54395dd913d1da1ecb6bda98d71e1762.ppt
- Количество слайдов: 27

Building a Chemical Informatics Grid Marlon Pierce Community Grids Laboratory Indiana University
CICC Project Information n n “Chemical Informatics and Cyberinfrastructure Collaboratory” is an NIH and MS-funded research project to combine the CI’s. Project web site and more information q q n Team members include q q q n www. chembiogrid. org/wiki Computer Science: Geoffrey Fox (PI), Dennis Gannon, Beth Plale, Marlon Pierce, Yuqing (Melanie) Wu, Malika Mahoui, Jake Kim Chemical Informatics and Chemistry: Gary Wiggins, Mu-Hyun (Mookie) Baik, David Wild, Rajarshi Guha, Kevin Gilbert I have stolen slides and content from these fine people. We collaborate with several groups q q q Peter Murray Rust’s group at University of Cambridge University of Michigan’s MACE group. Chemistry Development Kit (CDK) project DTP NIC at NIH Scripps High Throughput Screening Center
Chemical Informatics and Cyberinfrastructure Building Blocks n Chemical Informatics Resources: q Deluge of experimental data n n q Chemical databases maintained by various groups n q q n NIH Pub. Chem, NIH DTP Chemical informatics and computational chemistry n q > 100, 000 compounds screened by 10 publicly funded high throughput screening centers using various assay techniques (molecular to cellular) Molecular Libraries Screening Center Network Data clustering, data mining, descriptor calculations, toxicity prediction, docking, molecular modeling, and quantum chemistry Visualization tools Web resources: journal articles, etc. A Chemical Informatics Grid will need to integrate these into a common, loosely coupled, open, distributed computing environment.
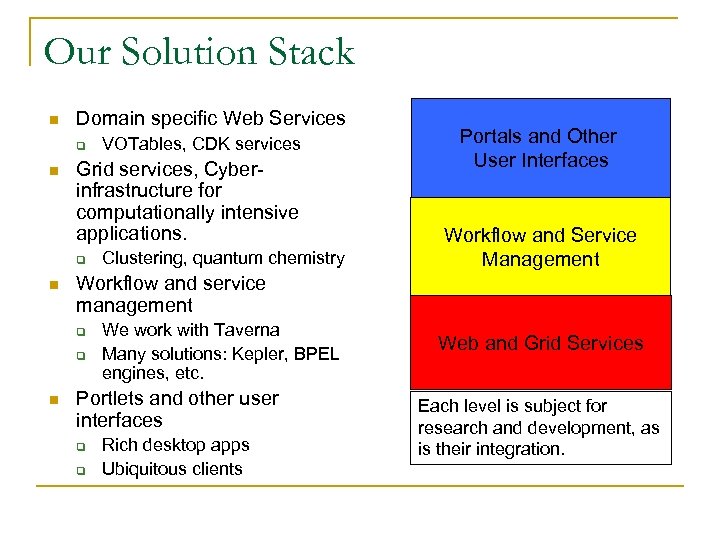
Our Solution Stack n Domain specific Web Services q n Grid services, Cyberinfrastructure for computationally intensive applications. q n Clustering, quantum chemistry Portals and Other User Interfaces Workflow and Service Management Workflow and service management q q n VOTables, CDK services We work with Taverna Many solutions: Kepler, BPEL engines, etc. Portlets and other user interfaces q q Rich desktop apps Ubiquitous clients Web and Grid Services Each level is subject for research and development, as is their integration.

