52b3af6ba2da0db860525331eca666bc.ppt
- Количество слайдов: 67

Building a Global Map of (Human) Gene Expression Misha Kapushesky European Bioinformatics Institute, EMBL St. Petersburg, Russia May, 2010
From one genome to many biological states • While there is only one genome sequence, different genes are expressed in many different cell types and tissues, different developmental or disease states • The size and structure of this “expression space” is still largely unknown • Most individual experiments are looking at small regions • We would like to build a map of the global human gene expression space

Mapping the human transcriptome A microarray experiment Traditional research The map we want to build Everest Kathmandu Lhasa

How to build such a global map • This space is huge - There are thousands of potentially different states – cell types, tissue types, developmental stages, disease states, systems under various treatments (drugs, radiation, stress, …) – • It is not feasible to study them all in a single laboratory experiment (costs, rare samples, …) • However thousands of gene expression experiments are performed every year (microarrays, new generation sequencing) • Can we use the published data to build the global expression map?





Array. Express • www. ebi. ac. uk/arrayexpress • Data from over 280, 000 assays and over 10, 000 independent studies (microarrays, sequencing, …) • Gene expression and other functional genomics assays • Over 200 species • Data collection and exchange from GEO

Can we integrate these data to answer questions that go beyond what was done in the individual studies? • On a quantitative level - data on only the same microarray platform can be integrated
A global map of human gene expression • Angela Gonzales (EBI) • Misha Kapushesky (EBI) • Janne Nikkila (Helsinki University of Technology) • Helen Parkinson (EBI), • Wolfgang Huber (EMBL) • Esko Ukkonen (University of Helsinki) Margus Lukk et al, Nature Biotechnology, 28, p 322 -324 (April, 2010)

The most popular gene expression microarray platform: Affymetrix U 133 A • We collected over 9000 raw data files from Affymetrix U 133 A from GEO and Array. Express • Applying strict quality controls, removing the duplicates • Data on 5372 samples remained from 206 different studies generated in 163 different laboratories grouped in 369 different biological ‘conditions’ (tissue types, diseases, various cell lines, etc) • The 369 conditions grouped in different larger ‘metagroups’

Different metagroupings (4 and 15):
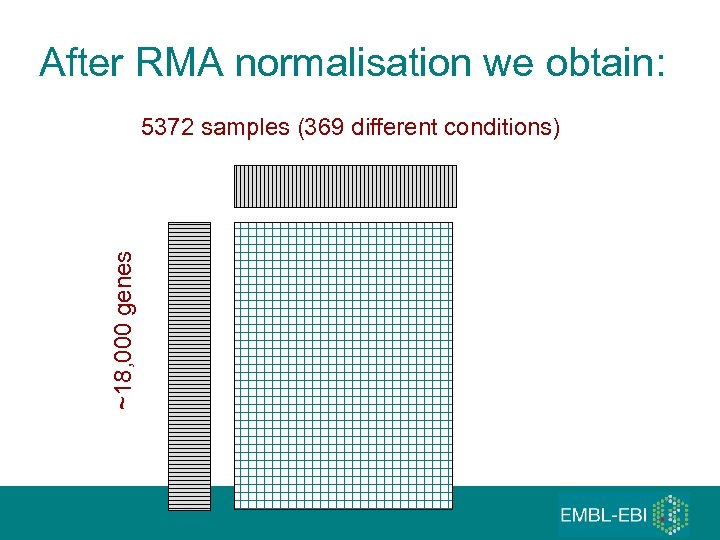
After RMA normalisation we obtain: ~18, 000 genes 5372 samples (369 different conditions)

