623d77f469a01f2e07fbc2888f32498b.ppt
- Количество слайдов: 97

Biomedical IE Heng Ji jih@rpi. edu
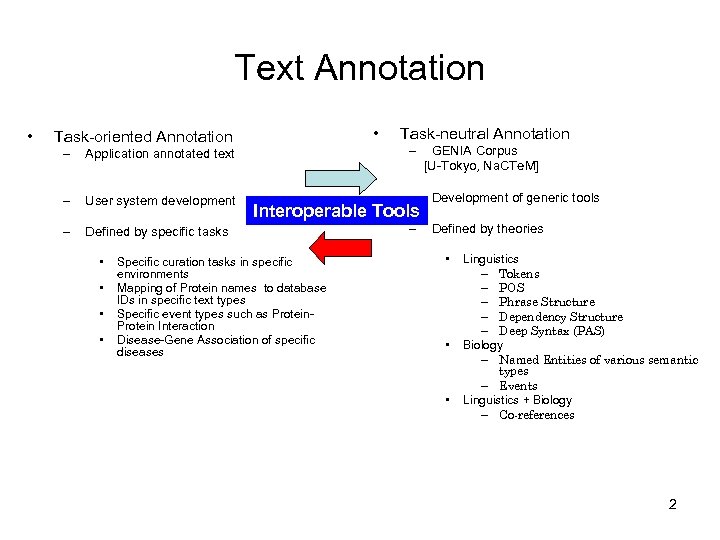
Text Annotation • • Task-oriented Annotation Task-neutral Annotation – Application annotated text – – User system development – Development of generic tools – Defined by specific tasks – Defined by theories • • Interoperable Tools Specific curation tasks in specific environments Mapping of Protein names to database IDs in specific text types Specific event types such as Protein Interaction Disease-Gene Association of specific diseases GENIA Corpus [U-Tokyo, Na. CTe. M] • • • Linguistics – Tokens – POS – Phrase Structure – Dependency Structure – Deep Syntax (PAS) Biology – Named Entities of various semantic types – Events Linguistics + Biology – Co-references 2
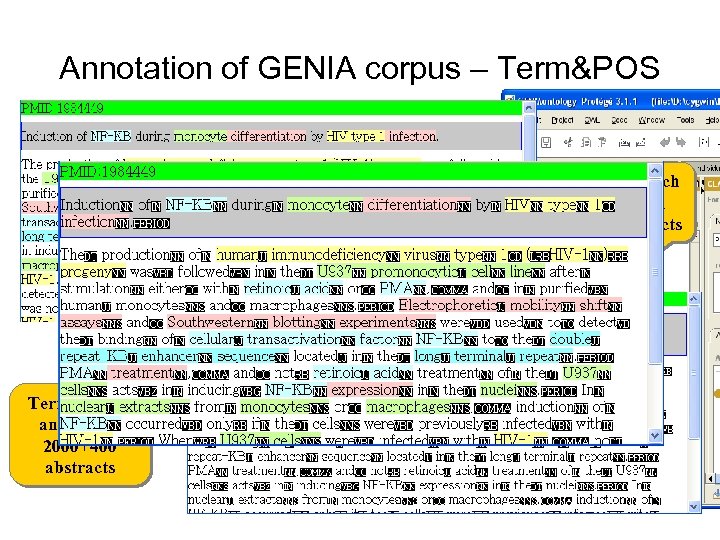
Annotation of GENIA corpus – Term&POS Part-of-speech annotation 2, 000 abstracts Term (entity) annotation 2000+400 abstracts 3
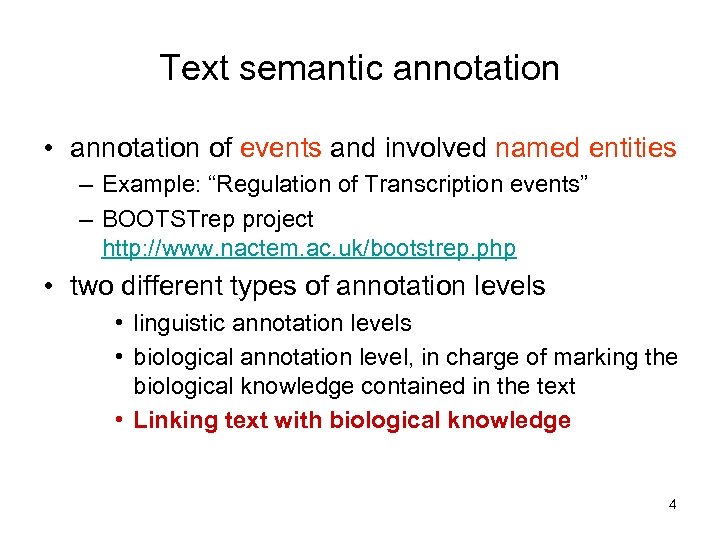
Text semantic annotation • annotation of events and involved named entities – Example: “Regulation of Transcription events” – BOOTSTrep project http: //www. nactem. ac. uk/bootstrep. php • two different types of annotation levels • linguistic annotation levels • biological annotation level, in charge of marking the biological knowledge contained in the text • Linking text with biological knowledge 4