A Library of Chemical Informatics Web Services

All Services Great and Small n Like most Grids, a Chemical Informatics Grid will have the classic styles: q q n But we also need many additional services q q n Data Grid Services: these provide access to data sources like Pub. Chem, etc. Execution Grid Services: used for running cluster analysis programs, molecular modeling codes, etc, on Tera. Grid and similar places. Handling format conversions (In. Ch. I<->SMILES) Shipping and manipulating tabular data Determining toxicity of compounds Generating batch 2 D images So one of our core activities is “build lots of services”
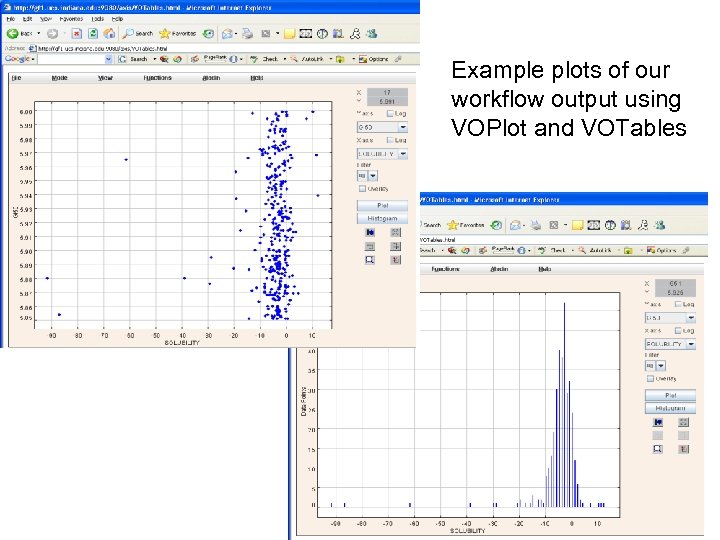
VOTables: Handling Tabular Data n n n Developed by the Virtual Observatory community for encoding astronomy data. The VOTable format is an XML representation of the tabular data (data coming from BCI, NIH DTP databases, and so on). VOTables-compatible tools have been built q n We just inherit them. SAVOT and JAVOT JAVA Parser APIs for VOTable allow us to easily build VOTable-based applications q q q Web Services Spread sheet Plotting applications. n VOPlot and Top. Cat are two
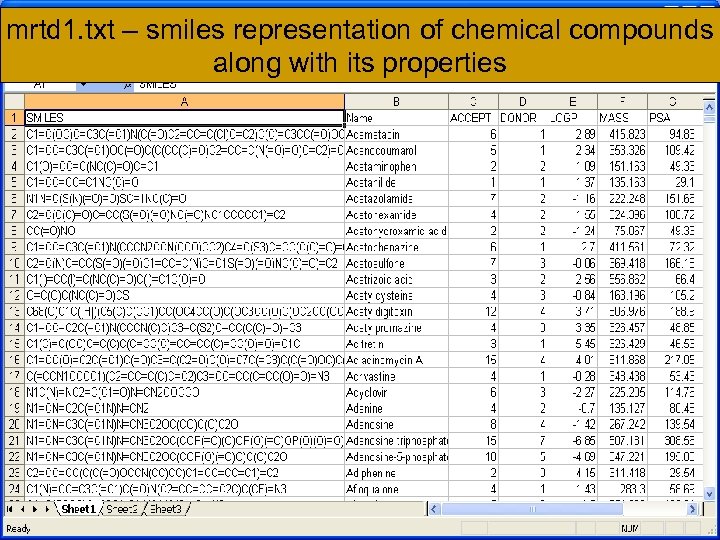
mrtd 1. txt – smiles representation of chemical compounds along with its properties

Votable. xml : xml representation of mrtd 1. txt file
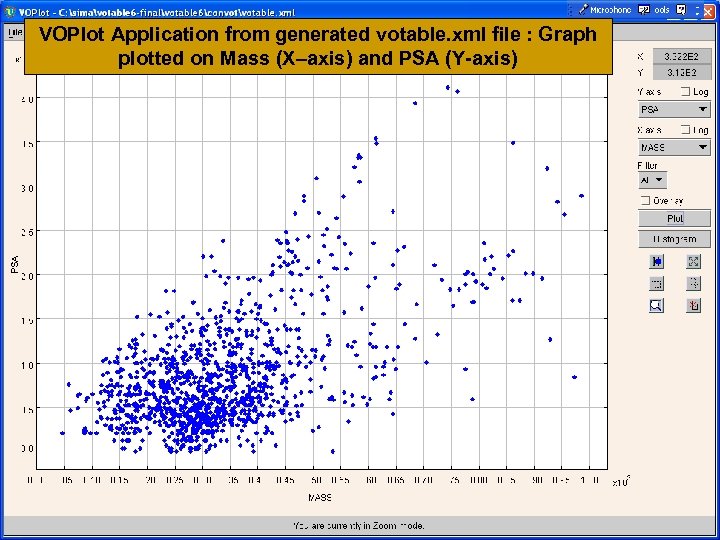
VOPlot Application from generated votable. xml file : Graph plotted on Mass (X–axis) and PSA (Y-axis)
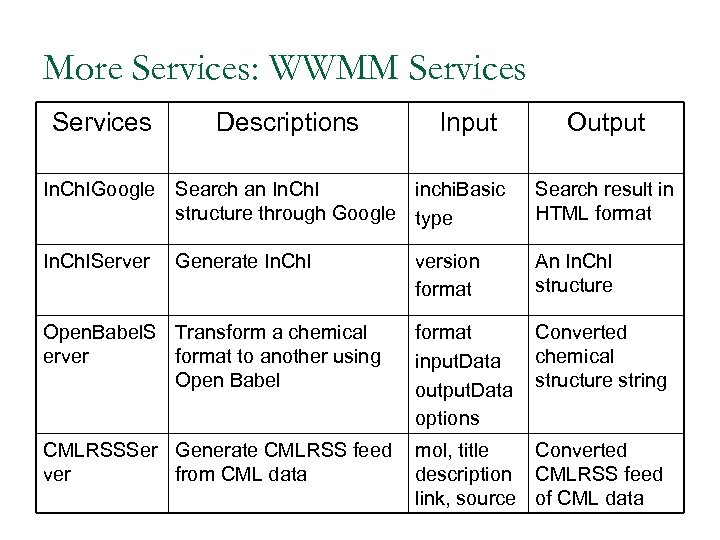