2 nd Principal Component Analysis – each dot is one of the 5372 samples 1 st

2 nd 1 st 16 19/03/2018 Human gene expression map
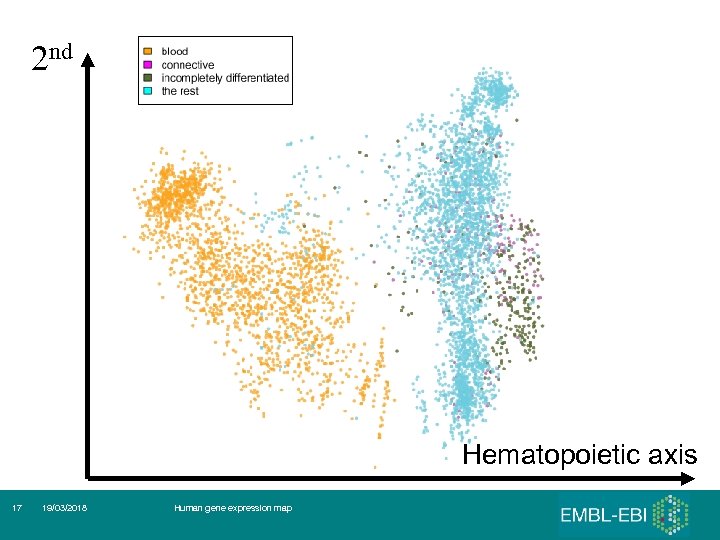
2 nd Hematopoietic axis 17 19/03/2018 Human gene expression map
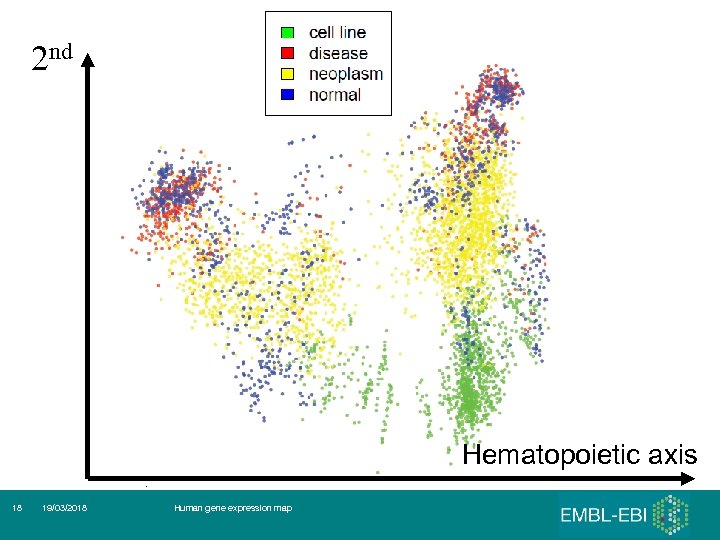
2 nd Hematopoietic axis 18 19/03/2018 Human gene expression map
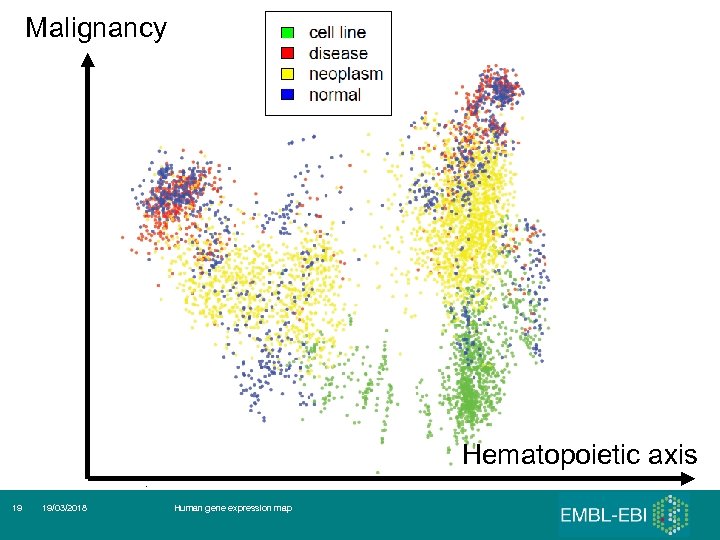
Malignancy Hematopoietic axis 19 19/03/2018 Human gene expression map

