6dbd955f9983aad248cfb27c0876da98.ppt
- Количество слайдов: 40
Biozon A unified knowledge resource on DNA sequences, proteins, complexes and cellular pathways Golan Yona Department of Computer Science Cornell University Golan Yona, Cornell University Ipam 04
The human genome project The first step - The quaternary code of the cell (organism) It codes proteins and RNA molecules – the basic procedures (many of them have unknown functions) Proteins, RNA and DNA can interact (modules) They form pathways – complex programs with input and output Golan Yona, Cornell University Ipam 04

The unknown(s) Can we decipher the “meaning” of our genome: Can we identify the role of the basic procedures (proteins) Can we predict the interactions between them Can we identify the complex programs (pathways) Can we find regularities? Global principles? (the way proteins are organized into families (p-table), the methods used to compile complex programs from the basic procedures. ) Golan Yona, Cornell University Ipam 04

A very active field Sequence and structure databases (Swiss. Prot, PDB, Gen. Bank) Databases of protein domains and families (Prosite, Pfam, Inter. Pro) Databases of interactions (Bind, DIP) Databases of pathways (Kegg, Meta. Cyc) Golan Yona, Cornell University Ipam 04
The challenge of data integration • The biochemical function of genes depends on their extended biological context – their relations to other genes, the set of interactions they form, the pathways they participate in, their subcellular location and so on. • This broader biological context is important for the characterization of new and existing genes, interactions and pathways. • There is a strong need to corroborate and integrate data from different resources and different aspects of biological systems for the effective analysis of genes and other biological entities (from complexes to protein families and biochemical pathways) Golan Yona, Cornell University Ipam 04