More Services: WWMM Services Descriptions Input Output In. Ch. IGoogle Search an In. Ch. I inchi. Basic structure through Google type Search result in HTML format In. Ch. IServer version format An In. Ch. I structure Open. Babel. S Transform a chemical erver format to another using Open Babel format input. Data output. Data options Converted chemical structure string CMLRSSSer Generate CMLRSS feed ver from CML data mol, title Converted description CMLRSS feed link, source of CML data Generate In. Ch. I
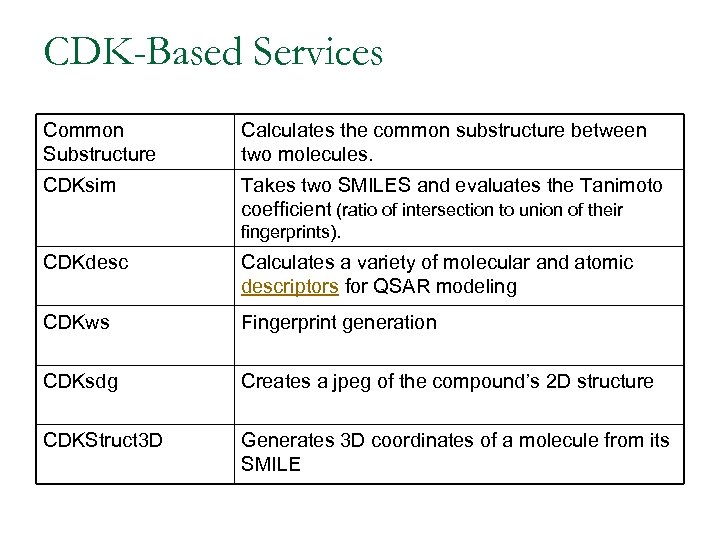
CDK-Based Services Common Substructure Calculates the common substructure between two molecules. CDKsim Takes two SMILES and evaluates the Tanimoto coefficient (ratio of intersection to union of their fingerprints). CDKdesc Calculates a variety of molecular and atomic descriptors for QSAR modeling CDKws Fingerprint generation CDKsdg Creates a jpeg of the compound’s 2 D structure CDKStruct 3 D Generates 3 D coordinates of a molecule from its SMILE
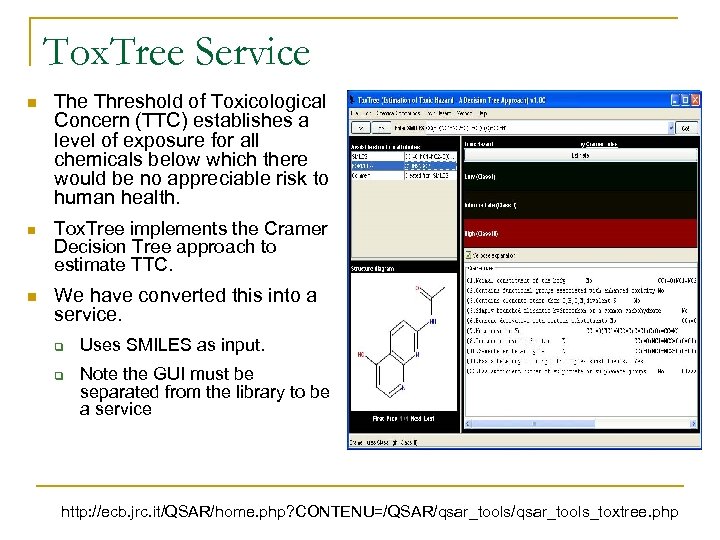
Tox. Tree Service n The Threshold of Toxicological Concern (TTC) establishes a level of exposure for all chemicals below which there would be no appreciable risk to human health. n Tox. Tree implements the Cramer Decision Tree approach to estimate TTC. n We have converted this into a service. q q Uses SMILES as input. Note the GUI must be separated from the library to be a service http: //ecb. jrc. it/QSAR/home. php? CONTENU=/QSAR/qsar_tools_toxtree. php