Events and variables • Biological events can be centred on: – verbs, e. g. activate, – nouns with verb-like meanings (nominalised verbs), e. g. transcription • Different parts of sentence correspond to different types of variables in the event e. g. – What caused event • The nar. L gene product activates the nitrate reductase operon – What was affected by event • Analysis of mutants … – Where event took place • These fusions were formed on plasmid cloning vectors
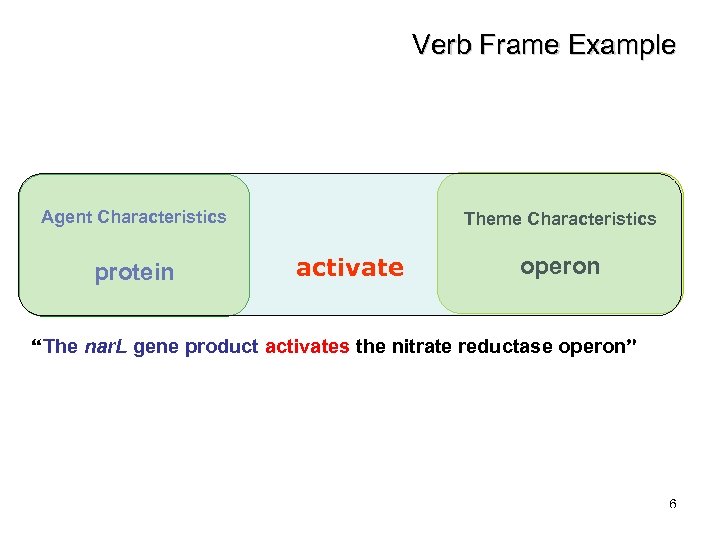
Verb Frame Example Agent Characteristics protein Theme Characteristics activate operon “The nar. L gene product activates the nitrate reductase operon” 6
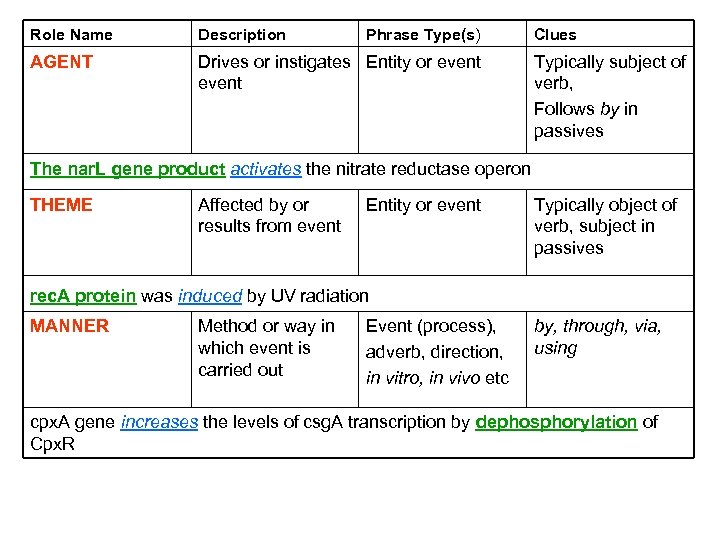
Role Name Description Phrase Type(s) AGENT Drives or instigates Entity or event Clues Typically subject of verb, Follows by in passives The nar. L gene product activates the nitrate reductase operon THEME Affected by or results from event Entity or event Typically object of verb, subject in passives rec. A protein was induced by UV radiation MANNER Method or way in which event is carried out Event (process), adverb, direction, in vitro, in vivo etc by, through, via, using cpx. A gene increases the levels of csg. A transcription by dephosphorylation of Cpx. R
