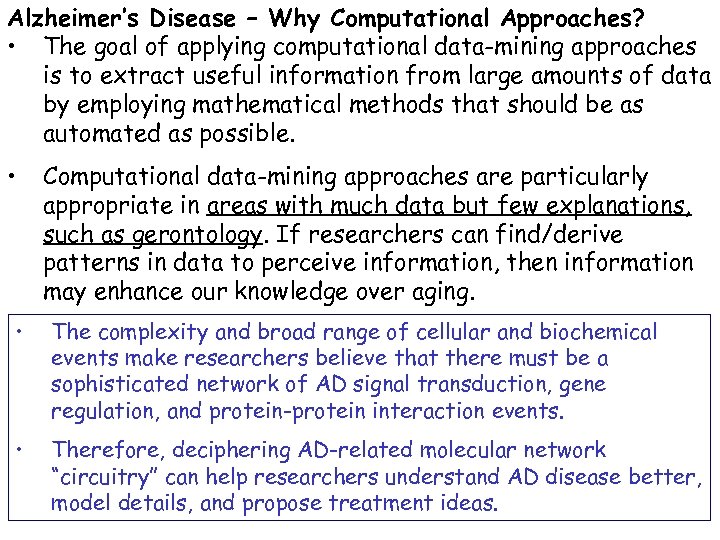
 A simplistic picture Frontal Lobe Temporal Lobe Hippocampus Cerebral Cortex Astrocytes Basal Nucleus of Meynert Cerebrum Brain and Nervous System A 2 M APOE Microglia Alzheimer Disease ALOX 12 ABCA 1 ABCA 2 NME 1 Neurons NEF 3 PARK 2 STH APP
A simplistic picture Frontal Lobe Temporal Lobe Hippocampus Cerebral Cortex Astrocytes Basal Nucleus of Meynert Cerebrum Brain and Nervous System A 2 M APOE Microglia Alzheimer Disease ALOX 12 ABCA 1 ABCA 2 NME 1 Neurons NEF 3 PARK 2 STH APP
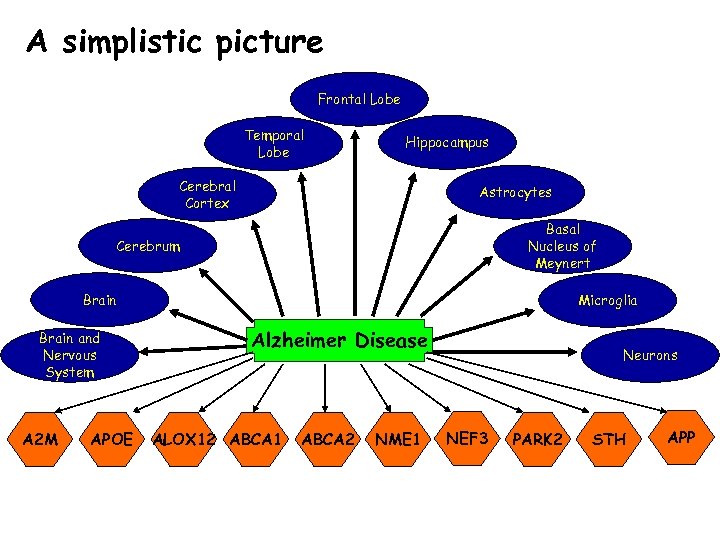
 Frontal Lobe Temporal Lobe Hippocampus Cerebral Cortex Astrocytes Basal Nucleus of Meynert Cerebrum Brain and Nervous System A 2 M APOE Microglia Alzheimer Disease ALOX 12 ABCA 1 ABCA 2 NME 1 Neurons NEF 3 PARK 2 STH APP
Frontal Lobe Temporal Lobe Hippocampus Cerebral Cortex Astrocytes Basal Nucleus of Meynert Cerebrum Brain and Nervous System A 2 M APOE Microglia Alzheimer Disease ALOX 12 ABCA 1 ABCA 2 NME 1 Neurons NEF 3 PARK 2 STH APP
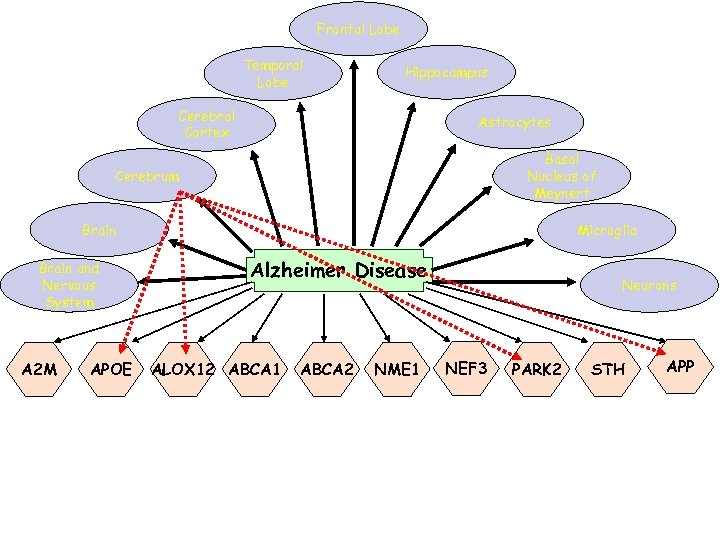
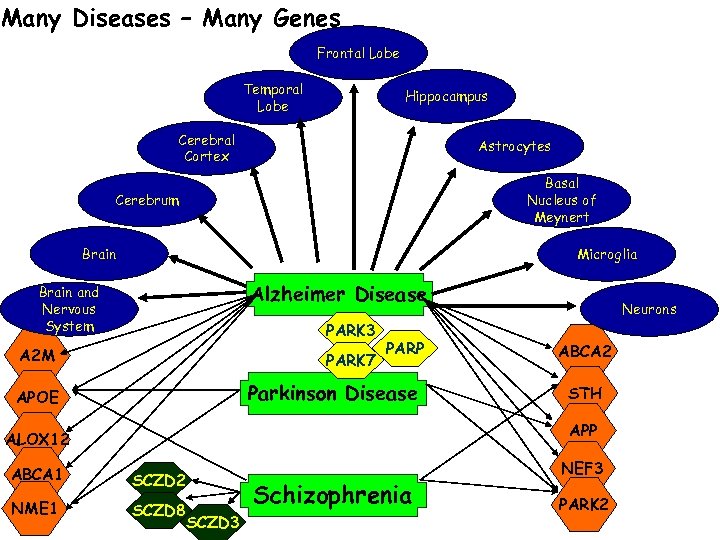 Many Diseases – Many Genes Frontal Lobe Temporal Lobe Hippocampus Cerebral Cortex Astrocytes Basal Nucleus of Meynert Cerebrum Brain Microglia Alzheimer Disease Brain and Nervous System PARK 3 A 2 M PARK 7 PARP Parkinson Disease APOE Neurons ABCA 2 STH APP ALOX 12 ABCA 1 SCZD 2 NME 1 SCZD 8 SCZD 3 Schizophrenia NEF 3 PARK 2
Many Diseases – Many Genes Frontal Lobe Temporal Lobe Hippocampus Cerebral Cortex Astrocytes Basal Nucleus of Meynert Cerebrum Brain Microglia Alzheimer Disease Brain and Nervous System PARK 3 A 2 M PARK 7 PARP Parkinson Disease APOE Neurons ABCA 2 STH APP ALOX 12 ABCA 1 SCZD 2 NME 1 SCZD 8 SCZD 3 Schizophrenia NEF 3 PARK 2
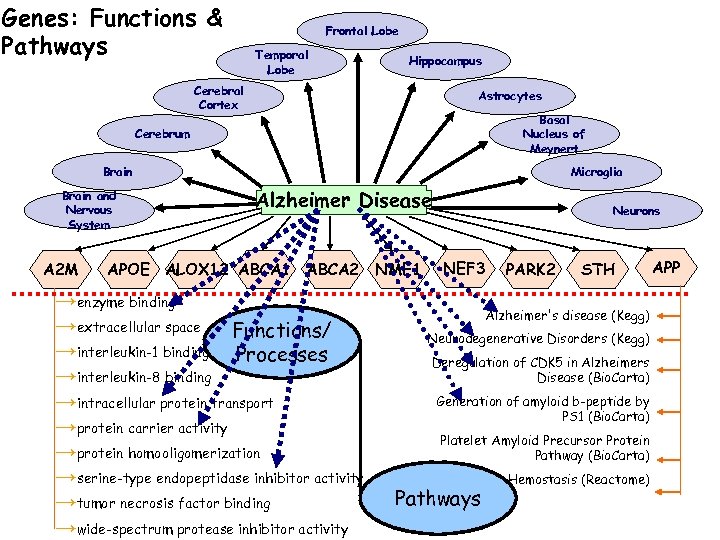 Genes: Functions & Pathways Frontal Lobe Temporal Lobe Hippocampus Cerebral Cortex Astrocytes Basal Nucleus of Meynert Cerebrum Brain and Nervous System A 2 M Microglia Alzheimer Disease APOE ALOX 12 ABCA 1 ABCA 2 →enzyme binding →extracellular space Functions/ →interleukin-1 binding Processes →interleukin-8 binding →intracellular protein transport →protein carrier activity →protein homooligomerization →serine-type endopeptidase inhibitor activity →tumor necrosis factor binding →wide-spectrum protease inhibitor activity NME 1 Neurons NEF 3 PARK 2 STH Alzheimer's disease (Kegg) Neurodegenerative Disorders (Kegg) Deregulation of CDK 5 in Alzheimers Disease (Bio. Carta) Generation of amyloid b-peptide by PS 1 (Bio. Carta) Platelet Amyloid Precursor Protein Pathway (Bio. Carta) Pathways Hemostasis (Reactome) APP
