cd8d26dd3a526131337906d5f5328933.ppt
- Количество слайдов: 20

Bionformatics and the protein folding problem: sequence analysis and structure comparison of the SH 3 domain Stefan M. Larson 1, Ariel Di Nardo 2, Alan R. Davidson 2, 3 1 Biophysics Program, Department of Structural Biology, Stanford University 2 Department of Biochemistry, University of Toronto 3 Department of Molecular and Medical Genetics, University of Toronto
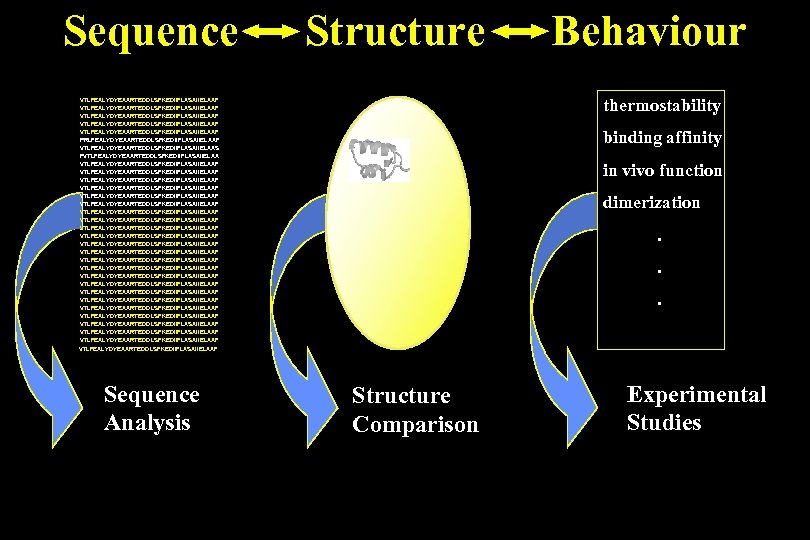
Sequence Structure thermostability VTLFEALYDYEAARTEDDLSFKEDIIFLASAIIELAAF VTLFEALYDYEAARTEDDLSFKEDIIFLASAIIELAAF PRLFEALYDYEAARTEDDLSFKEDIIFLASAIIELAAF VTLFEALYDYEAARTEDDLSFKEDIIFLASAIIELAAS FVTLFEALYDYEAARTEDDLSFKEDIIFLASAIIELAAF VTLFEALYDYEAARTEDDLSFKEDIIFLASAIIELAAF VTLFEALYDYEAARTEDDLSFKEDIIFLASAIIELAAF VTLFEALYDYEAARTEDDLSFKEDIIFLASAIIELAAF VTLFEALYDYEAARTEDDLSFKEDIIFLASAIIELAAF VTLFEALYDYEAARTEDDLSFKEDIIFLASAIIELAAF VTLFEALYDYEAARTEDDLSFKEDIIFLASAIIELAAF Sequence Analysis Behaviour binding affinity in vivo function dimerization. . . Structure Comparison Experimental Studies
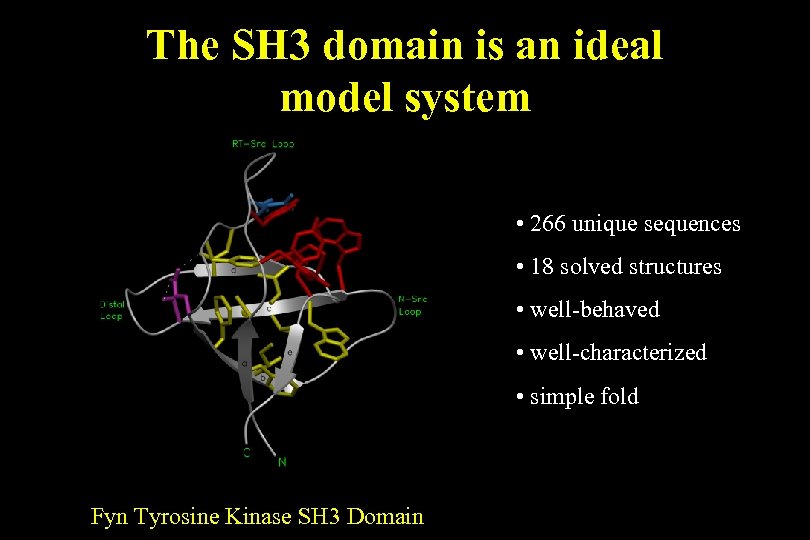
The SH 3 domain is an ideal model system • 266 unique sequences • 18 solved structures • well-behaved • well-characterized • simple fold Fyn Tyrosine Kinase SH 3 Domain
Aims of the study 1. Assemble a complete and accurate alignment of all available SH 3 sequences. 2. Analyze residue frequencies and conservation patterns in the sequence alignment to quantitate sequence variation in the SH 3 fold. 3. Develop an algorithm for covariation analysis which detects meaningful residue interactions within the SH 3 domain. 4. Interpret conservation and covariation patterns to identify residues and interactions critical for stability and function of the SH 3 domain. 5. Align and rigorously compare all available SH 3 structures to quantitate the structural variation in the SH 3 domain. 6. Compare the results of sequence alignment analysis and structure comparison to provide insight in the sequence-structure relationship.