Searching for data • • • Swiss. Prot protein … PDB structure Interactions Pathways DNA sequence The domain structure Similar structures and sequences Protein families Expression data Maybe for a few genes – but on a large scale… Golan Yona, Cornell University Ipam 04

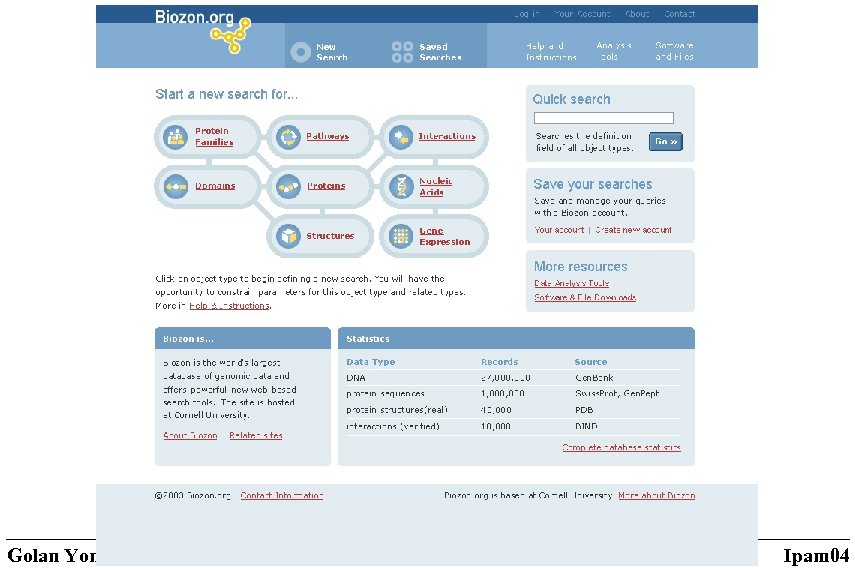
Aaron Birkland The Biozon project A unified biological knowledge resource • An efficient system for storage, retrieval manipulation and exploration of biological data (both at the macro-molecular and the cellular level) • Integration of multiple sources (tens of databases plus inhouse computations) – the keys are the physical objects • Emphasis on multi-feature protein and DNA characterization and classification. Golan Yona, Cornell University Ipam 04
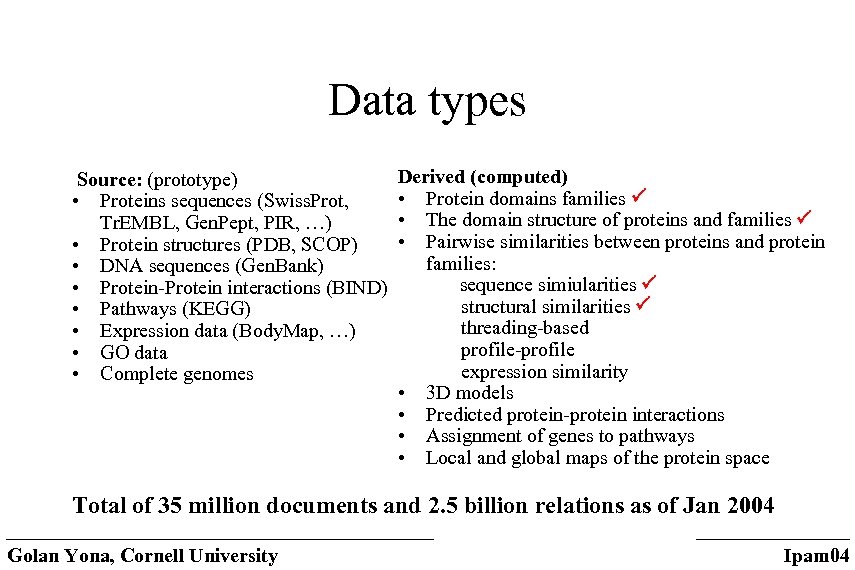
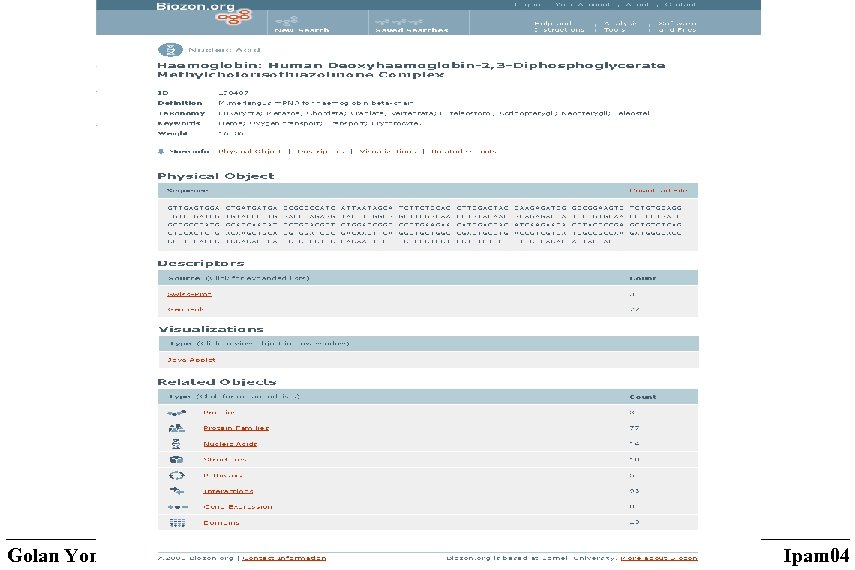
Data types Source: (prototype) • Proteins sequences (Swiss. Prot, Tr. EMBL, Gen. Pept, PIR, …) • Protein structures (PDB, SCOP) • DNA sequences (Gen. Bank) • Protein-Protein interactions (BIND) • Pathways (KEGG) • Expression data (Body. Map, …) • GO data • Complete genomes Derived (computed) • Protein domains families • The domain structure of proteins and families • Pairwise similarities between proteins and protein families: sequence simiularities structural similarities threading-based profile-profile expression similarity • 3 D models • Predicted protein-protein interactions • Assignment of genes to pathways • Local and global maps of the protein space Total of 35 million documents and 2. 5 billion relations as of Jan 2004 Golan Yona, Cornell University Ipam 04