Genes: Functions & Pathways Frontal Lobe Temporal Lobe Hippocampus Cerebral Cortex Astrocytes Basal Nucleus of Meynert Cerebrum Brain and Nervous System A 2 M Microglia Alzheimer Disease APOE ALOX 12 ABCA 1 ABCA 2 →enzyme binding →extracellular space Functions/ →interleukin-1 binding Processes →interleukin-8 binding →intracellular protein transport →protein carrier activity →protein homooligomerization →serine-type endopeptidase inhibitor activity →tumor necrosis factor binding →wide-spectrum protease inhibitor activity NME 1 Neurons NEF 3 PARK 2 STH Alzheimer's disease (Kegg) Neurodegenerative Disorders (Kegg) Deregulation of CDK 5 in Alzheimers Disease (Bio. Carta) Generation of amyloid b-peptide by PS 1 (Bio. Carta) Platelet Amyloid Precursor Protein Pathway (Bio. Carta) Pathways Hemostasis (Reactome) APP
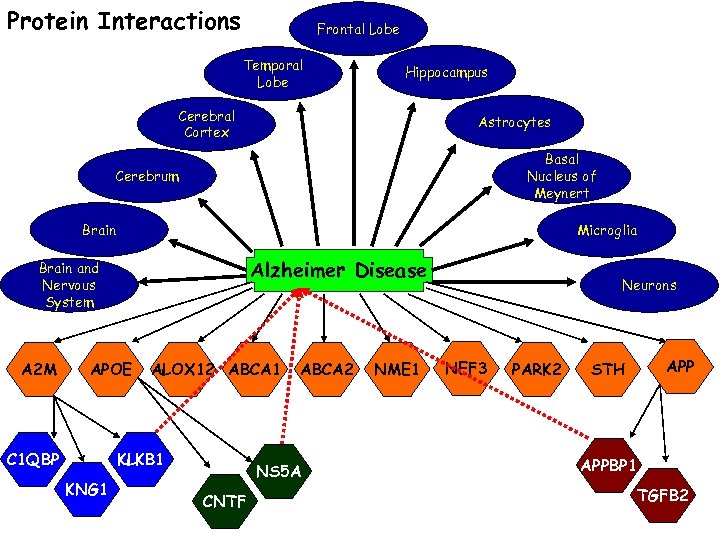 Protein Interactions Frontal Lobe Temporal Lobe Hippocampus Cerebral Cortex Astrocytes Basal Nucleus of Meynert Cerebrum Brain Microglia Alzheimer Disease Brain and Nervous System A 2 M APOE C 1 QBP ALOX 12 ABCA 1 KLKB 1 KNG 1 ABCA 2 NS 5 A CNTF NME 1 Neurons NEF 3 PARK 2 APP STH APPBP 1 TGFB 2
Protein Interactions Frontal Lobe Temporal Lobe Hippocampus Cerebral Cortex Astrocytes Basal Nucleus of Meynert Cerebrum Brain Microglia Alzheimer Disease Brain and Nervous System A 2 M APOE C 1 QBP ALOX 12 ABCA 1 KLKB 1 KNG 1 ABCA 2 NS 5 A CNTF NME 1 Neurons NEF 3 PARK 2 APP STH APPBP 1 TGFB 2
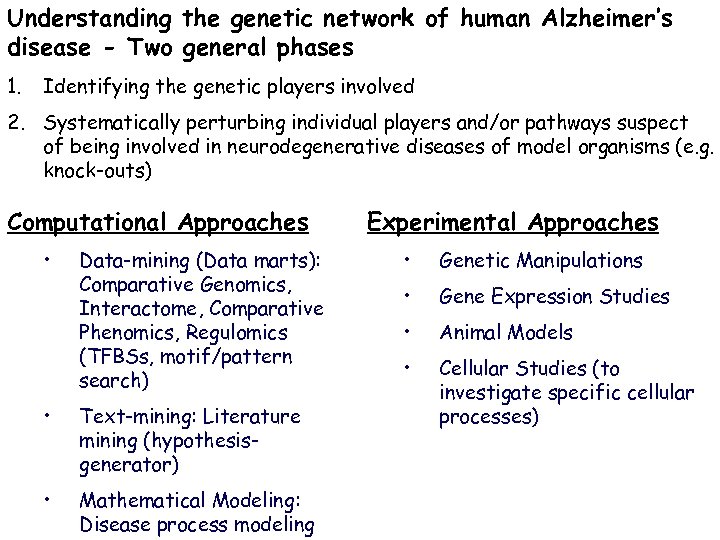 Understanding the genetic network of human Alzheimer’s disease - Two general phases 1. Identifying the genetic players involved 2. Systematically perturbing individual players and/or pathways suspect of being involved in neurodegenerative diseases of model organisms (e. g. knock-outs) Computational Approaches • Data-mining (Data marts): Comparative Genomics, Interactome, Comparative Phenomics, Regulomics (TFBSs, motif/pattern search) • Text-mining: Literature mining (hypothesisgenerator) • Mathematical Modeling: Disease process modeling Experimental Approaches • Genetic Manipulations • Gene Expression Studies • Animal Models • Cellular Studies (to investigate specific cellular processes)
Understanding the genetic network of human Alzheimer’s disease - Two general phases 1. Identifying the genetic players involved 2. Systematically perturbing individual players and/or pathways suspect of being involved in neurodegenerative diseases of model organisms (e. g. knock-outs) Computational Approaches • Data-mining (Data marts): Comparative Genomics, Interactome, Comparative Phenomics, Regulomics (TFBSs, motif/pattern search) • Text-mining: Literature mining (hypothesisgenerator) • Mathematical Modeling: Disease process modeling Experimental Approaches • Genetic Manipulations • Gene Expression Studies • Animal Models • Cellular Studies (to investigate specific cellular processes)
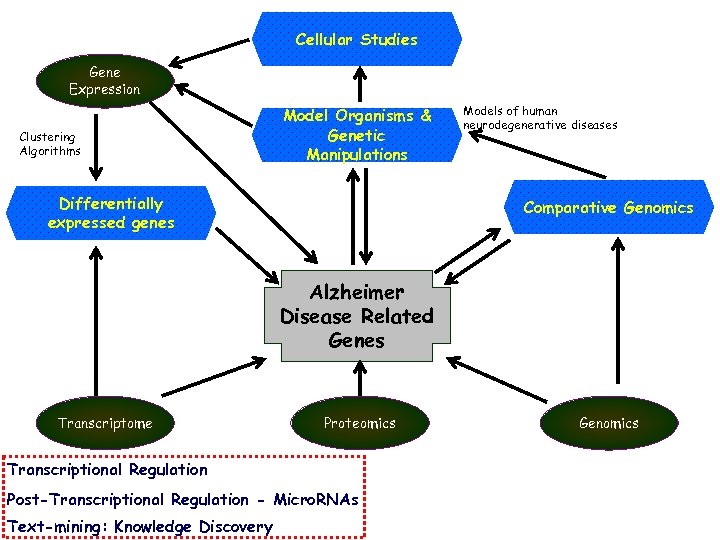 Cellular Studies Gene Expression Clustering Algorithms Model Organisms & Genetic Manipulations Differentially expressed genes Models of human neurodegenerative diseases Comparative Genomics Alzheimer Disease Related Genes Transcriptome Proteomics Transcriptional Regulation Post-Transcriptional Regulation - Micro. RNAs Text-mining: Knowledge Discovery Genomics