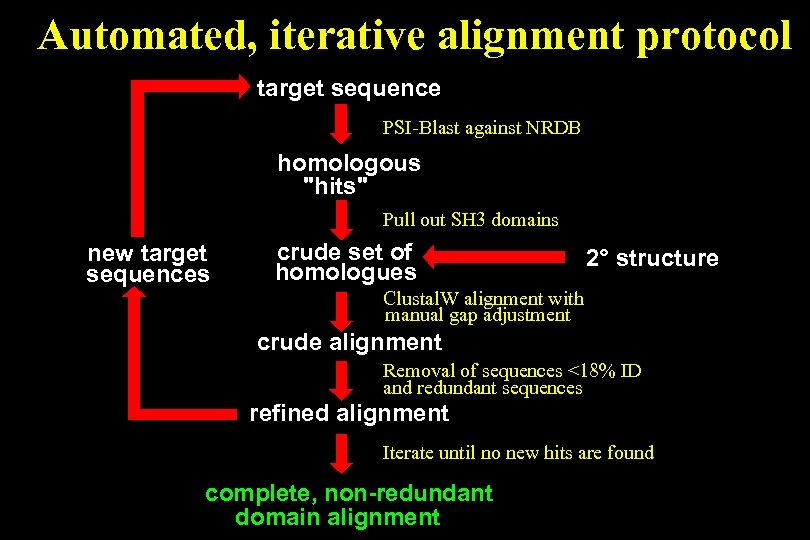
Automated, iterative alignment protocol target sequence PSI-Blast against NRDB homologous "hits" Pull out SH 3 domains new target sequences crude set of homologues 2° structure Clustal. W alignment with manual gap adjustment crude alignment Removal of sequences <18% ID and redundant sequences refined alignment Iterate until no new hits are found complete, non-redundant domain alignment
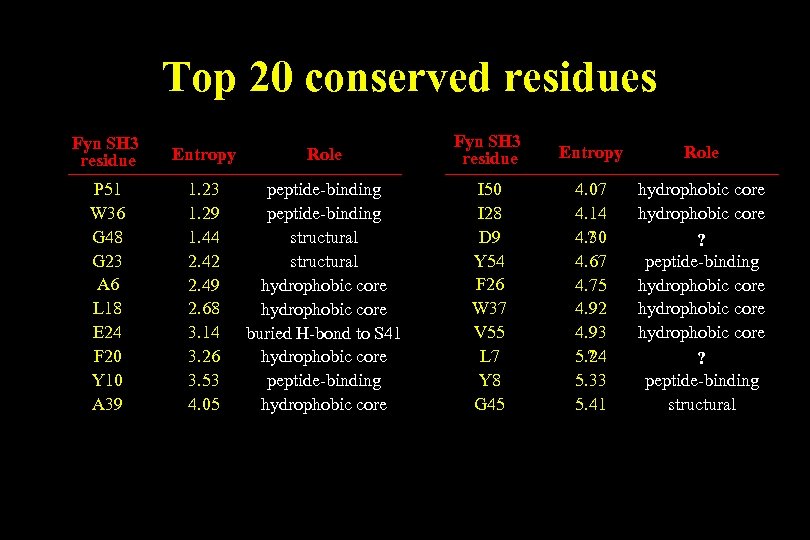
Top 20 conserved residues Fyn SH 3 residue Entropy P 51 W 36 G 48 G 23 A 6 L 18 E 24 F 20 Y 10 A 39 1. 23 1. 29 1. 44 2. 42 2. 49 2. 68 3. 14 3. 26 3. 53 4. 05 Role Fyn SH 3 residue Entropy Role peptide-binding structural hydrophobic core buried H-bond to S 41 hydrophobic core peptide-binding hydrophobic core I 50 I 28 D 9 Y 54 F 26 W 37 V 55 L 7 Y 8 G 45 4. 07 4. 14 4. 30 ? 4. 67 4. 75 4. 92 4. 93 5. 24 ? 5. 33 5. 41 hydrophobic core ? peptide-binding hydrophobic core ? peptide-binding structural
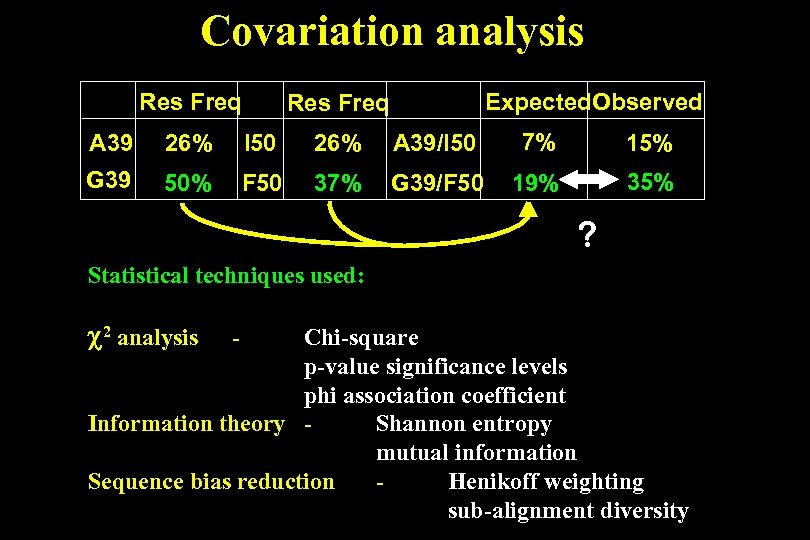