Golan Yona, Cornell University Ipam 04
Golan Yona, Cornell University Ipam 04
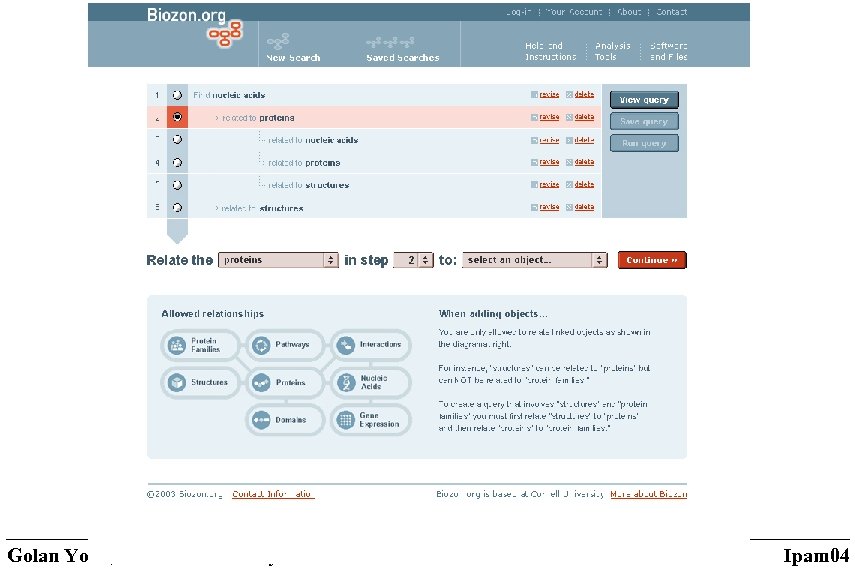
Querying data • Complex queries – span different data types – All proteins with known structures that participate in known interactions – All pathways that contain proteins with solved structures – The structures of these proteins – DNA sequences that encode for proteins kinases Golan Yona, Cornell University Ipam 04
Golan Yona, Cornell University Ipam 04
Expandable model • Current: semi-automatic updates • Stability: consistency and integrity through the use of triggers, time stamps • Scalability: The database was designed so as to allow easy integration of other data types and other databases. The Amazon. com model • Optimality: Using a concise representation of computed distances, recovery upon retrieval (optimal results as opposed to heuristics) Warehouse of computed data Golan Yona, Cornell University Ipam 04
Methods Part 1: First identify the functions of the basic procedures (proteins): • • A) Identify the evolutionary building blocks of proteins. B) Develop new representations for proteins and methods to measure similarity. Statistical models for protein families. C) Embedding techniques to study the geometry of the protein universe. Grammars to study the derivation rules D) Unsupervised learning techniques to find the statistical regularities Part 2: Machine learning techniques to predict interactions Part 3: Algorithms for pathway prediction Part 4: Learning algorithms to identify the elements of cellular computations Part 5: The whole picture … BIOZON Golan Yona, Cornell University Ipam 04