Cellular Studies Gene Expression Clustering Algorithms Model Organisms & Genetic Manipulations Differentially expressed genes Models of human neurodegenerative diseases Comparative Genomics Alzheimer Disease Related Genes Transcriptome Proteomics Transcriptional Regulation Post-Transcriptional Regulation - Micro. RNAs Text-mining: Knowledge Discovery Genomics
![NCBI Entrez Gene Query: (alzheimer[Disease/Phenotype] OR alzheimer[All fields]) AND NCBI Entrez Gene Query: (alzheimer[Disease/Phenotype] OR alzheimer[All fields]) AND](https://present5.com/presentation/0adc0ff243138239ed47505cce7c3f4e/image-51.jpg) NCBI Entrez Gene Query: (alzheimer[Disease/Phenotype] OR alzheimer[All fields]) AND "homo sapiens"[Organism] 143 Genes A 2 M CD 40 FAS ABCA 1 MME RABGAP 1 L CDC 2 FASLG ABCA 2 MPO RTN 4 CDK 5 FRAP 1 ABCB 1 MRE 11 A SERPINA 3 CDK 5 R 1 FYN ABL 1 MSI 1 SFRS 12 CDK 5 R 2 GABBR 1 ACE MTRR SLC 1 A 2 CHAT GAL AD 5 NACA SLC 6 A 3 CHRNA 4 GAPDH AD 6 NCAM 1 SLC 6 A 4 CHRNA 7 GFAP AD 7 NCSTN SNCB CLU GRIA 1 AD 8 NDRG 2 SORL 1 COL 18 A 1 GRIA 2 AD 9 NES TFAM COL 25 A 1 GRIA 3 ADAM 10 NGFR TGFB 1 COX 10 GRIN 2 A AGER NME 1 TNF CRH GRIN 2 B AHSG NME 2 TUBB 3 CTCF GSK 3 B APBA 1 NOS 3 UBQLN 1 CTNNA 3 HADH 2 APBB 2 NRG 1 VSNL 1 CTSB HPCAL 1 APH 1 A OLR 1 CTSD HTR 2 A APOC 1 P 18 SRP CXCR 3 IDE APOD PARK 7 CYP 46 A 1 IFNG APOE PAXIP 1 DHCR 24 IGF 2 R APOM PCSK 1 DLST IL 1 B APP PCSK 2 DSCR 1 ITM 2 B ASAHL PCSK 9 E 2 F 1 KCNC 4 ATF 2 PIN 1 EEF 2 KLK 10 BACE 1 PLAU EEF 2 K KLK 7 BACE 2 PON 1 EIF 2 AK 2 LAMA 1 BAX PRDX 1 EIF 4 E LAMC 1 BCHE PRDX 2 EIF 4 EBP 1 LOC 644264 BCL 2 PRDX 3 ENO 1 LRP 8 BCL 2 L 2 PRNP ERBB 4 MAP 2 K 1 BLMH PSEN 1 ESR 1 MAPT CBS PSEN 2 FALZ MEOX 2 RPS 3 A Mining Interactome
NCBI Entrez Gene Query: (alzheimer[Disease/Phenotype] OR alzheimer[All fields]) AND "homo sapiens"[Organism] 143 Genes A 2 M CD 40 FAS ABCA 1 MME RABGAP 1 L CDC 2 FASLG ABCA 2 MPO RTN 4 CDK 5 FRAP 1 ABCB 1 MRE 11 A SERPINA 3 CDK 5 R 1 FYN ABL 1 MSI 1 SFRS 12 CDK 5 R 2 GABBR 1 ACE MTRR SLC 1 A 2 CHAT GAL AD 5 NACA SLC 6 A 3 CHRNA 4 GAPDH AD 6 NCAM 1 SLC 6 A 4 CHRNA 7 GFAP AD 7 NCSTN SNCB CLU GRIA 1 AD 8 NDRG 2 SORL 1 COL 18 A 1 GRIA 2 AD 9 NES TFAM COL 25 A 1 GRIA 3 ADAM 10 NGFR TGFB 1 COX 10 GRIN 2 A AGER NME 1 TNF CRH GRIN 2 B AHSG NME 2 TUBB 3 CTCF GSK 3 B APBA 1 NOS 3 UBQLN 1 CTNNA 3 HADH 2 APBB 2 NRG 1 VSNL 1 CTSB HPCAL 1 APH 1 A OLR 1 CTSD HTR 2 A APOC 1 P 18 SRP CXCR 3 IDE APOD PARK 7 CYP 46 A 1 IFNG APOE PAXIP 1 DHCR 24 IGF 2 R APOM PCSK 1 DLST IL 1 B APP PCSK 2 DSCR 1 ITM 2 B ASAHL PCSK 9 E 2 F 1 KCNC 4 ATF 2 PIN 1 EEF 2 KLK 10 BACE 1 PLAU EEF 2 K KLK 7 BACE 2 PON 1 EIF 2 AK 2 LAMA 1 BAX PRDX 1 EIF 4 E LAMC 1 BCHE PRDX 2 EIF 4 EBP 1 LOC 644264 BCL 2 PRDX 3 ENO 1 LRP 8 BCL 2 L 2 PRNP ERBB 4 MAP 2 K 1 BLMH PSEN 1 ESR 1 MAPT CBS PSEN 2 FALZ MEOX 2 RPS 3 A Mining Interactome
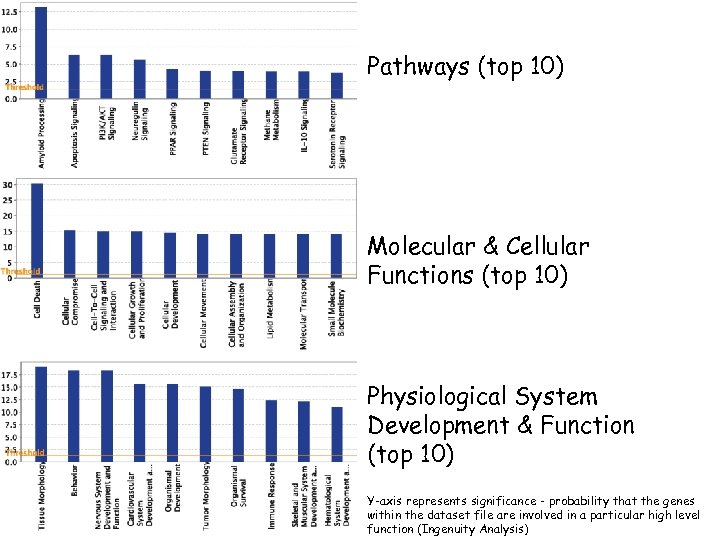 Pathways (top 10) Molecular & Cellular Functions (top 10) Physiological System Development & Function (top 10) Y-axis represents significance - probability that the genes within the dataset file are involved in a particular high level function (Ingenuity Analysis)