Covariation analysis Res Freq Expected. Observed Res Freq A 39 26% I 50 26% A 39/I 50 7% 15% G 39 50% F 50 37% G 39/F 50 19% 35% ? Statistical techniques used: 2 analysis - Chi-square p-value significance levels phi association coefficient Information theory Shannon entropy mutual information Sequence bias reduction Henikoff weighting sub-alignment diversity
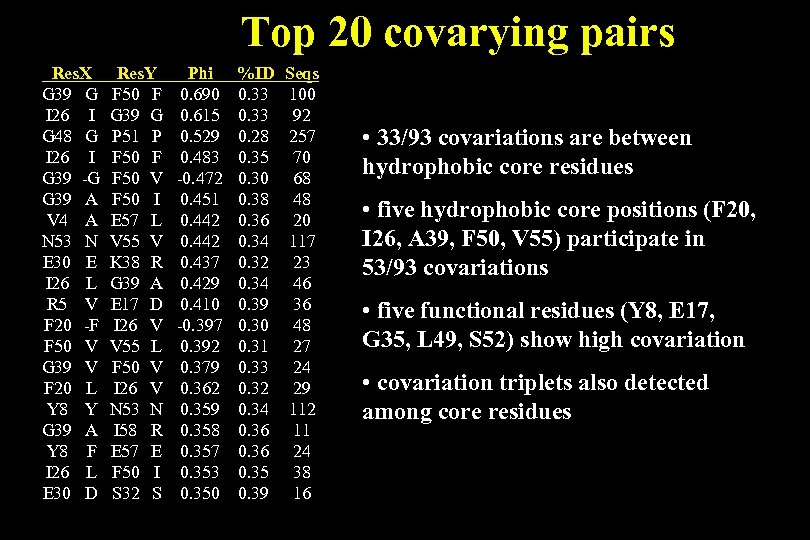
Top 20 covarying pairs Res. X G 39 G I 26 I G 48 G I 26 I G 39 -G G 39 A V 4 A N 53 N E 30 E I 26 L R 5 V F 20 -F F 50 V G 39 V F 20 L Y 8 Y G 39 A Y 8 F I 26 L E 30 D Res. Y F 50 F G 39 G P 51 P F 50 F F 50 V F 50 I E 57 L V 55 V K 38 R G 39 A E 17 D I 26 V V 55 L F 50 V I 26 V N 53 N I 58 R E 57 E F 50 I S 32 S Phi 0. 690 0. 615 0. 529 0. 483 -0. 472 0. 451 0. 442 0. 437 0. 429 0. 410 -0. 397 0. 392 0. 379 0. 362 0. 359 0. 358 0. 357 0. 353 0. 350 %ID 0. 33 0. 28 0. 35 0. 30 0. 38 0. 36 0. 34 0. 32 0. 34 0. 39 0. 30 0. 31 0. 33 0. 32 0. 34 0. 36 0. 35 0. 39 Seqs 100 92 257 70 68 48 20 117 23 46 36 48 27 24 29 112 11 24 38 16 • 33/93 covariations are between hydrophobic core residues • five hydrophobic core positions (F 20, I 26, A 39, F 50, V 