Hematopoietic and malignancy axes Lukk et al, Nature Biotechnology, 28: 322

2 nd 1 st 3 rd
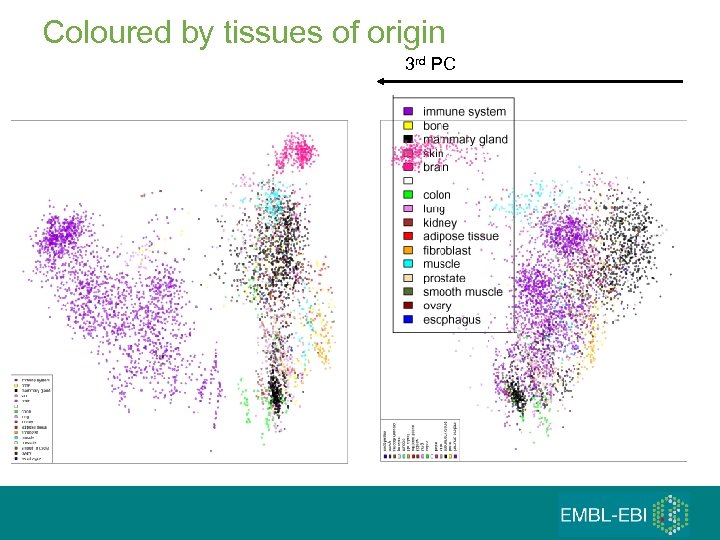
Coloured by tissues of origin 3 rd PC
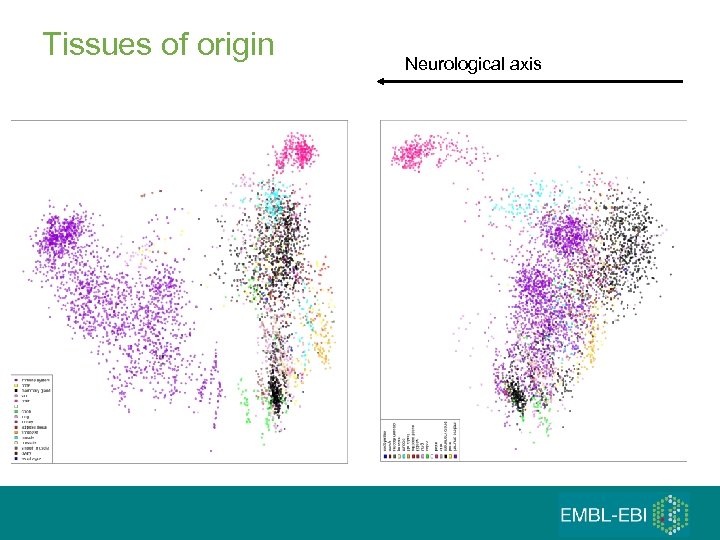
Tissues of origin Neurological axis

First 3 (5) principal components 1. 2. 3. 4. 5. Hematopoietic axis – blood, ‘solid tissues’, ‘incompletely differentiated cells and connective tissues’ Malignancy axis - Cell lines – cancer – normals and other diseases Neurological axis – nervous system / the rest RNA degradation Samples seem to ‘cluster’ by the tissues of origin


Hierarchical clustering of 97 groups with at least 10 replicates each 26 19/03/2018 Human gene expression map

Comparison of the 97 larger sample groups to the rest Incompletely differentiated cell type and connective tissue group

Conclusions so far • We have identified 6 major transcription profile classes in these data: 1. cell lines 2. incompletely differentiated cells and connective tissues 3. neoplasms 4. blood 5. brain 6. muscle • Cell lines cluster together!



Gene expression across the 5372 samples • The expression of most genes is relatively constant • There are only 1034 probesets (mapping to less than 900) genes where normalised signal variability has standard deviation > 2

Clustering of 97 sample groups and 1000 most variable probesets (about 900 genes) 1 1. 2. 3. 4. 5. 6. 7. 8. 9. 10. 11. 2 3 4 5 Immune repsonse Nervous system development Lipid raft Mitosis Neurotransmitter uptake Cytoskeletal protein binding Extracellular matrix Extracellular regions Extracellular matirx Extracellular region Mitosis 6 7 8 9 10 11 12 12. 13. 14. 15. 16. 17. 18. 19. 13 14 16 17 18 19 15 Defence response Nervous system development Actin cytoskeleton organisation and biogenesis Protein carrier activity No significant resout Antigen presentation, exogenous antigen Trans – 1, 2 -dyhydrobenzene, 1, 2 -dyhydrogenase activity S 100 alpha binding
Clustering based on subset of these genes produce similar results • Clustering based on 350 most variable probesets gives almost the same result • Even clustering based on 30 most variable probesets is very close