Pathways (top 10) Molecular & Cellular Functions (top 10) Physiological System Development & Function (top 10) Y-axis represents significance - probability that the genes within the dataset file are involved in a particular high level function (Ingenuity Analysis)
![NCBI Entrez Gene Query: (alzheimer[Disease/Phenotype] OR alzheimer[All fields]) AND NCBI Entrez Gene Query: (alzheimer[Disease/Phenotype] OR alzheimer[All fields]) AND](https://present5.com/presentation/0adc0ff243138239ed47505cce7c3f4e/image-55.jpg) NCBI Entrez Gene Query: (alzheimer[Disease/Phenotype] OR alzheimer[All fields]) AND "homo sapiens"[Organism] 143 AD-associated genes Mining about 800 gene expression datasets http: //depts. washington. edu/l 2 l/
NCBI Entrez Gene Query: (alzheimer[Disease/Phenotype] OR alzheimer[All fields]) AND "homo sapiens"[Organism] 143 AD-associated genes Mining about 800 gene expression datasets http: //depts. washington. edu/l 2 l/
 Text-mining Med. Line Abstracts • Data Source: Gene. RIF – Gene reference into function – Manually entered/curated sentences. • Gene. RIF: “Abstract of Abstracts” • NLP - Meta. Map and GATE (General Architecture for Text Engineering) • Keywords: MESH and UMLS concepts for Alzheimer’s disease (AD, Alzheimer’s dementia, Alzheimer disease, etc. ) 299 unique genes associated with Alzheimer’s disease
Text-mining Med. Line Abstracts • Data Source: Gene. RIF – Gene reference into function – Manually entered/curated sentences. • Gene. RIF: “Abstract of Abstracts” • NLP - Meta. Map and GATE (General Architecture for Text Engineering) • Keywords: MESH and UMLS concepts for Alzheimer’s disease (AD, Alzheimer’s dementia, Alzheimer disease, etc. ) 299 unique genes associated with Alzheimer’s disease
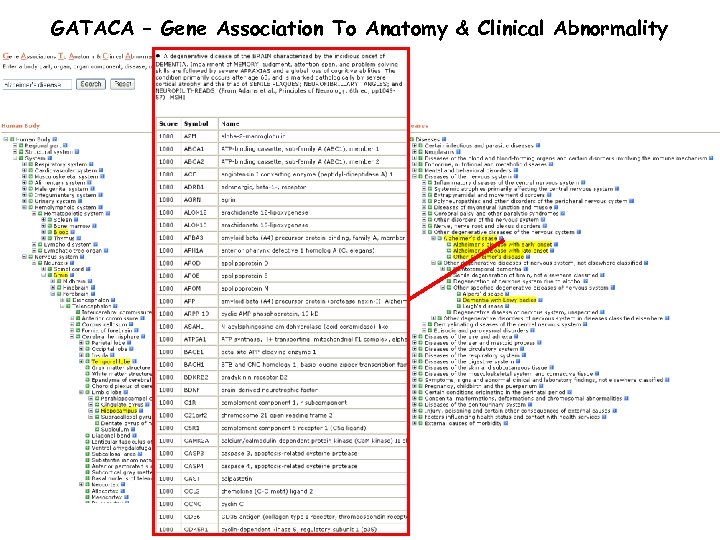 GATACA – Gene Association To Anatomy & Clinical Abnormality
GATACA – Gene Association To Anatomy & Clinical Abnormality
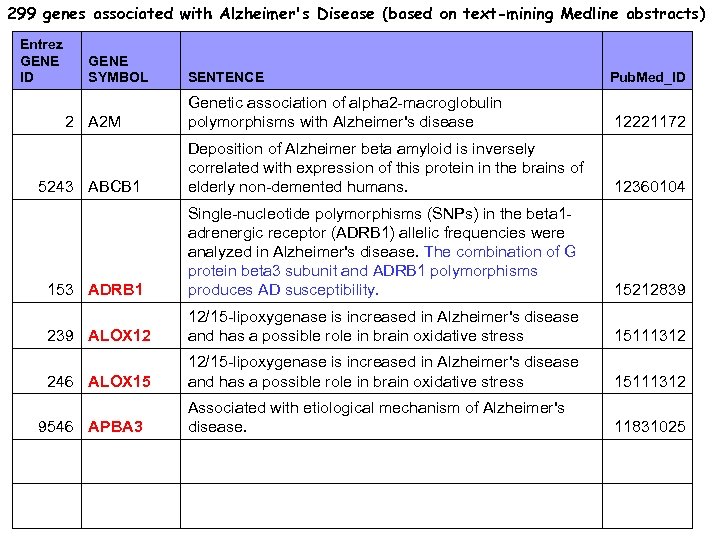 299 genes associated with Alzheimer's Disease (based on text-mining Medline abstracts) Entrez GENE ID GENE SYMBOL SENTENCE Pub. Med_ID Genetic association of alpha 2 -macroglobulin polymorphisms with Alzheimer's disease 12221172 Deposition of Alzheimer beta amyloid is inversely correlated with expression of this protein in the brains of elderly non-demented humans. 12360104 153 ADRB 1 Single-nucleotide polymorphisms (SNPs) in the beta 1 adrenergic receptor (ADRB 1) allelic frequencies were analyzed in Alzheimer's disease. The combination of G protein beta 3 subunit and ADRB 1 polymorphisms produces AD susceptibility. 15212839 239 ALOX 12 12/15 -lipoxygenase is increased in Alzheimer's disease and has a possible role in brain oxidative stress 15111312 246 ALOX 15 12/15 -lipoxygenase is increased in Alzheimer's disease and has a possible role in brain oxidative stress 15111312 Associated with etiological mechanism of Alzheimer's disease. 11831025 2 A 2 M 5243 ABCB 1 9546 APBA 3