55) participate in 53/93 covariations • five functional residues (Y 8, E 17, G 35, L 49, S 52) show high covariation • covariation triplets also detected among core residues
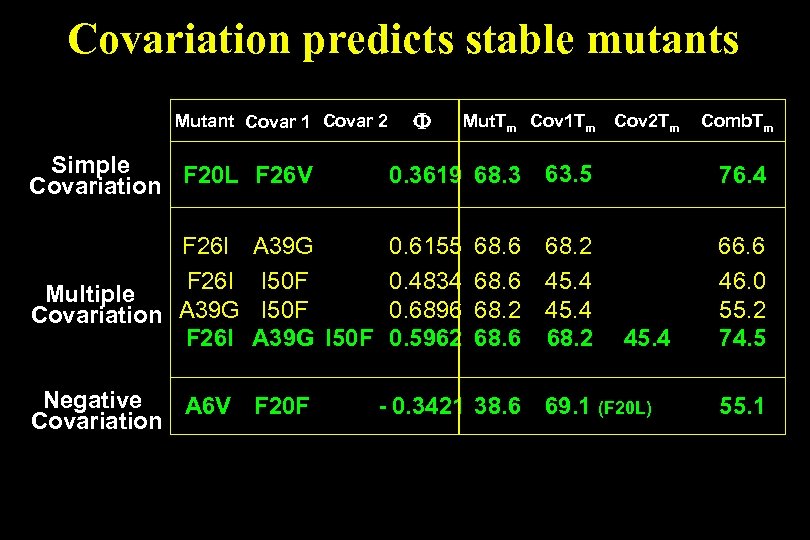
Covariation predicts stable mutants Mutant Covar 1 Covar 2 Mut. Tm Cov 1 Tm Cov 2 Tm Comb. Tm Simple Covariation F 20 L F 26 V 0. 3619 68. 3 63. 5 76. 4 F 26 I Multiple Covariation A 39 G F 26 I 0. 6155 0. 4834 0. 6896 0. 5962 68. 2 45. 4 68. 2 66. 6 46. 0 55. 2 74. 5 A 39 G I 50 F Negative A 6 V F 20 F Covariation 68. 6 68. 2 68. 6 45. 4 - 0. 3421 38. 6 69. 1 (F 20 L) 55. 1
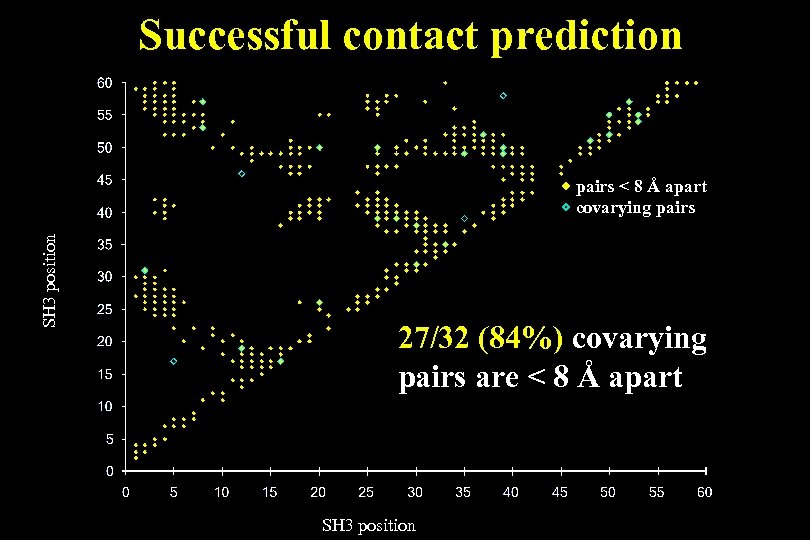
Successful contact prediction SH 3 position pairs < 8 Å apart covarying pairs 27/32 (84%) covarying pairs are < 8 Å apart SH 3 position
Structural variation in SH 3 domains
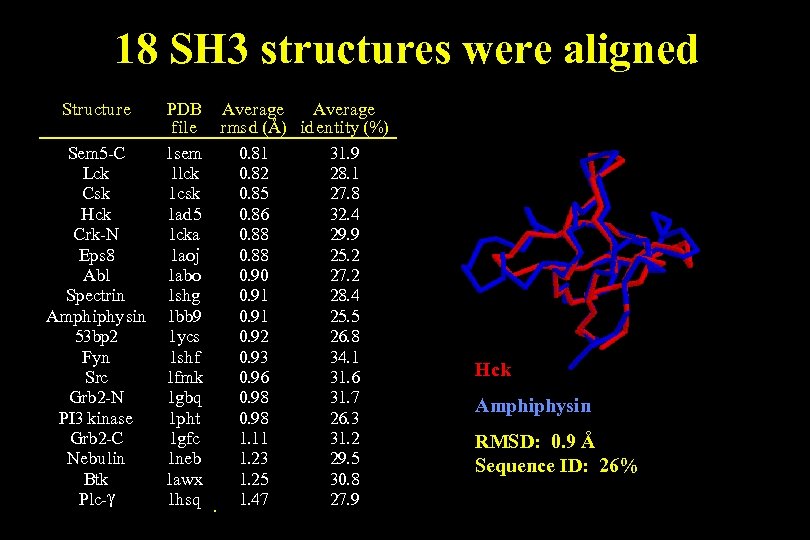
18 SH 3 structures were aligned Structure Sem 5 -C Lck Csk Hck Crk-N Eps 8 Abl Spectrin Amphiphysin 53 bp 2 Fyn Src Grb 2 -N PI 3 kinase Grb 2 -C Nebulin Btk Plc-g PDB Average file rmsd (Å) identity (%) 1 sem 0. 81 31. 9 1 lck 0. 82 28. 1 1 csk 0. 