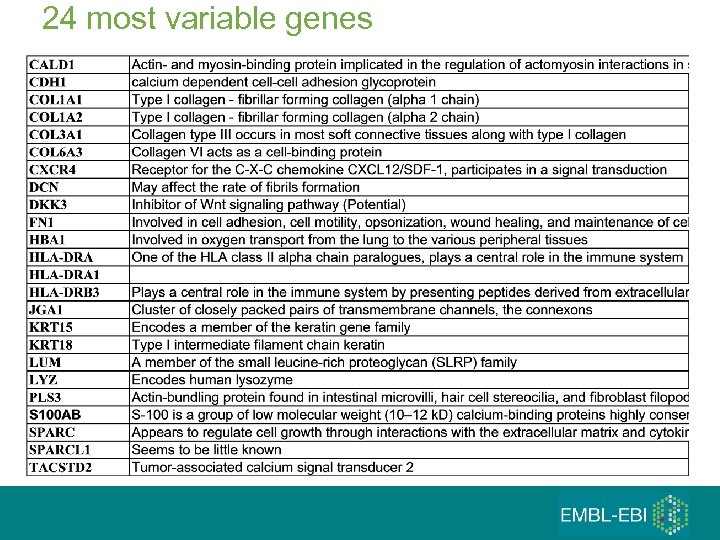
24 most variable genes
www. ebi. ac. uk/gxa/human/U 133 a

Can we go beyond the 6 major classes?
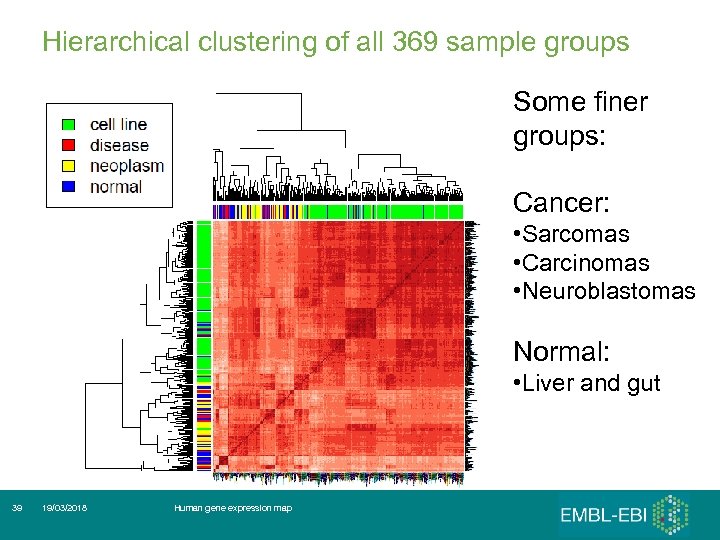
Hierarchical clustering of all 369 sample groups Some finer groups: Cancer: • Sarcomas • Carcinomas • Neuroblastomas Normal: • Liver and gut 39 19/03/2018 Human gene expression map

Normal blood and blood non-neoplastic disease Leukemia Other blood neoplasm Blood cell lines
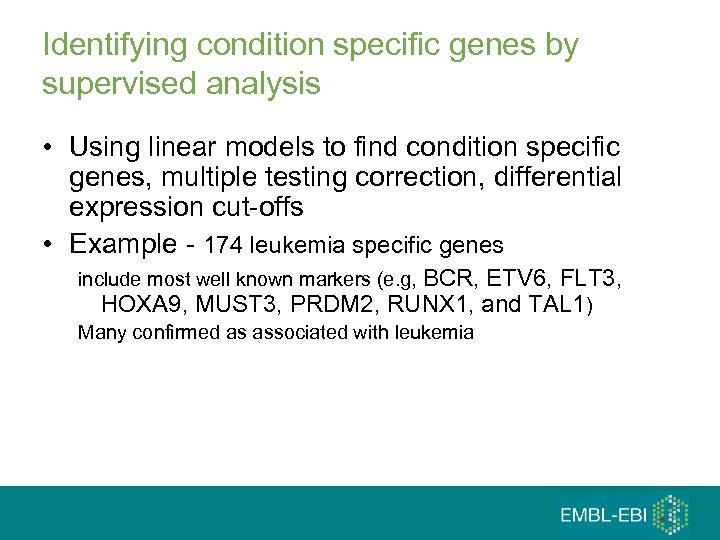
Identifying condition specific genes by supervised analysis • Using linear models to find condition specific genes, multiple testing correction, differential expression cut-offs • Example - 174 leukemia specific genes include most well known markers (e. g, BCR, ETV 6, FLT 3, HOXA 9, MUST 3, PRDM 2, RUNX 1, and TAL 1) Many confirmed as associated with leukemia