299 genes associated with Alzheimer's Disease (based on text-mining Medline abstracts) Entrez GENE ID GENE SYMBOL SENTENCE Pub. Med_ID Genetic association of alpha 2 -macroglobulin polymorphisms with Alzheimer's disease 12221172 Deposition of Alzheimer beta amyloid is inversely correlated with expression of this protein in the brains of elderly non-demented humans. 12360104 153 ADRB 1 Single-nucleotide polymorphisms (SNPs) in the beta 1 adrenergic receptor (ADRB 1) allelic frequencies were analyzed in Alzheimer's disease. The combination of G protein beta 3 subunit and ADRB 1 polymorphisms produces AD susceptibility. 15212839 239 ALOX 12 12/15 -lipoxygenase is increased in Alzheimer's disease and has a possible role in brain oxidative stress 15111312 246 ALOX 15 12/15 -lipoxygenase is increased in Alzheimer's disease and has a possible role in brain oxidative stress 15111312 Associated with etiological mechanism of Alzheimer's disease. 11831025 2 A 2 M 5243 ABCB 1 9546 APBA 3
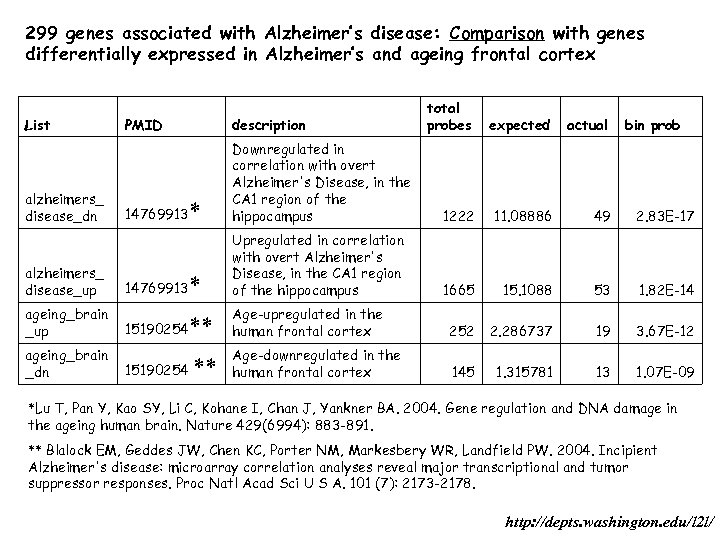 299 genes associated with Alzheimer’s disease: Comparison with genes differentially expressed in Alzheimer’s and ageing frontal cortex List alzheimers_ disease_dn alzheimers_ disease_up total probes expected actual PMID description 14769913 * Downregulated in correlation with overt Alzheimer's Disease, in the CA 1 region of the hippocampus 1222 11. 08886 49 2. 83 E-17 * Upregulated in correlation with overt Alzheimer's Disease, in the CA 1 region of the hippocampus 1665 15. 1088 53 1. 82 E-14 252 2. 286737 19 3. 67 E-12 145 1. 315781 13 1. 07 E-09 14769913 ageing_brain _up 15190254 ** Age-upregulated in the human frontal cortex ageing_brain _dn 15190254 ** Age-downregulated in the human frontal cortex bin prob *Lu T, Pan Y, Kao SY, Li C, Kohane I, Chan J, Yankner BA. 2004. Gene regulation and DNA damage in the ageing human brain. Nature 429(6994): 883 -891. ** Blalock EM, Geddes JW, Chen KC, Porter NM, Markesbery WR, Landfield PW. 2004. Incipient Alzheimer's disease: microarray correlation analyses reveal major transcriptional and tumor suppressor responses. Proc Natl Acad Sci U S A. 101 (7): 2173 -2178. http: //depts. washington. edu/l 2 l/
299 genes associated with Alzheimer’s disease: Comparison with genes differentially expressed in Alzheimer’s and ageing frontal cortex List alzheimers_ disease_dn alzheimers_ disease_up total probes expected actual PMID description 14769913 * Downregulated in correlation with overt Alzheimer's Disease, in the CA 1 region of the hippocampus 1222 11. 08886 49 2. 83 E-17 * Upregulated in correlation with overt Alzheimer's Disease, in the CA 1 region of the hippocampus 1665 15. 1088 53 1. 82 E-14 252 2. 286737 19 3. 67 E-12 145 1. 315781 13 1. 07 E-09 14769913 ageing_brain _up 15190254 ** Age-upregulated in the human frontal cortex ageing_brain _dn 15190254 ** Age-downregulated in the human frontal cortex bin prob *Lu T, Pan Y, Kao SY, Li C, Kohane I, Chan J, Yankner BA. 2004. Gene regulation and DNA damage in the ageing human brain. Nature 429(6994): 883 -891. ** Blalock EM, Geddes JW, Chen KC, Porter NM, Markesbery WR, Landfield PW. 2004. Incipient Alzheimer's disease: microarray correlation analyses reveal major transcriptional and tumor suppressor responses. Proc Natl Acad Sci U S A. 101 (7): 2173 -2178. http: //depts. washington. edu/l 2 l/