OSCAR 3 Service n n Oscar 3 is a tool for shallow, chemistry-specific natural language parsing of chemical documents (i. e. journal articles). It identifies (or attempts to identify): q q q n n Results are exported as an XML file. There is a larger effort, Sci. Borg, in this area q n Chemical names: singular nouns, plurals, verbs etc. , also formulae and acronyms. Chemical data: Spectra, melting/boiling point, yield etc. in experimental sections. Other entities: Things like N(5)-C(3) and so on. http: //www. cl. cam. ac. uk/~aac 10/escience/sciborg. html It also has potentially very interesting Workflows http: //wwmm. ch. cam. ac. uk/wikis/wwmm/index. php/Oscar 3

Use Cases and Workflows Putting data and clustering together in a distributed environment.
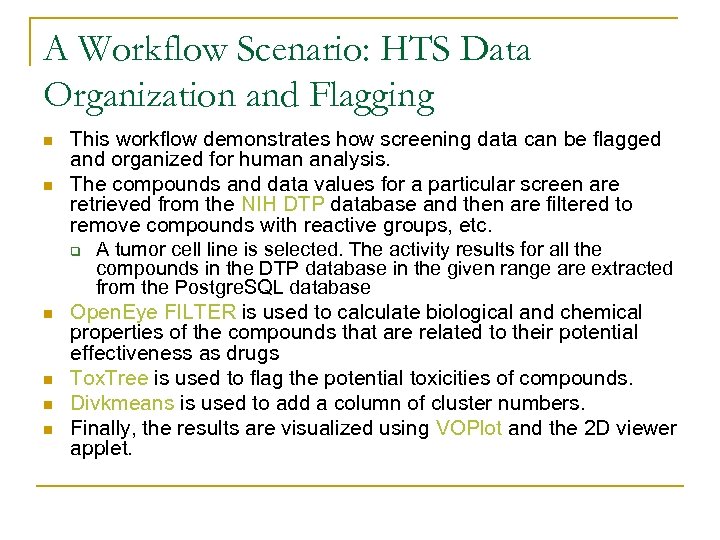
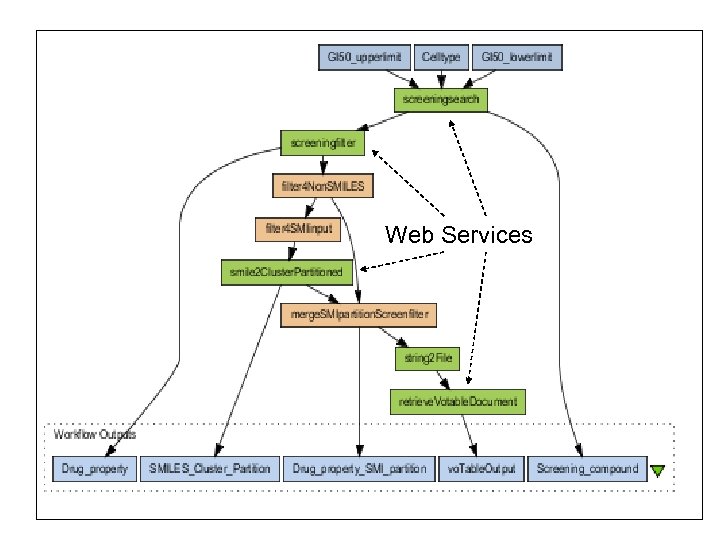
A Workflow Scenario: HTS Data Organization and Flagging n n n This workflow demonstrates how screening data can be flagged and organized for human analysis. The compounds and data values for a particular screen are retrieved from the NIH DTP database and then are filtered to remove compounds with reactive groups, etc. q A tumor cell line is selected. The activity results for all the compounds in the DTP database in the given range are extracted from the Postgre. SQL database Open. Eye FILTER is used to calculate biological and chemical properties of the compounds that are related to their potential effectiveness as drugs Tox. Tree is used to flag the potential toxicities of compounds. Divkmeans is used to add a column of cluster numbers. Finally, the results are visualized using VOPlot and the 2 D viewer applet.