Methods Part 1: First identify the functions of the basic procedures (proteins): • • A) Identify the evolutionary building blocks of proteins. B) Develop new representations for proteins and methods to measure similarity. Statistical models for protein families. C) Embedding techniques to study the geometry of the protein universe. Grammars to study the derivation rules D) Unsupervised learning techniques to find the statistical regularities Part 2: Machine learning techniques to predict interactions Part 3: Algorithms for pathway prediction Part 4: Learning algorithms to identify the elements of cellular computations Part 5: The whole picture … BIOZON Golan Yona, Cornell University Ipam 04
The domain structure of a protein (with Niranjan Nagarajan) • A domain is considered the fundamental unit of protein structure, folding, function, evolution and design. • Compact • Stable • Folds independently? • Has a specific function Golan Yona, Cornell University Ipam 04
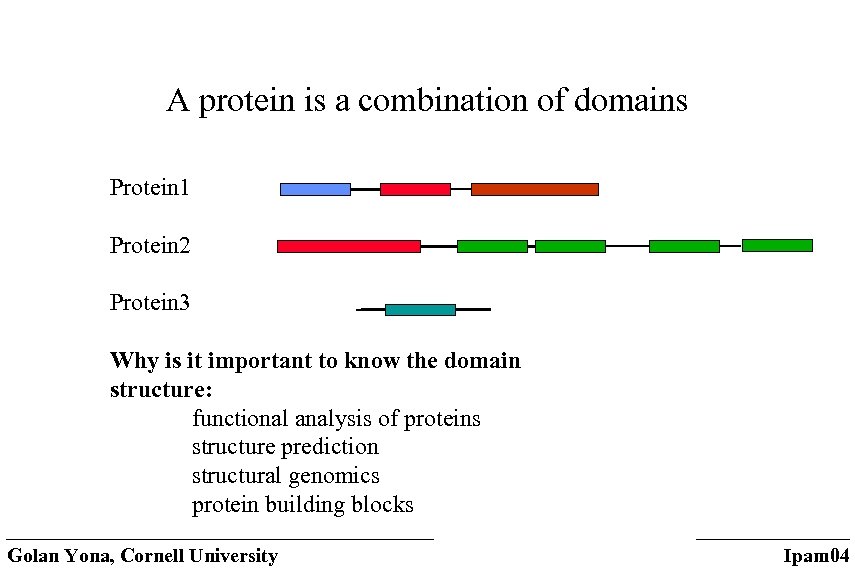
A protein is a combination of domains Protein 1 Protein 2 Protein 3 Why is it important to know the domain structure: functional analysis of proteins structure prediction structural genomics protein building blocks Golan Yona, Cornell University Ipam 04
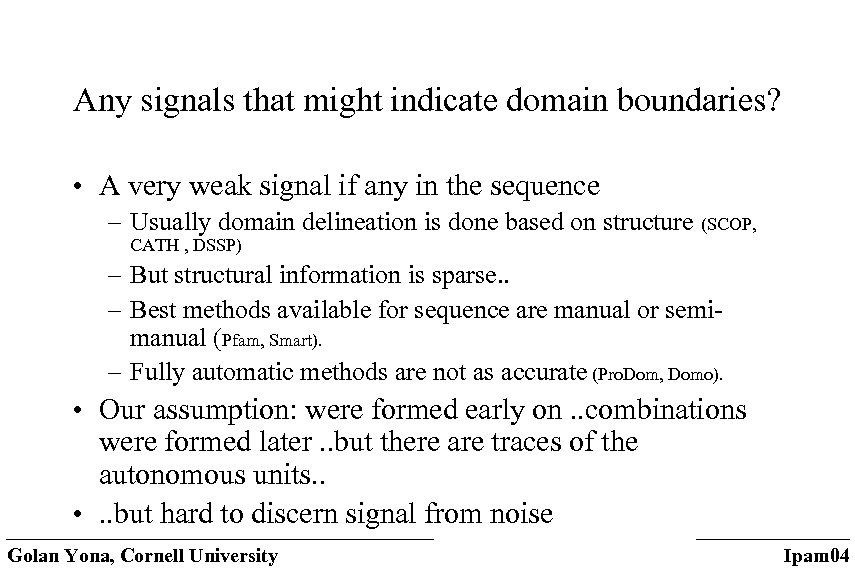
Any signals that might indicate domain boundaries? • A very weak signal if any in the sequence – Usually domain delineation is done based on structure (SCOP, CATH , DSSP) – But structural information is sparse. . – Best methods available for sequence are manual or semimanual (Pfam, Smart). – Fully automatic methods are not as accurate (Pro. Dom, Domo). • Our assumption: were formed early on. . combinations were formed later. . but there are traces of the autonomous units. . • . . but hard to discern signal from noise Golan Yona, Cornell University Ipam 04