• Beyond the major 6 classes the ‘signal’ becomes weak • The problem may be lab effects The large biological effects are stronger than the lab effects However, when we zoom into particular subclasses, the lab effects may be taking precedence

Mapping the human transcriptome Our current view on global A microarray experiment transcriptome Traditional research The map we want to build Everest Kathmandu Lhasa
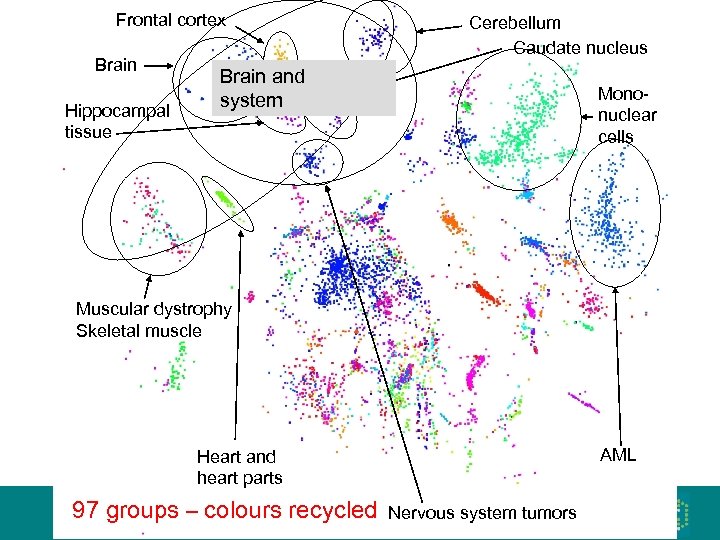
Frontal cortex Brain Hippocampal tissue Cerebellum Caudate nucleus Brain and nervous system Mononuclear cells Muscular dystrophy Skeletal muscle AML Heart and heart parts 97 groups – colours recycled Nervous system tumors

Second approach • Integrating data on statistics level

Gene Expression Atlas • • • Ele Holloway Ibrahim Emam Pavel Kurnosov Helen Parkinson Anrey Zorin Tony Burdett Gabriella Rustici Eleanor William Andrew Tikhonov
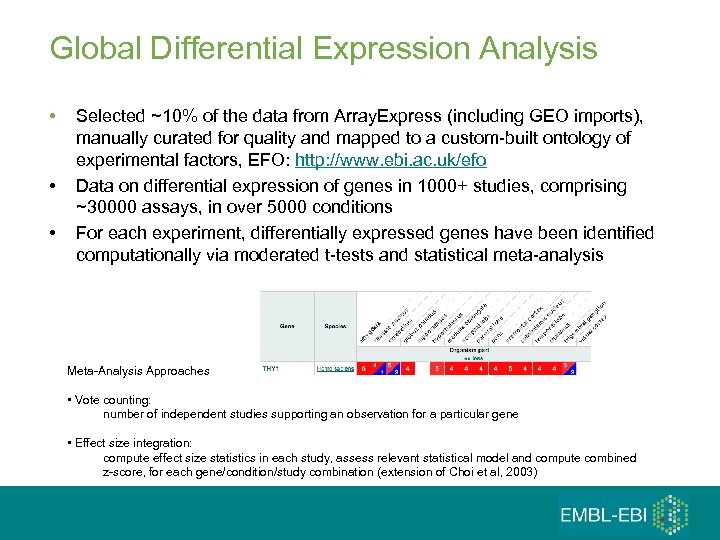
Global Differential Expression Analysis • • • Selected ~10% of the data from Array. Express (including GEO imports), manually curated for quality and mapped to a custom-built ontology of experimental factors, EFO: http: //www. ebi. ac. uk/efo Data on differential expression of genes in 1000+ studies, comprising ~30000 assays, in over 5000 conditions For each experiment, differentially expressed genes have been identified computationally via moderated t-tests and statistical meta-analysis Meta-Analysis Approaches • Vote counting: number of independent studies supporting an observation for a particular gene • Effect size integration: compute effect size statistics in each study, assess relevant statistical model and compute combined z-score, for each gene/condition/study combination (extension of Choi et al, 2003)