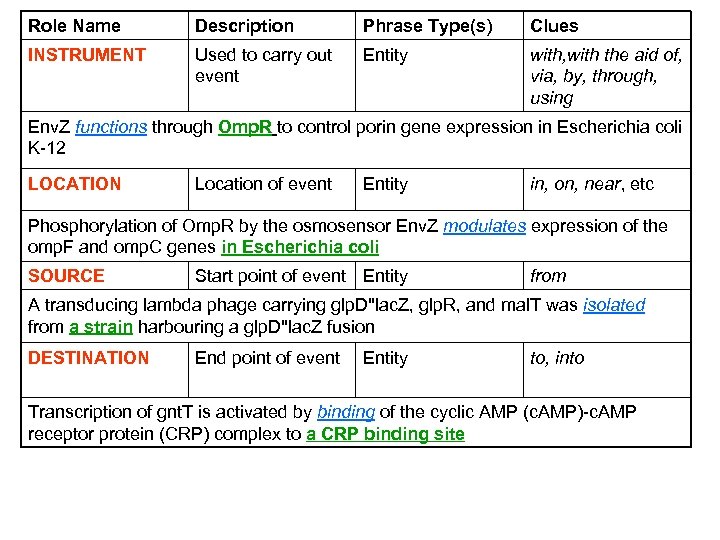
Role Name Description Phrase Type(s) Clues INSTRUMENT Used to carry out event Entity with, with the aid of, via, by, through, using Env. Z functions through Omp. R to control porin gene expression in Escherichia coli K-12 LOCATION Location of event Entity in, on, near, etc Phosphorylation of Omp. R by the osmosensor Env. Z modulates expression of the omp. F and omp. C genes in Escherichia coli SOURCE Start point of event Entity from A transducing lambda phage carrying glp. D''lac. Z, glp. R, and mal. T was isolated from a strain harbouring a glp. D''lac. Z fusion DESTINATION End point of event Entity to, into Transcription of gnt. T is activated by binding of the cyclic AMP (c. AMP)-c. AMP receptor protein (CRP) complex to a CRP binding site
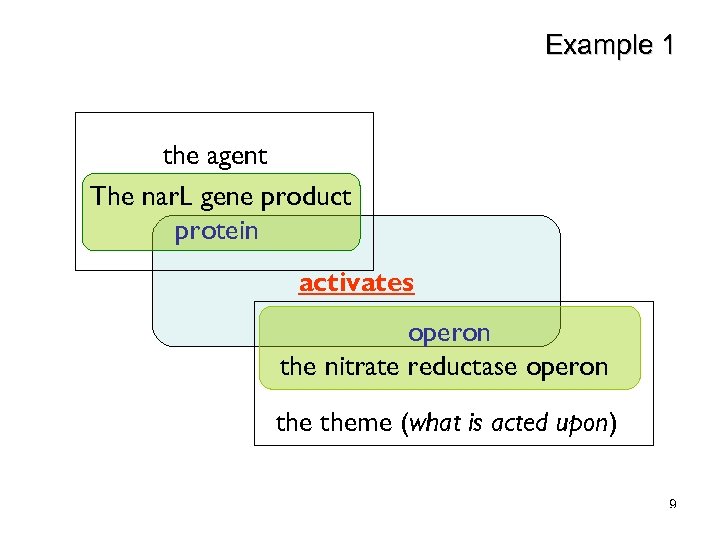
Example 1 the agent The nar. L gene product protein activates operon the nitrate reductase operon theme (what is acted upon) 9