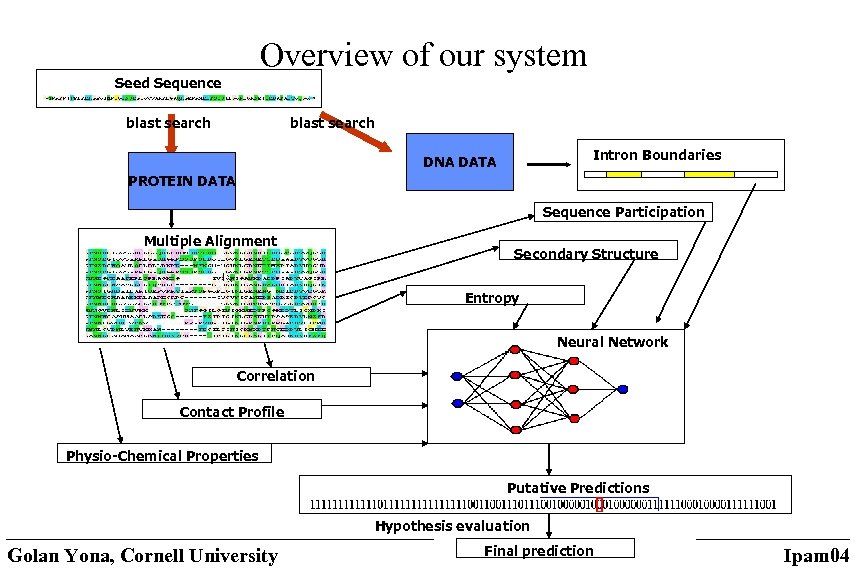
Overview of our system Seed Sequence blast search Intron Boundaries DNA DATA PROTEIN DATA Sequence Participation Multiple Alignment Secondary Structure Entropy Neural Network Correlation Contact Profile Physio-Chemical Properties Putative Predictions Hypothesis evaluation Golan Yona, Cornell University Final prediction Ipam 04
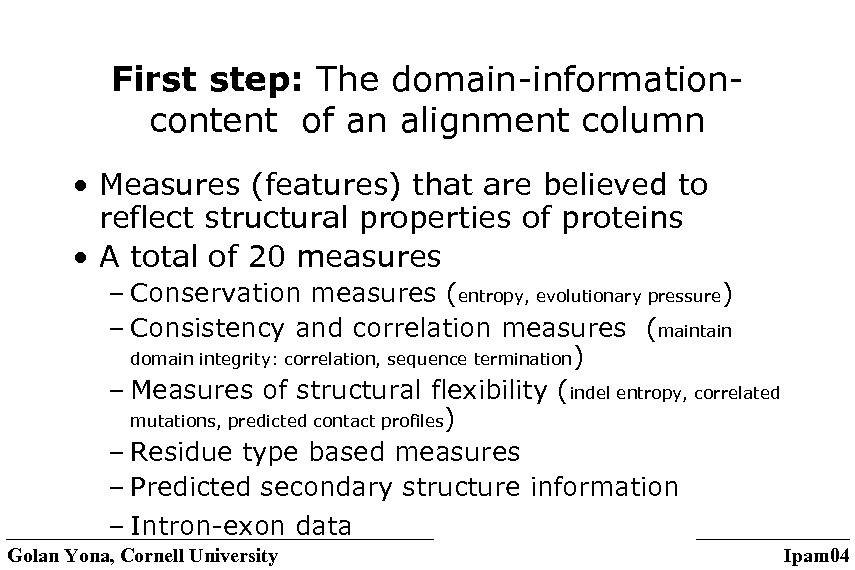
First step: The domain-informationcontent of an alignment column • Measures (features) that are believed to reflect structural properties of proteins • A total of 20 measures – Conservation measures (entropy, evolutionary pressure) – Consistency and correlation measures (maintain domain integrity: correlation, sequence termination) – Measures of structural flexibility (indel entropy, correlated mutations, predicted contact profiles) – Residue type based measures – Predicted secondary structure information – Intron-exon data Golan Yona, Cornell University Ipam 04
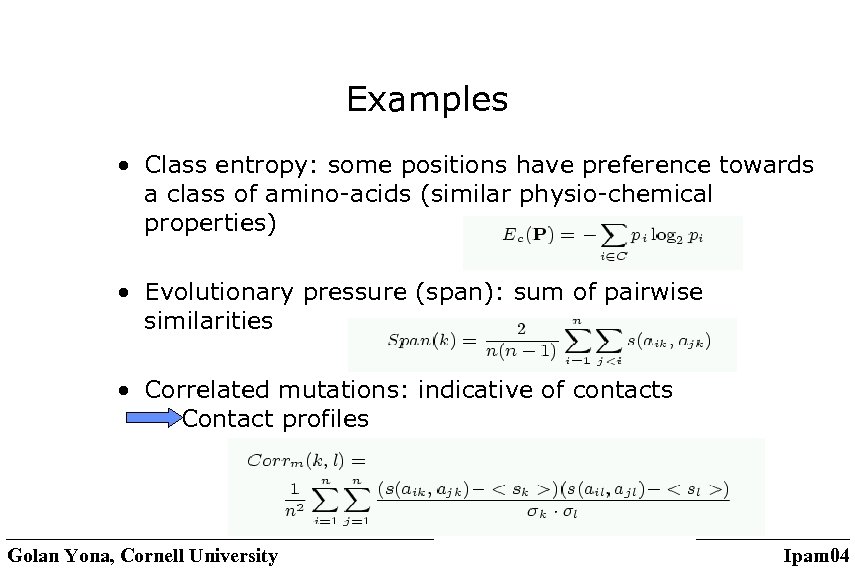
Examples • Class entropy: some positions have preference towards a class of amino-acids (similar physio-chemical properties) • Evolutionary pressure (span): sum of pairwise similarities • Correlated mutations: indicative of contacts Contact profiles Golan Yona, Cornell University Ipam 04
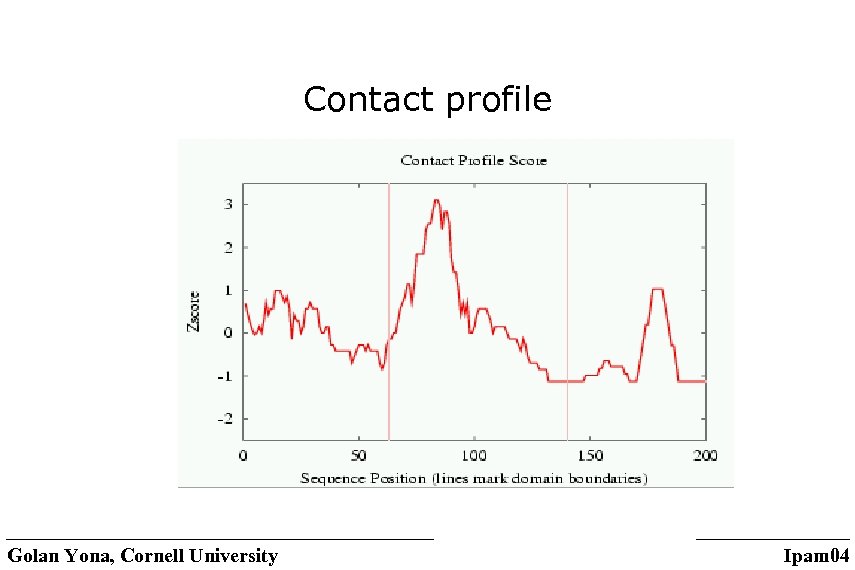
Contact profile Golan Yona, Cornell University Ipam 04
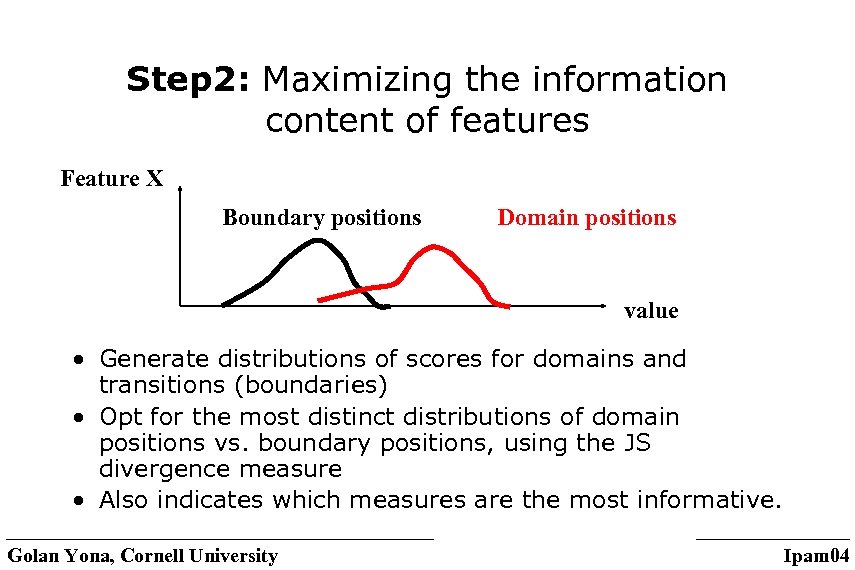