85 27. 8 1 ad 5 0. 86 32. 4 1 cka 0. 88 29. 9 1 aoj 0. 88 25. 2 1 abo 0. 90 27. 2 1 shg 0. 91 28. 4 1 bb 9 0. 91 25. 5 1 ycs 0. 92 26. 8 1 shf 0. 93 34. 1 1 fmk 0. 96 31. 6 1 gbq 0. 98 31. 7 1 pht 0. 98 26. 3 1 gfc 1. 11 31. 2 1 neb 1. 23 29. 5 1 awx 1. 25 30. 8 1 hsq 1. 47 27. 9 Hck Amphiphysin RMSD: 0. 9 Å Sequence ID: 26%
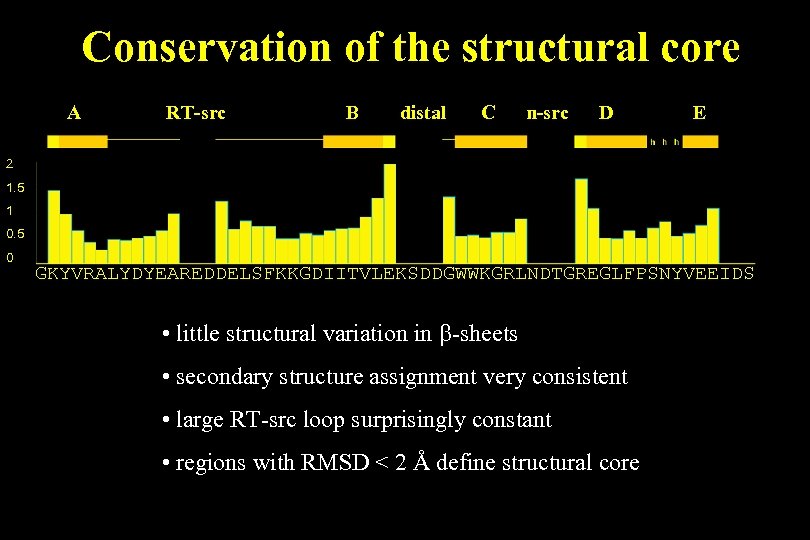
Conservation of the structural core A RT-src B distal C n-src D E 2 1. 5 1 0. 5 0 GKYVRALYDYEAREDDELSFKKGDIITVLEKSDDGWWKGRLNDTGREGLFPSNYVEEIDS • little structural variation in b-sheets • secondary structure assignment very consistent • large RT-src loop surprisingly constant • regions with RMSD < 2 Å define structural core
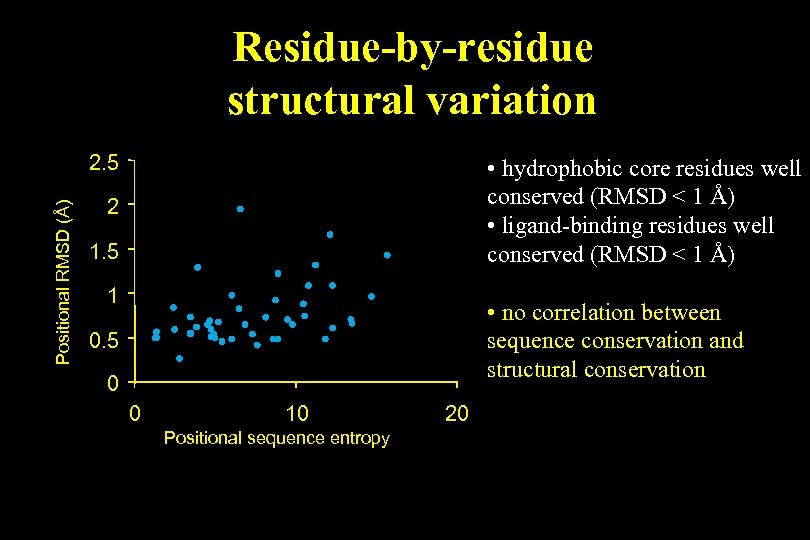
Residue-by-residue structural variation Positional RMSD (Å) 2. 5 • hydrophobic core residues well conserved (RMSD < 1 Å) • ligand-binding residues well conserved (RMSD < 1 Å) 2 1. 5 1 • no correlation between sequence conservation and structural conservation 0. 5 0 0 10 Positional sequence entropy 20
Function of SH 3 domains
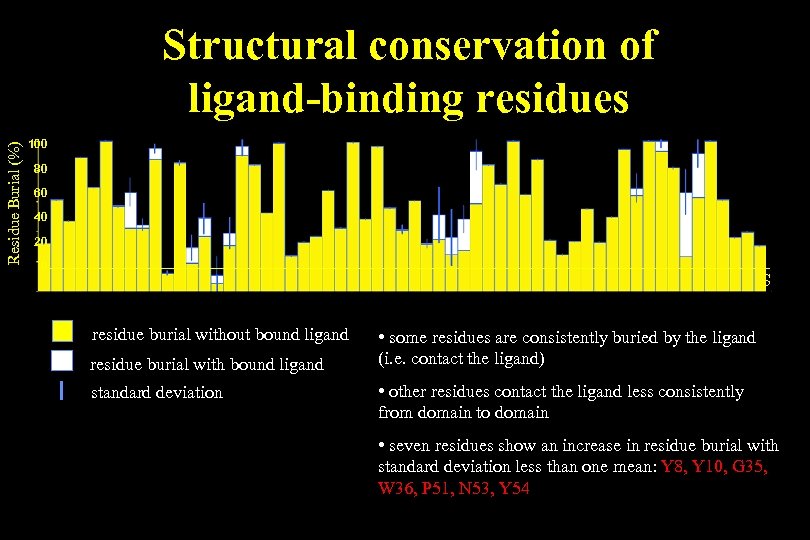