Linguistically Annotated Corpora • GENIA – Domain • Mesh term: Human, Blood Cells, and Transcription Factors. – Annotation: POS, named entity, parse tree • Penn Bio. IE – Domain • the molecular genetics of oncology • the inhibition of enzymes of the CYP 450 class. – Annotation: POS, named entity, parse tree • Yapex • GENETAG a corpus of 20 K MEDLINE® sentences for gene/protein NER 10

The GENIA annotation • Linguistic annotation – Reveals linguistic structures behind the text • Part-of-speech annotation – annotates for the syntactic category of each word. • Syntactic Tree annotation – annotates for the syntactic structure of sentences. • Semantic annotation – Reveals knowledge pieces delivered by the text. • Term annotation – annotates domain-specific terms • Event annotation – annotates events on biological entities. Ontology-driven annotation 11

What about existing resources? • Ontologies important for knowledge discovery – They form the link between terms in texts and biological databases – Can be used to add meaning, semantic annotation of texts 12
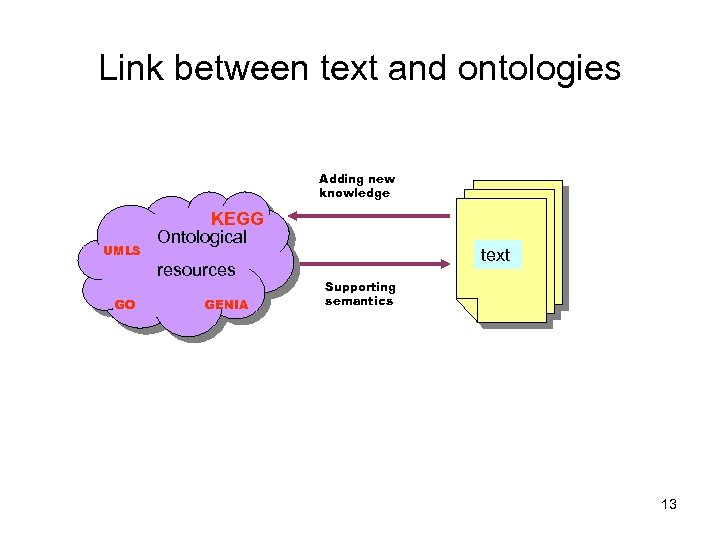
Link between text and ontologies Adding new knowledge UMLS KEGG Ontological resources GO GENIA text Supporting semantics 13
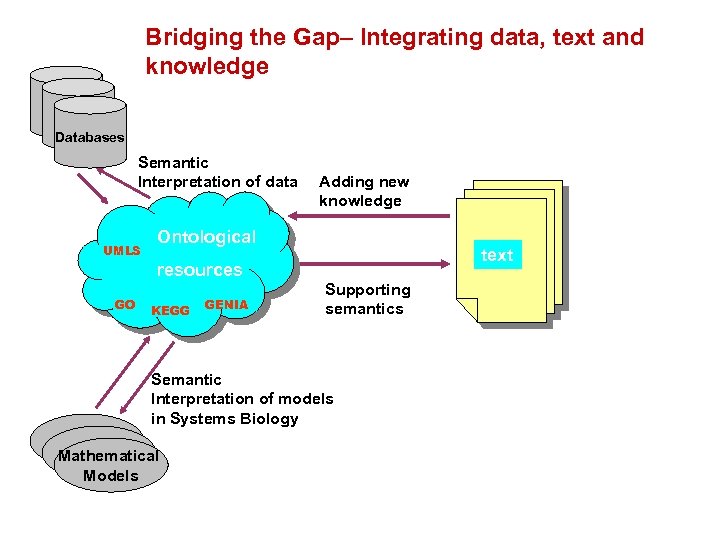
Bridging the Gap– Integrating data, text and knowledge Databases Semantic Interpretation of data UMLS Adding new knowledge Ontological text resources GO KEGG GENIA Supporting semantics Semantic Interpretation of models in Systems Biology Mathematical Models