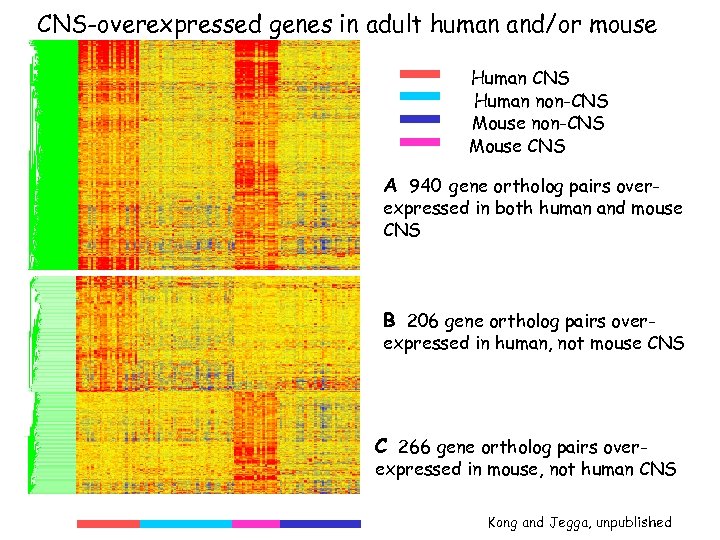 CNS-overexpressed genes in adult human and/or mouse Human CNS Human non-CNS Mouse CNS A 940 gene ortholog pairs over- expressed in both human and mouse CNS B 206 gene ortholog pairs over- expressed in human, not mouse CNS C 266 gene ortholog pairs over- expressed in mouse, not human CNS Kong and Jegga, unpublished
CNS-overexpressed genes in adult human and/or mouse Human CNS Human non-CNS Mouse CNS A 940 gene ortholog pairs over- expressed in both human and mouse CNS B 206 gene ortholog pairs over- expressed in human, not mouse CNS C 266 gene ortholog pairs over- expressed in mouse, not human CNS Kong and Jegga, unpublished
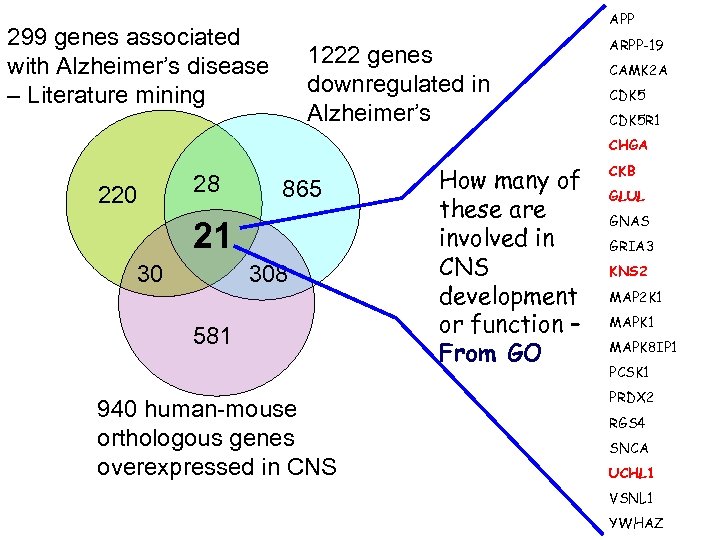 APP 299 genes associated with Alzheimer’s disease – Literature mining 1222 genes downregulated in Alzheimer’s ARPP-19 CAMK 2 A CDK 5 R 1 CHGA 220 28 865 21 30 308 581 940 human-mouse orthologous genes overexpressed in CNS How many of these are involved in CNS development or function – From GO CKB GLUL GNAS GRIA 3 KNS 2 MAP 2 K 1 MAPK 8 IP 1 PCSK 1 PRDX 2 RGS 4 SNCA UCHL 1 VSNL 1 YWHAZ
APP 299 genes associated with Alzheimer’s disease – Literature mining 1222 genes downregulated in Alzheimer’s ARPP-19 CAMK 2 A CDK 5 R 1 CHGA 220 28 865 21 30 308 581 940 human-mouse orthologous genes overexpressed in CNS How many of these are involved in CNS development or function – From GO CKB GLUL GNAS GRIA 3 KNS 2 MAP 2 K 1 MAPK 8 IP 1 PCSK 1 PRDX 2 RGS 4 SNCA UCHL 1 VSNL 1 YWHAZ
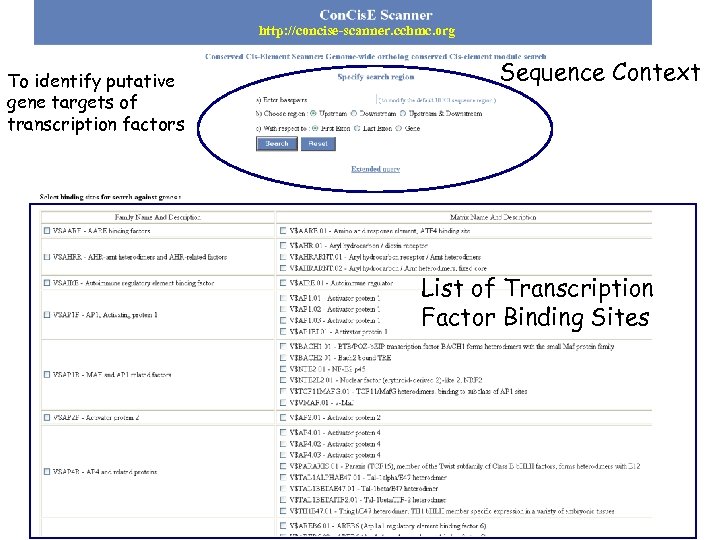 http: //concise-scanner. cchmc. org To identify putative gene targets of transcription factors Sequence Context List of Transcription Factor Binding Sites
http: //concise-scanner. cchmc. org To identify putative gene targets of transcription factors Sequence Context List of Transcription Factor Binding Sites
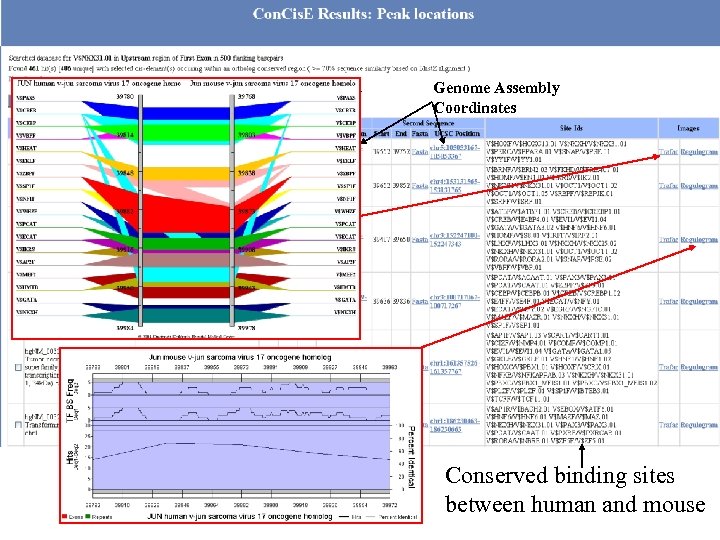 Human Mouse Genome. Trafac Coordinates Genome Assembly Coordinates Conserved binding sites between human and mouse
Human Mouse Genome. Trafac Coordinates Genome Assembly Coordinates Conserved binding sites between human and mouse
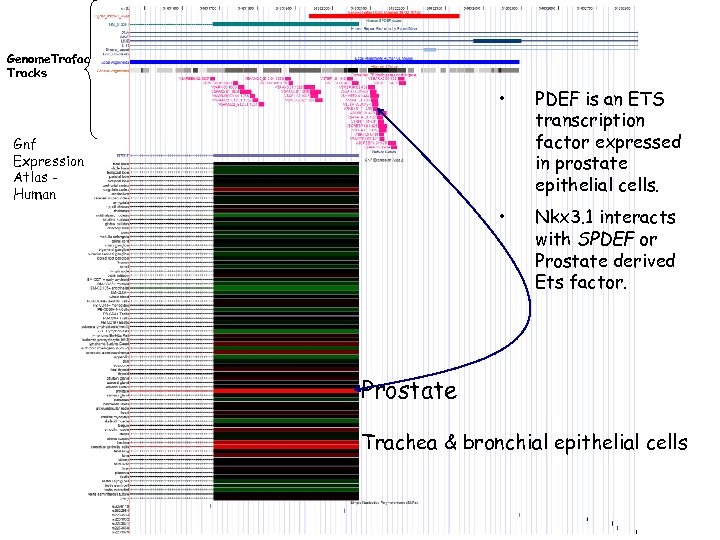 Genome. Trafac Tracks • • Gnf Expression Atlas Human PDEF is an ETS transcription factor expressed in prostate epithelial cells. Nkx 3. 1 interacts with SPDEF or Prostate derived Ets factor. Prostate Trachea & bronchial epithelial cells
Genome. Trafac Tracks • • Gnf Expression Atlas Human PDEF is an ETS transcription factor expressed in prostate epithelial cells. Nkx 3. 1 interacts with SPDEF or Prostate derived Ets factor. Prostate Trachea & bronchial epithelial cells