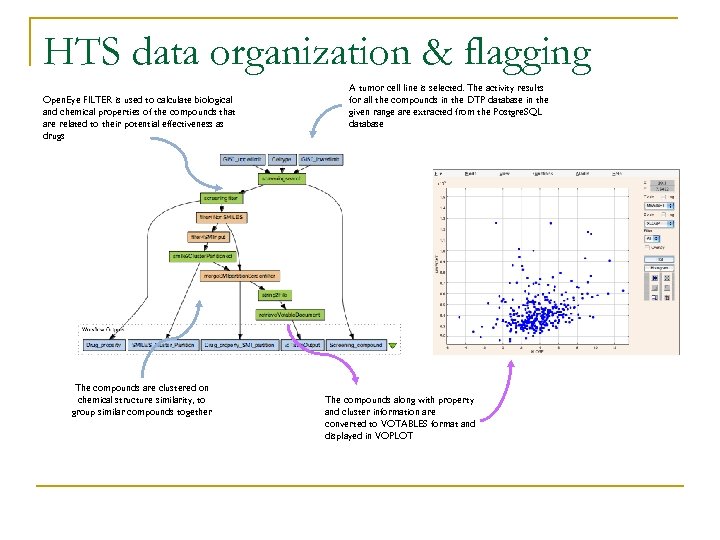
HTS data organization & flagging Open. Eye FILTER is used to calculate biological and chemical properties of the compounds that are related to their potential effectiveness as drugs The compounds are clustered on chemical structure similarity, to group similar compounds together A tumor cell line is selected. The activity results for all the compounds in the DTP database in the given range are extracted from the Postgre. SQL database The compounds along with property and cluster information are converted to VOTABLES format and displayed in VOPLOT
Web Services
Example plots of our workflow output using VOPlot and VOTables
Chemical Informatics and the Tera. Grid
A Workflow for IU’s Big Red Demo n Pub. Med abstracts q 555, 007 Pub. Med abstracts of 2005 – 2006 (part) q 1, 000 abstracts per node n n 511 nodes X 1, 000 input abstracts used for the demo OSCAR 3 q Extracts chemical information from text and produces an XML instance highlighting the chemical information SMILES extraction q Extracting SMILES elements from OSCAR’s XML output files q Unique SMILES list within a batch Use this to drive docking and molecular modeling applications.
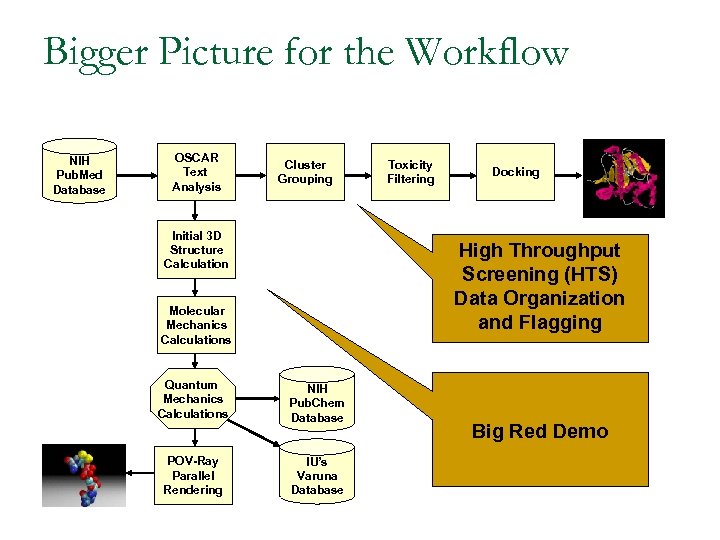
Bigger Picture for the Workflow NIH Pub. Med Database OSCAR Text Analysis Cluster Grouping Initial 3 D Structure Calculation Toxicity Filtering Docking High Throughput Screening (HTS) Data Organization and Flagging Molecular Mechanics Calculations Quantum Mechanics Calculations NIH Pub. Chem Database POV-Ray Parallel Rendering IU’s Varuna Database Big Red Demo