Resources for Bio-Text Mining • Lexical / terminological resources – SPECIALIST lexicon, Metathesaurus (UMLS) – Lists of terms / lexical entries (hierarchical relations) • Ontological resources – Metathesaurus, Semantic Network, GO, SNOMED CT, etc – Encode relations among entities Bodenreider, O. “Lexical, Terminological, and Ontological Resources for Biological Text Mining”, Chapter 3, Text Mining for Biology and Biomedicine, pp. 43 -66 15

SPECIALIST lexicon – UMLS specialist lexicon http: //SPECIALIST. nlm. nih. gov • Each lexical entry contains morphological (e. g. cauterize, cauterizes, cauterized, cauterizing), syntactic (e. g. complementation patterns for verbs, nouns, adjectives), orthographic information (e. g. esophagus – oesophagus) • General language lexicon with many biomedical terms (over 180, 000 records) • Lexical programs include variation (spelling), base form, inflection, acronyms 16
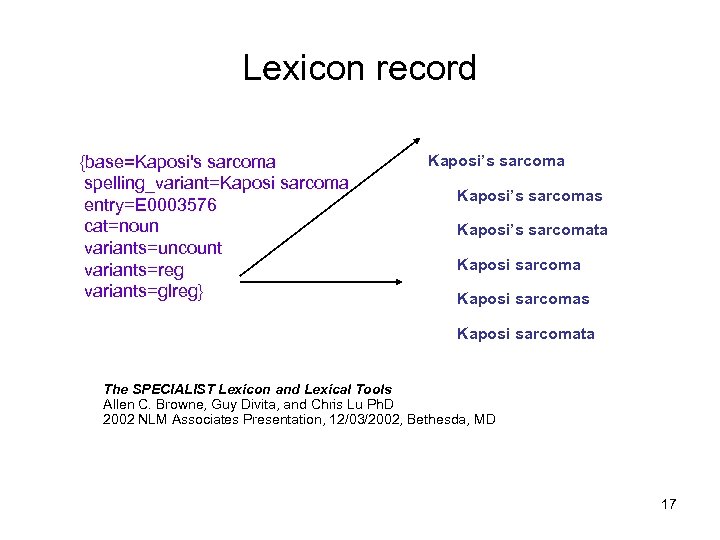
Lexicon record {base=Kaposi's sarcoma spelling_variant=Kaposi sarcoma entry=E 0003576 cat=noun variants=uncount variants=reg variants=glreg} Kaposi’s sarcomas Kaposi’s sarcomata Kaposi sarcomas Kaposi sarcomata The SPECIALIST Lexicon and Lexical Tools Allen C. Browne, Guy Divita, and Chris Lu Ph. D 2002 NLM Associates Presentation, 12/03/2002, Bethesda, MD 17
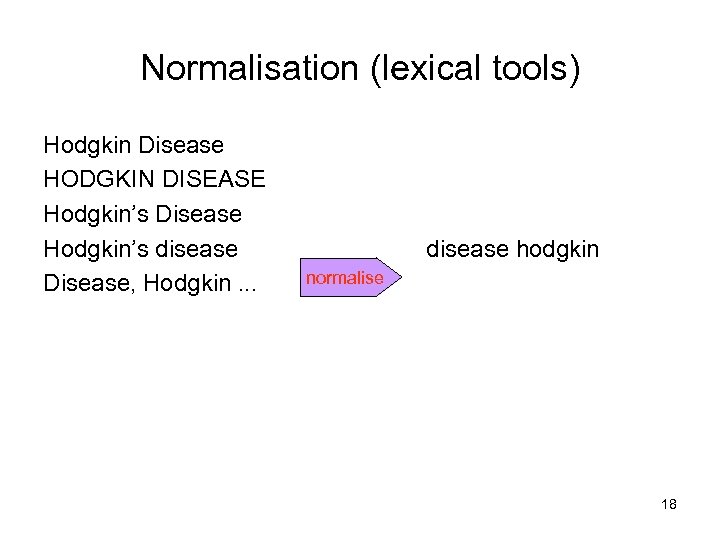
Normalisation (lexical tools) Hodgkin Disease HODGKIN DISEASE Hodgkin’s Disease Hodgkin’s disease Disease, Hodgkin. . . disease hodgkin normalise 18