Analysing each contributing dataset separately: AML CML normal genes one-way ANOVA
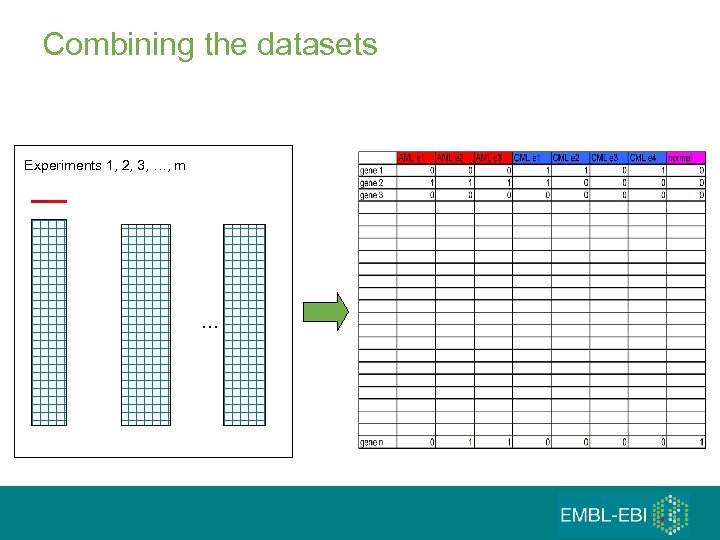
Combining the datasets Experiments 1, 2, 3, …, m …

Effect size-based meta-analysis • We have for each gene in each experiment/condition: p-value for significance simulaneous t-statistics & confidence intervals d. e. label (“up” or “down”) • However, we would like to: Measure of strength of d. e. effect size Ability to combine d. e. findings statistically • Effect Size Standardized mean difference or similar (e. g. , correlation coef. )

Meta-analysis Procedure • For each gene-experiment-condition combination Compute effect size from simultaneous d. e. t-statistics • Combine effect sizes across multiple studies Using fixed-effects or randomeffects models Obtain for each gene-condition combination: • Mean effect size estimate • Combined z-score • Overall p-value

Long tail of annotations…

Annotating data with ontologies • Diverse nature of annotations on data • Need to support complex queries which contain semantic information E. g. which genes are under-expressed in brain cancer samples in human or mouse • If we annotate with adenocarcinoma do we get this data? James Malone
Decoupling knowledge from data Atlas/AE James Malone

Semantically-enriched Queries with EFO

We can use the ontology structure We can perform effect size meta-analysis on a hierarchy, if we follow several rules:

Increased statistical power

Condition-specificity through EFO

Condition-specific Gene Expression

www. ebi. ac. uk/gxa Query for genes species Query for conditions The ‘advanced query’ option allows building more complex queries http: //www. ebi. ac. uk/gxa