Step 2: Maximizing the information content of features Feature X Boundary positions Domain positions value • Generate distributions of scores for domains and transitions (boundaries) • Opt for the most distinct distributions of domain positions vs. boundary positions, using the JS divergence measure • Also indicates which measures are the most informative. Golan Yona, Cornell University Ipam 04
Step 3: The learning system • A neural network is trained to model the complex decision boundary surface • Predicts correctly 94% of domain positions and 88% of the transitions in the test set Golan Yona, Cornell University Ipam 04
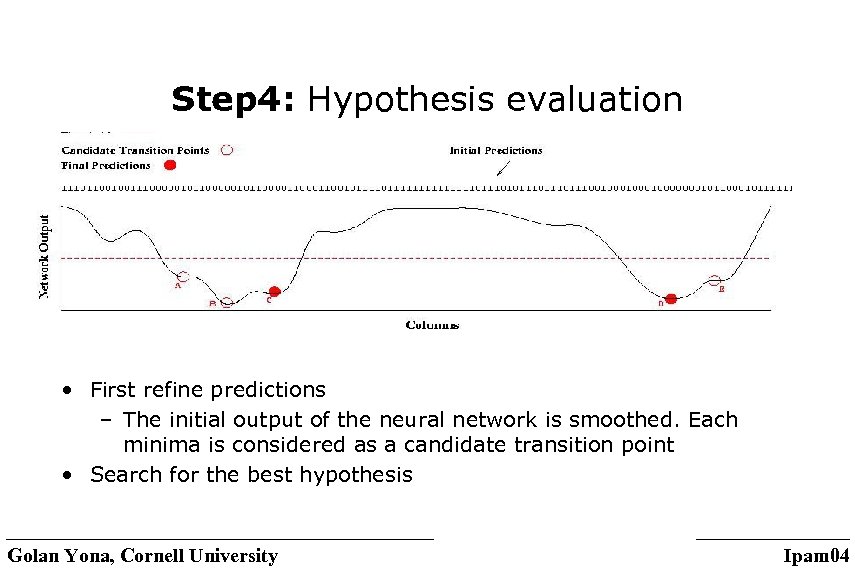
Step 4: Hypothesis evaluation • First refine predictions – The initial output of the neural network is smoothed. Each minima is considered as a candidate transition point • Search for the best hypothesis Golan Yona, Cornell University Ipam 04
The domain generator model • Finds the best of all possible hypotheses • We assume a model: random generator that moves repeatedly between a domain state and a linker state and emits one domain or transition at a time according to different source probability distributions. • Total probability is the product Golan Yona, Cornell University Ipam 04
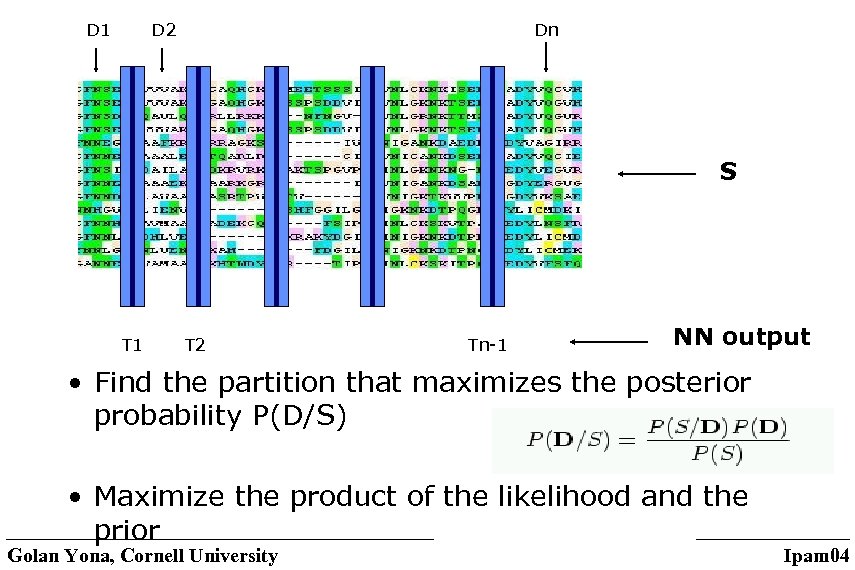
D 1 D 2 Dn S T 1 T 2 Tn-1 NN output • Find the partition that maximizes the posterior probability P(D/S) • Maximize the product of the likelihood and the prior Golan Yona, Cornell University Ipam 04
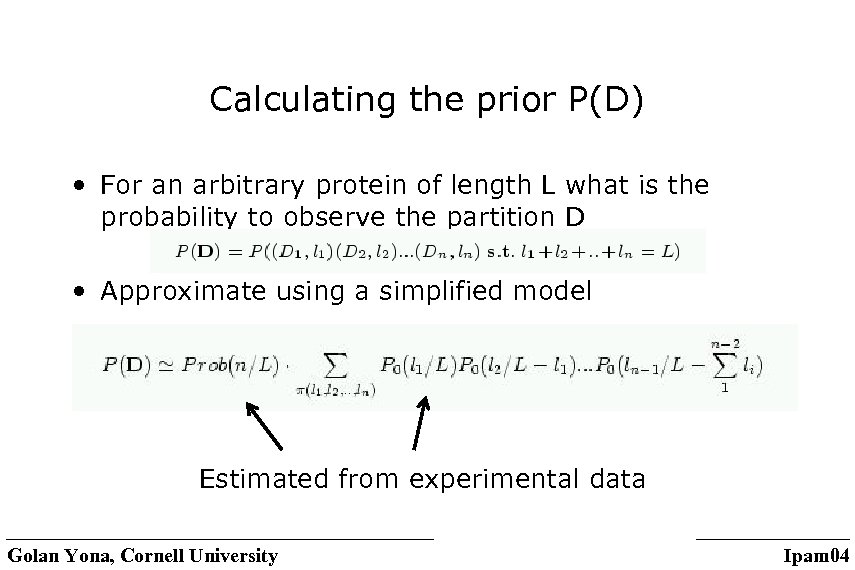
Calculating the prior P(D) • For an arbitrary protein of length L what is the probability to observe the partition D • Approximate using a simplified model Estimated from experimental data Golan Yona, Cornell University Ipam 04
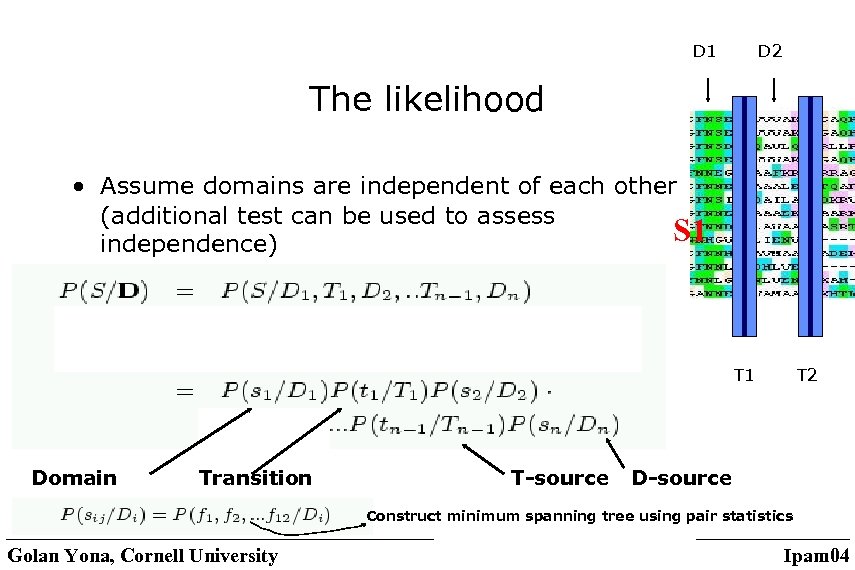