Steps of Norm Remove genitive Hodgkin’s Diseases Replace punctuation with spaces Hodgkin Diseases Remove stop words Hodgkin Diseases Lowercase hodgkin diseases Uninflect each word hodgkin disease Word order sort disease hodgkin Lexical tools of the UMLS http: //lexsrv 3. nlm. nih. gov/SPECIALIST/index. html 19

The Gene Ontology (GO) • Controlled vocabulary for the annotation of gene products http: //www. geneontology. org/ 19, 468 terms. 95. 3% with definitions 10391 biological_process 1681 cellular_component 7396 molecular_function 20
Gene Ontology • GOA database (http: //www. ebi. ac. uk/GOA/) assigns gene products to the Gene Ontology • GO terms follow certain conventions of creation, have synonyms such as: – ornithine cycle is an exact synonym of urea cycle – cell division is a broad synonym of cytokinesis – cytochrome bc 1 complex is a related synonym of ubiquinol-cytochrome-c reductase activity 21

GO terms, definitions and ontologies in OBO id: GO: 0000002 name: mitochondrial genome maintenance namespace: biological_process def: "The maintenance of the structure and integrity of the mitochondrial genome. “ [GOC: ai] is_a: GO: 0007005 ! mitochondrion organization and biogenesis 22
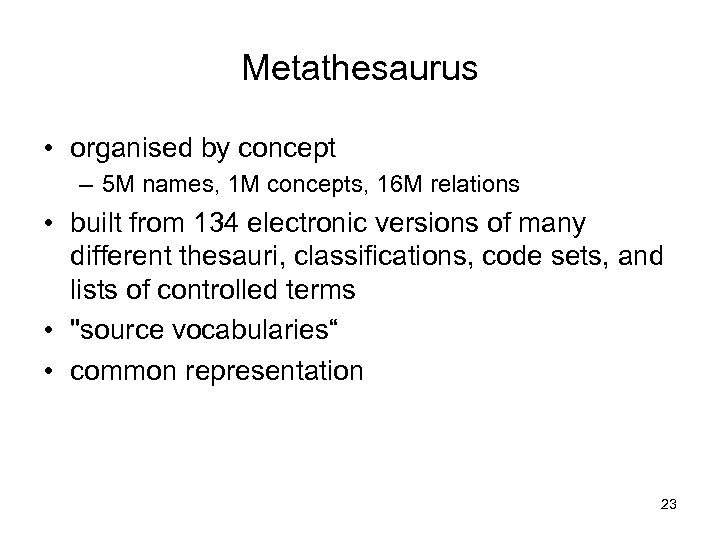
Metathesaurus • organised by concept – 5 M names, 1 M concepts, 16 M relations • built from 134 electronic versions of many different thesauri, classifications, code sets, and lists of controlled terms • "source vocabularies“ • common representation 23

Are the existing knowledge resources sufficient for TM? No! Why? l Limited lexical & terminological coverage of biological sub-domains l Resources focused on human specialists GO, UMLS, Uni. Prot ontology concept names frequently confused with terms 24