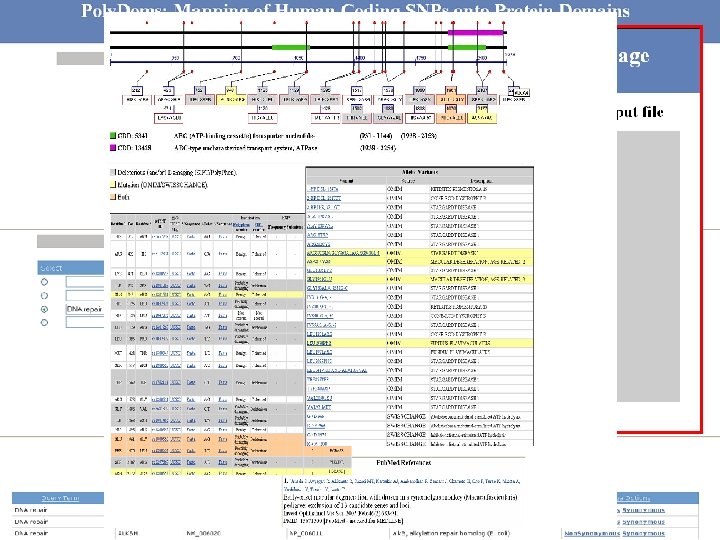 http: //polydoms. cchmc. org
http: //polydoms. cchmc. org
 Goals – Summary……… • Enable discovery of novel disease-gene relationships • Facilitate discovery of disease-pathway relationships • Enable discovery of novel pathways and targets and associate them with disease processes • Help researchers generate testable hypotheses • Support efforts to prioritize research • Facilitate meta-analyses
Goals – Summary……… • Enable discovery of novel disease-gene relationships • Facilitate discovery of disease-pathway relationships • Enable discovery of novel pathways and targets and associate them with disease processes • Help researchers generate testable hypotheses • Support efforts to prioritize research • Facilitate meta-analyses
 New/Future Directions……. Computational • Semantic Web (SW): “A vision for the next generation web in which data from multiple sources described with rich semantics are integrated to enable human processing by humans as well as software agents” (SW Life Sciences) • Semantic Web Languages – RDF (Resource Description Framework) – RDFS (RDF schema) and – OWL (Ontology Web Language) – SPARQL (semantic web querying language) • Prioritization and Ranking entities on novel Gene Networks and Inferencing Biological/Genomics • Gene regulation by micro. RNAs (mi. RNAs): – ~22 bp non-coding nucleotide RNAs that primarily act posttranscriptionally by suppressing m. RNAs – At least 1% of the transcripts in the genome code for mi. RNAs – mi. RNA have at least 20 -30% of the coding genes as their targets – mi. RNAs are implicated in various cellular processes, such as cell fate determination, cell death, and tumorigenesis (Bartel 2004). – E. g. : CREB-regulated mi. RNA regulates neuronal morphogenesis (Vo et al 2005)
New/Future Directions……. Computational • Semantic Web (SW): “A vision for the next generation web in which data from multiple sources described with rich semantics are integrated to enable human processing by humans as well as software agents” (SW Life Sciences) • Semantic Web Languages – RDF (Resource Description Framework) – RDFS (RDF schema) and – OWL (Ontology Web Language) – SPARQL (semantic web querying language) • Prioritization and Ranking entities on novel Gene Networks and Inferencing Biological/Genomics • Gene regulation by micro. RNAs (mi. RNAs): – ~22 bp non-coding nucleotide RNAs that primarily act posttranscriptionally by suppressing m. RNAs – At least 1% of the transcripts in the genome code for mi. RNAs – mi. RNA have at least 20 -30% of the coding genes as their targets – mi. RNAs are implicated in various cellular processes, such as cell fate determination, cell death, and tumorigenesis (Bartel 2004). – E. g. : CREB-regulated mi. RNA regulates neuronal morphogenesis (Vo et al 2005)