D 1 D 2 The likelihood • Assume domains are independent of each other (additional test can be used to assess S 1 independence) T 1 Domain Transition T-source T 2 D-source Construct minimum spanning tree using pair statistics Golan Yona, Cornell University Ipam 04
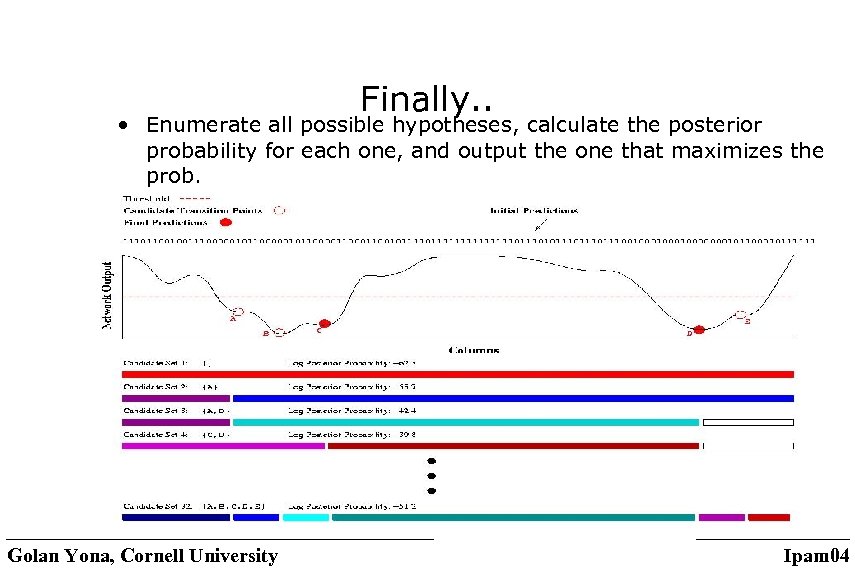
Finally. . • Enumerate all possible hypotheses, calculate the posterior probability for each one, and output the one that maximizes the prob. Golan Yona, Cornell University Ipam 04
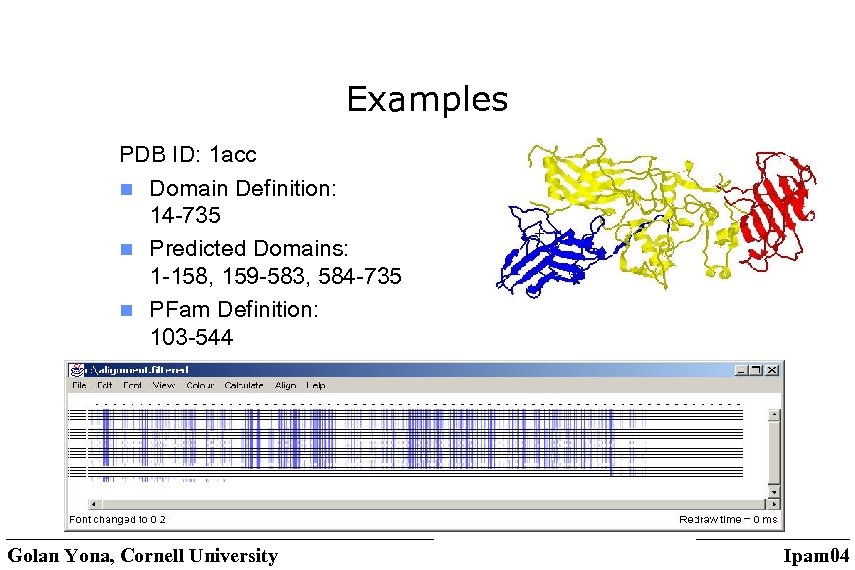
Examples PDB ID: 1 acc n Domain Definition: 14 -735 n Predicted Domains: 1 -158, 159 -583, 584 -735 n PFam Definition: 103 -544 Golan Yona, Cornell University Ipam 04
Methods Part 1: First identify the functions of the basic procedures (proteins): • • A) Identify the evolutionary building blocks of proteins. B) Develop new representations for proteins and methods to measure similarity. Statistical models for protein families. C) Embedding techniques to study the geometry of the protein universe. Grammars to study the derivation rules D) Unsupervised learning techniques to find the statistical regularities Part 2: Machine learning techniques to predict interactions Part 3: Algorithms for pathway prediction Part 4: Learning algorithms to identify the elements of cellular computations Part 5: The whole picture … BIOZON Golan Yona, Cornell University Ipam 04
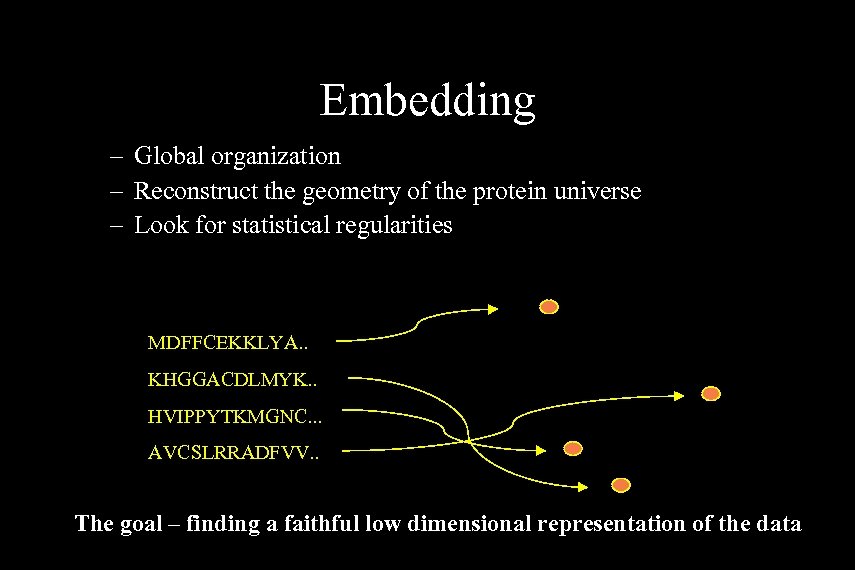
Embedding – Global organization – Reconstruct the geometry of the protein universe – Look for statistical regularities MDFFCEKKLYA. . KHGGACDLMYK. . HVIPPYTKMGNC. . . AVCSLRRADFVV. . The goal – finding a faithful low dimensional representation of the data Golan Yona, Cornell University Ipam 04
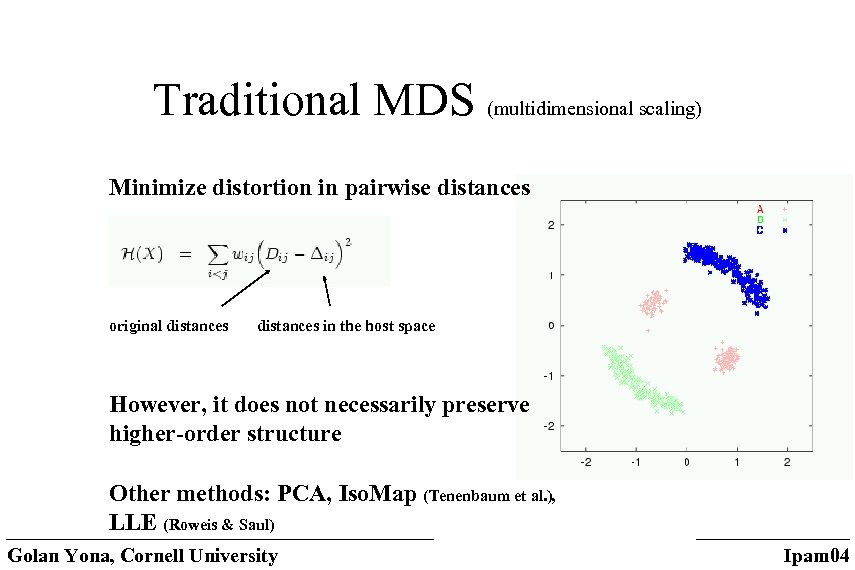