Naming conventions 3. Update and curation of resources – Fly. Base gene name coverage 31% (abstracts) to 84% (full texts) 4. Naming conventions and representation in heterogeneous resources – Term formation guidelines from formal bodies e. g. HUGO, IPI not uniformly used – Problems with integration of resources dystrophin used for 18 gene products “Dystrophin (muscular dystrophy, Duchenne and Becker types), included DXS 143, DXS 164, DXS 206, …” HUGO 25
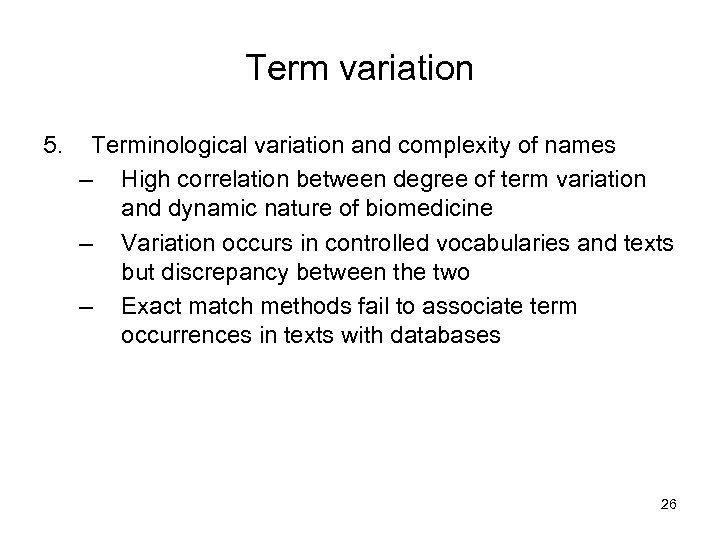
Term variation 5. Terminological variation and complexity of names – High correlation between degree of term variation and dynamic nature of biomedicine – Variation occurs in controlled vocabularies and texts but discrepancy between the two – Exact match methods fail to associate term occurrences in texts with databases 26

What’s in a name? • • • Breast cancer 1 (BRCA 1) p 53 Ribosomal protein S 27 Heat shock protein 110 Mitogen activated protein kinase 15 Mitogen activated protein kinase 5 From K. Cohen, NAACL 2007 27

Worst gene names • sema domain, seven thrombospondin repeats (type 1 and type 1 -like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5 A K. Cohen NAACL 2007 28

Worst gene names • sema domain, seven thrombospondin repeats (type 1 and type 1 -like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5 A K. Cohen NAACL 2007 29