 Take-home messages • Networks and integration of databases are keys to success in Bioinformatics. • Integration of data computation and data integration into a single cohesive whole will increase the efficiency of research effort – by reducing the serendipity & hit and miss nature of empirical research and – will provide valuable clues to the biomedical researchers on their choice of experiments limitations of funds, manpower and time. • Researchers/Users have to know what is available and how to access (what are the limitations), and use the resources they are offered or are available.
Take-home messages • Networks and integration of databases are keys to success in Bioinformatics. • Integration of data computation and data integration into a single cohesive whole will increase the efficiency of research effort – by reducing the serendipity & hit and miss nature of empirical research and – will provide valuable clues to the biomedical researchers on their choice of experiments limitations of funds, manpower and time. • Researchers/Users have to know what is available and how to access (what are the limitations), and use the resources they are offered or are available.
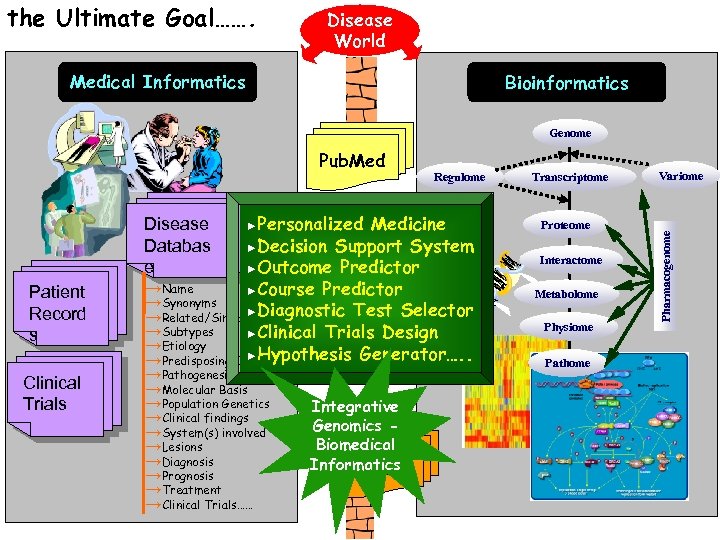 the Ultimate Goal……. Disease World Medical Informatics Bioinformatics Genome Medicine ►Decision Support System ►Outcome Predictor →Name ►Course Predictor →Synonyms ►Diagnostic Test Selector →Related/Similar Diseases →Subtypes ►Clinical Trials Design →Etiology ►Hypothesis Generator…. . →Predisposing Causes Disease Databas e Patient Record s Clinical Trials ►Personalized Regulome →Pathogenesis →Molecular Basis →Population Genetics →Clinical findings →System(s) involved →Lesions →Diagnosis →Prognosis →Treatment →Clinical Trials…… Integrative Genomics Biomedical Informatics OMIM Transcriptome Proteome Interactome Metabolome Physiome Pathome Variome Pharmacogenome Pub. Med
the Ultimate Goal……. Disease World Medical Informatics Bioinformatics Genome Medicine ►Decision Support System ►Outcome Predictor →Name ►Course Predictor →Synonyms ►Diagnostic Test Selector →Related/Similar Diseases →Subtypes ►Clinical Trials Design →Etiology ►Hypothesis Generator…. . →Predisposing Causes Disease Databas e Patient Record s Clinical Trials ►Personalized Regulome →Pathogenesis →Molecular Basis →Population Genetics →Clinical findings →System(s) involved →Lesions →Diagnosis →Prognosis →Treatment →Clinical Trials…… Integrative Genomics Biomedical Informatics OMIM Transcriptome Proteome Interactome Metabolome Physiome Pathome Variome Pharmacogenome Pub. Med
 http: //anil. cchmc. org (under presentations) “To him who devotes his life to science, nothing can give more happiness than increasing the number of discoveries, but his cup of joy is full when the results of his studies immediately find practical applications” Thank You! — Louis Pasteur http: //sbw. kgi. edu/
http: //anil. cchmc. org (under presentations) “To him who devotes his life to science, nothing can give more happiness than increasing the number of discoveries, but his cup of joy is full when the results of his studies immediately find practical applications” Thank You! — Louis Pasteur http: //sbw. kgi. edu/