Residue Burial (%) Structural conservation of ligand-binding residues 100 80 60 40 20 0 GKYVRALYDYEAREDDELSFKKGDIITVLEKSDDGWWKGRLNDTGREGLFPSNYVEEIDS residue burial without bound ligand residue burial with bound ligand standard deviation • some residues are consistently buried by the ligand (i. e. contact the ligand) • other residues contact the ligand less consistently from domain to domain • seven residues show an increase in residue burial with standard deviation less than one mean: Y 8, Y 10, G 35, W 36, P 51, N 53, Y 54
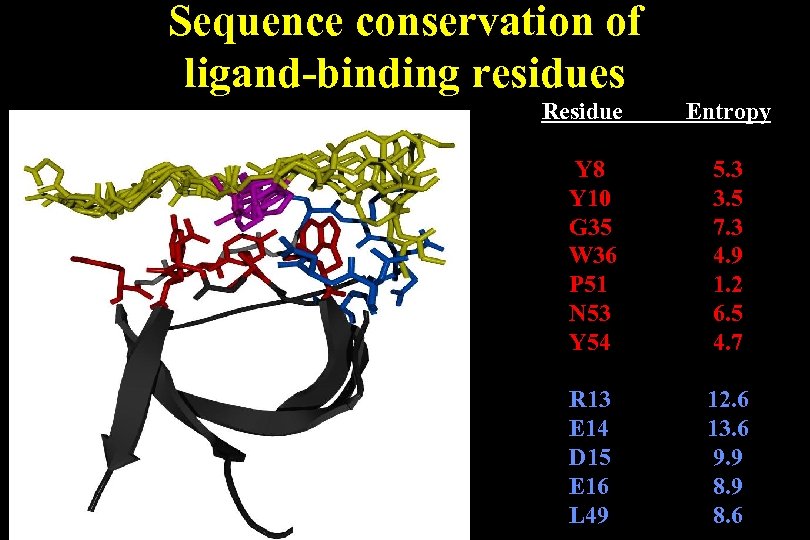
Sequence conservation of ligand-binding residues Residue Entropy Y 8 Y 10 G 35 W 36 P 51 N 53 Y 54 5. 3 3. 5 7. 3 4. 9 1. 2 6. 5 4. 7 R 13 E 14 D 15 E 16 L 49 12. 6 13. 6 9. 9 8. 6

Conclusions 1. Important sequence-structure relationships in the SH 3 domain are subtle, and are missed by studying only a single sequence and/or structure. 2. Covariation data was used to make accurate predictions about stabilizing mutations and residue contacts. 3. Residues participating in structurally conserved ligand contacts are more sequence conserved than residues contacting the ligand less consistently. This may be a source of binding specificity. 4. Bioinformatics was successfully used to gain valuable data in an already very well-characterized system

Acknowledgements Davidson Lab Dr. Alan Davidson Ariel Di Nardo Julian Northey Arianna Rath Supervisory Committee Dr. Richard Collins Dr. Chris Hogue

References Larson SM, Di Nardo AA, Davidson AR. (2000) "Analysis of covariation in an SH 3 domain sequence alignment: applications in tertiary contact prediction and the design of compensating hydrophobic core substitutions. "Journal of Molecular Biology 303(3): 443 -456 Larson SM & Davidson AR. (2000) "A comprehensive analysis of the sequences and structures comprising the SH 3 domain. " Protein Science (in press) Plaxco KW, Larson S, Ruczinski I, Riddle DS, Thayer EC, Buchwitz B, Davidson AR, Baker D. (2000) "Evolutionary conservation in protein folding kinetics. " Journal of Molecular Biology 298(2): 303 -312 http: //www. stanford. edu/~smlarson
cd8d26dd3a526131337906d5f5328933.ppt