Query results for gene ASPM Zoom into one of the ‘Glioblastoma’ studies. Each bar represents ASPM is downregulated in ‘normlal’ an expression condition in comparison to a disease in Upregulated in ‘Glioblastoma’ in 3 level a in 9 studies out of 10 particular sample indepnendent studies 61 Array. Express
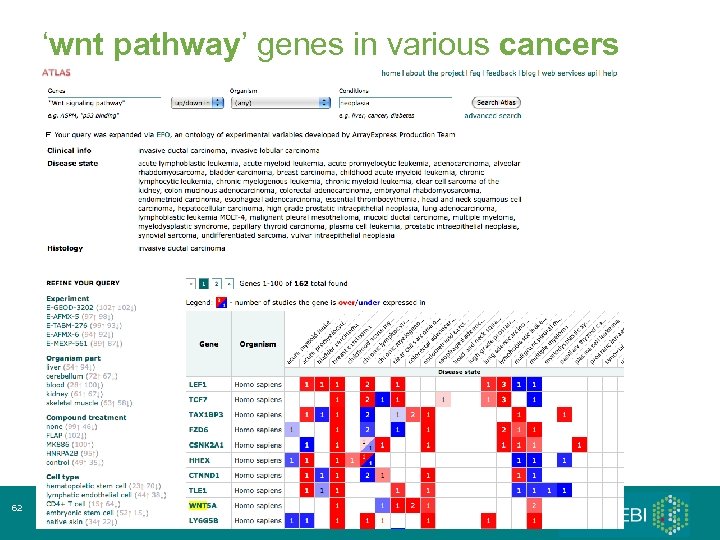
‘wnt pathway’ genes in various cancers 62 Array. Express

Integrating both approaches • First approach gives the global view, but obsucres the detail • The second approach gives detail, but doesn’t allow easily to integrate everything in one map • Can we combine both approaches?

Other data • RNAseq data • Proteomics data – Human Proteome Atlas from KTH in Stockholm (collaboration with Mathias Uhlen) • Time series – what states a cell goes through to become from an ESC to a mature cell?


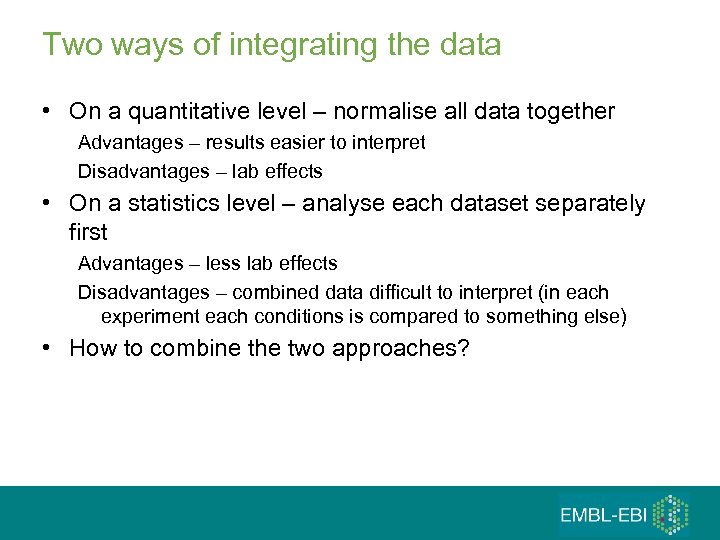
Two ways of integrating the data • On a quantitative level – normalise all data together Advantages – results easier to interpret Disadvantages – lab effects • On a statistics level – analyse each dataset separately first Advantages – less lab effects Disadvantages – combined data difficult to interpret (in each experiment each conditions is compared to something else) • How to combine the two approaches?

Acknowledgements • • • • • Margus Lukk Misha Kapushesky Angela Gonzales Helen Parkinson Gabriela Rustici Ugis Sarkans Ele Holloway Roby Mani Mohammadreza Shojatalab Nikolay Kolesnikov Niran Abeygunawardena Anjan Sharma Miroslaw Dylag Ekaterina Pilicheva Ibrahim Emam Pavel Kurnosov Andrew Tikhonov Andrey Zorin • • • • • Anna Farne Eleanor Williams Tony Burdett James Malone Holly Zheng Tomasz Adamusiak • Susanna-Assunta Sansone Philippe Rocca-Serra Natalija Sklyar Marco Brandizi Chris Taylor Eamonn Maguire Maria Krestyaninova Mikhail Gostev Johan Rung Natalja Kurbatova Katherine Lawler Nils Gehlenborg Lynn French Collaborators Audrey Kaufman (EBI) Wolfgang Huber (EBI) Sami Kaski (Helsinki) Morris Swertz (Groningen) … Funding European Commision • FELICS • Mol. PAGE • ENGAGE • Mu. GEN • SLING • DIAMONDS • EMERALD NIH (NHGRI) EMBL
52b3af6ba2da0db860525331eca666bc.ppt