Traditional MDS (multidimensional scaling) Minimize distortion in pairwise distances original distances in the host space However, it does not necessarily preserve higher-order structure Other methods: PCA, Iso. Map (Tenenbaum et al. ), LLE (Roweis & Saul) Golan Yona, Cornell University Ipam 04
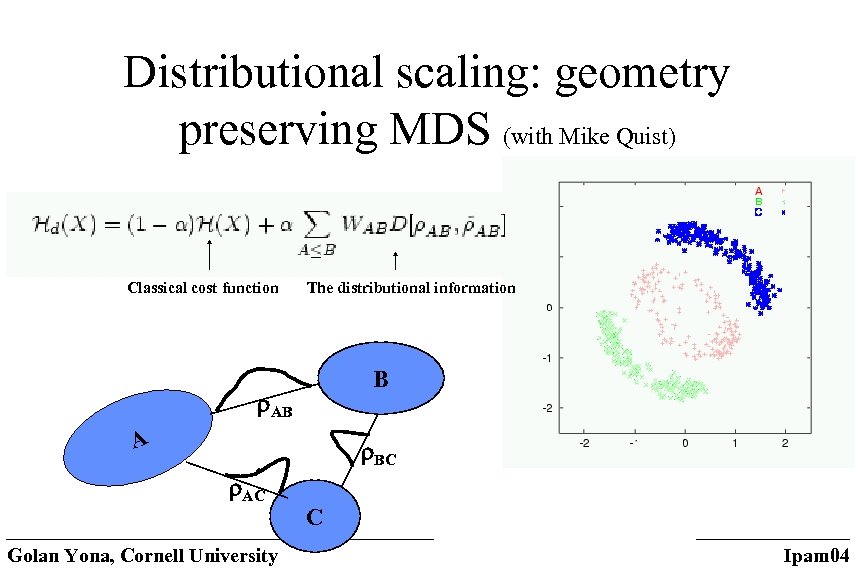
Distributional scaling: geometry preserving MDS (with Mike Quist) Classical cost function The distributional information B AB A BC AC Golan Yona, Cornell University C Ipam 04
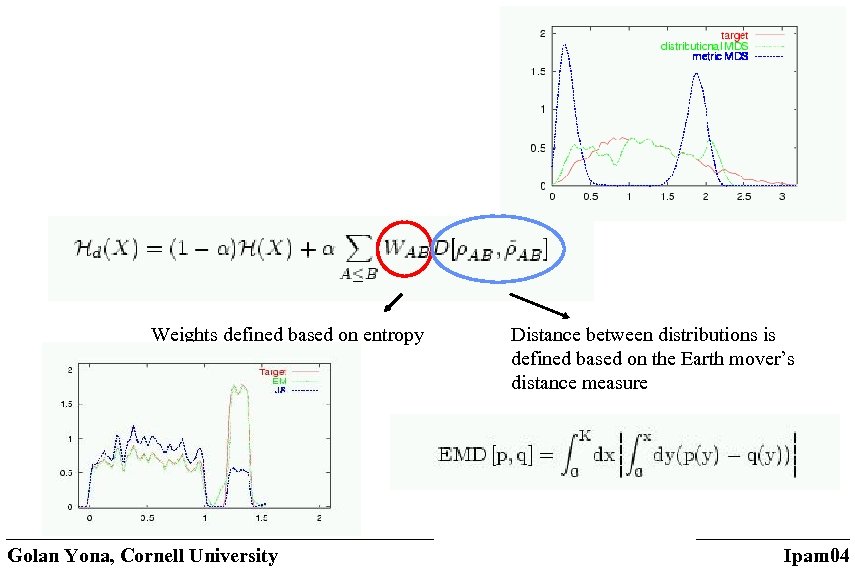
Weights defined based on entropy Golan Yona, Cornell University Distance between distributions is defined based on the Earth mover’s distance measure Ipam 04
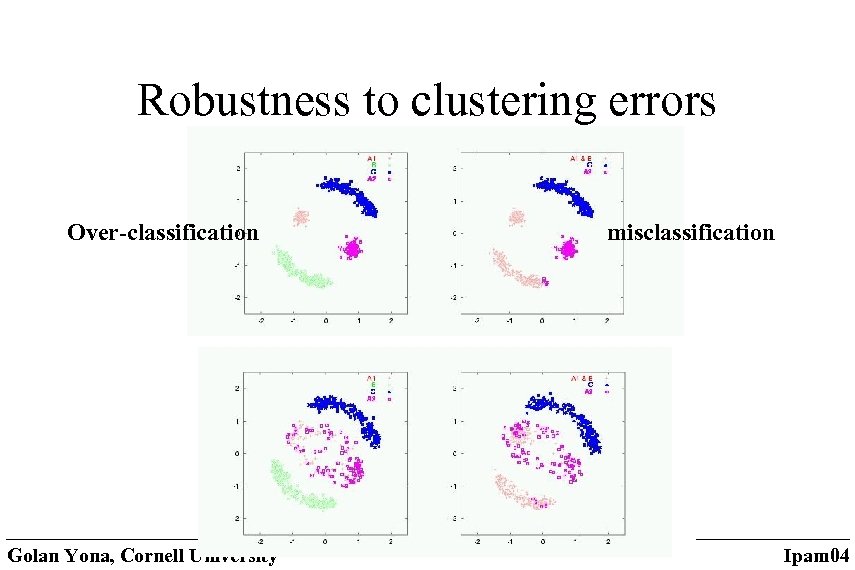
Robustness to clustering errors Over-classification Golan Yona, Cornell University misclassification Ipam 04
Global Map of the Protein Space Golan Yona, Cornell University Ipam 04

Acknowledgments • My students: Aaron Birkland - Biozon Niranjan Nagarajan – domain prediction, protein-protein interactions Umar Syed – the mixture model of stochastic decision trees, function prediction Mike Quist - embedding Bill Dirks – analysis of expression data Liviu Popescu – pathway prediction Jason Davis - protein-protein interactions Garmay Leung – structure comparison Richard Chung – structural profile-profile comparison Hugh Edwards, Chris Chau, Rob Cronin, Taruna Seth, Bo Fuld, Adi Alon, Arthur Kong, Wilmin Martono, Keith Jamison, John Tam, Allen Wang, Kuan Chang, William Yeh, Charitha Tillekeratne NEXT Golan Yona, Cornell University Ipam 04

Acknowledgments Collaborations: • Ran El-Yaniv • Klara Kedem • Dave Lin Funding: • NSF • SUN Microsystems Golan Yona, Cornell University Ipam 04
6dbd955f9983aad248cfb27c0876da98.ppt