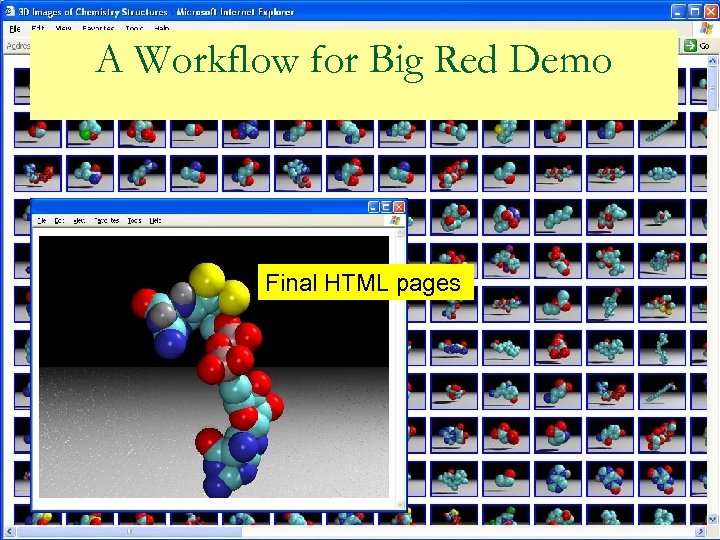
A Workflow for Big Red Demo Final HTML pages

VARUNA – Towards a Grid-based Molecular Modeling Environment Taking the Big Red demo from stunt to science.
Automatic Quantum Mechanical Curation of Structure Data (Auto. Ge. FF) n n n Chemical research logic is often driven by molecular structure Large-scale, small molecule DB’s (such as Pub. Chem and, through OSCAR, Pub. Med) have low-resolution structure data Often key properties are not consistently available: q n QM web-services will provide tools for generating high-resolution data q n e. g. : Rotation-barriers, Redox Potentials, Polarizabilities, IR frequencies, reactivity towards nucleophiles Produces a new, curated database of QM results These can then be combined with databases of proteins (PDB, MOAD, PDB-Bind) for docking and other detailed simulation studies.
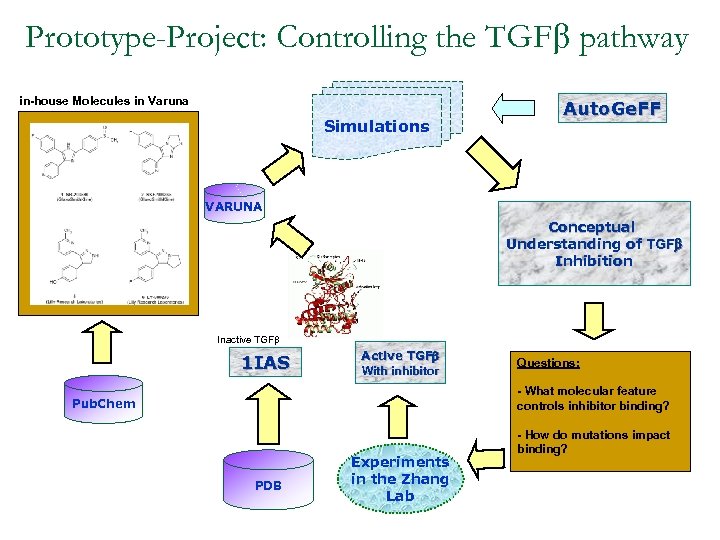
Prototype-Project: Controlling the TGFb pathway in-house Molecules in Varuna Simulations Auto. Ge. FF VARUNA Conceptual Understanding of TGFb Inhibition Inactive TGFb 1 IAS Active TGFb With inhibitor Questions: - What molecular feature controls inhibitor binding? Pub. Chem PDB Experiments in the Zhang Lab - How do mutations impact binding?
More Information n n Contact me: mpierce@cs. indiana. edu Website and wiki: q q n www. chembiogrid. org www. chemibiogrid. org/wiki We have project plus collaborator mailing lists if you really are interested.
54395dd913d1da1ecb6bda98d71e1762.ppt