Worst gene names • sema domain, seven thrombospondin repeats (type 1 and type 1 -like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5 A • SEMA 5 A K. Cohen NAACL 2007 30
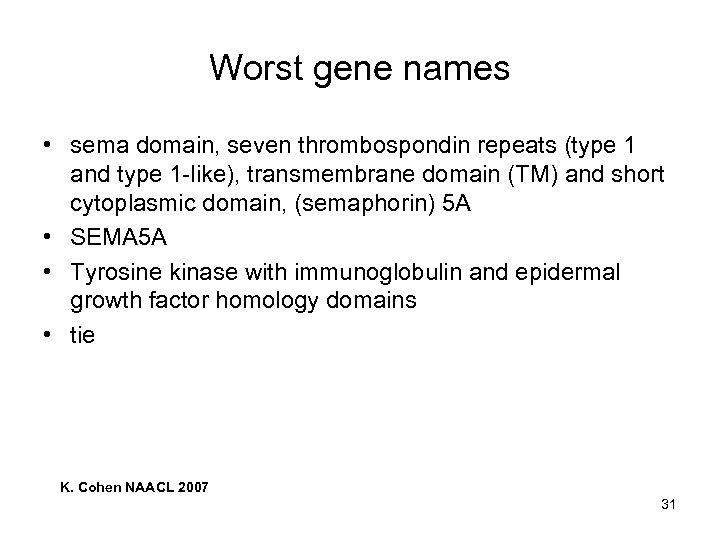
Worst gene names • sema domain, seven thrombospondin repeats (type 1 and type 1 -like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5 A • SEMA 5 A • Tyrosine kinase with immunoglobulin and epidermal growth factor homology domains • tie K. Cohen NAACL 2007 31
![Term ambiguity Neurofibromatosis 2 [disease] NF 2 Neurofibromin 2 [protein] Neurofibromatosis 2 gene [gene] Term ambiguity Neurofibromatosis 2 [disease] NF 2 Neurofibromin 2 [protein] Neurofibromatosis 2 gene [gene]](https://present5.com/presentation/623d77f469a01f2e07fbc2888f32498b/image-32.jpg)
Term ambiguity Neurofibromatosis 2 [disease] NF 2 Neurofibromin 2 [protein] Neurofibromatosis 2 gene [gene] O. Bodenreider, MIE 2005 tutorial http: //www. nactem. ac. uk/ 32
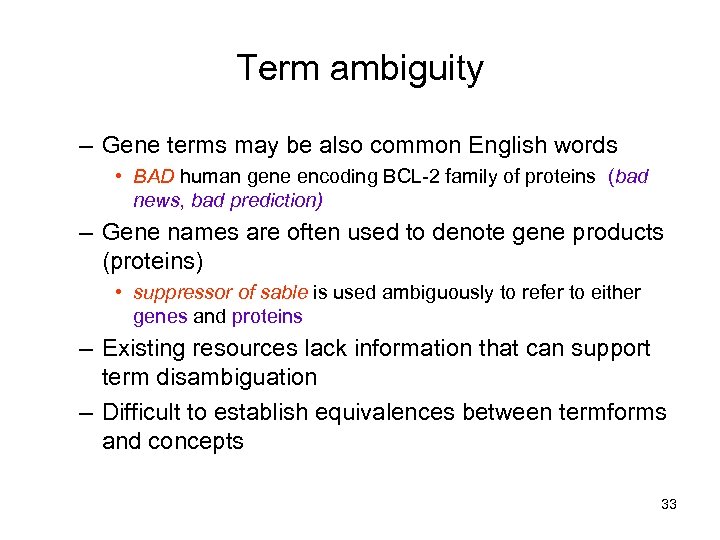
Term ambiguity – Gene terms may be also common English words • BAD human gene encoding BCL-2 family of proteins (bad news, bad prediction) – Gene names are often used to denote gene products (proteins) • suppressor of sable is used ambiguously to refer to either genes and proteins – Existing resources lack information that can support term disambiguation – Difficult to establish equivalences between termforms and concepts 33
Homologues • Cycline-dependent kinase inhibitor first introduced to represent a protein family p 27 – But it is used interchangeably with p 27 or p 27 kip 1, as the name of the individual protein and not as the name of the protein family (Morgan 2003). • NFKB 2 denotes the name of a family of 2 individual proteins with separate IDs in Swiss. Prot. – These proteins are homologues belonging to different species, homo sapiens & chicken. 34

Terms – Term: linguistic realisation of specialised concepts, e. g. genes, proteins, diseases – Terminology: collection of terms structured (hierarchy) denoting relationships among concepts, part-whole, is -a, specific, generic, etc. – Terms link text and ontologies – Mapping is not trivial (main challenge) 35
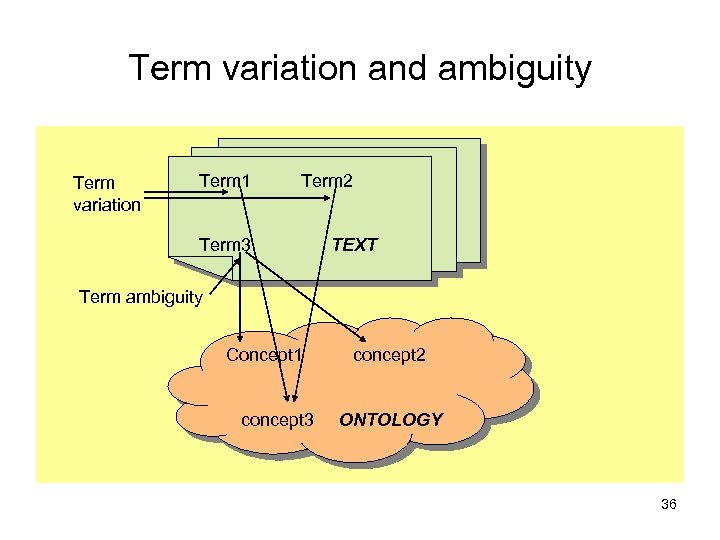
Term variation and ambiguity Term variation Term 1 Term 2 Term 3 TEXT Term ambiguity Concept 1 concept 3 concept 2 ONTOLOGY 36
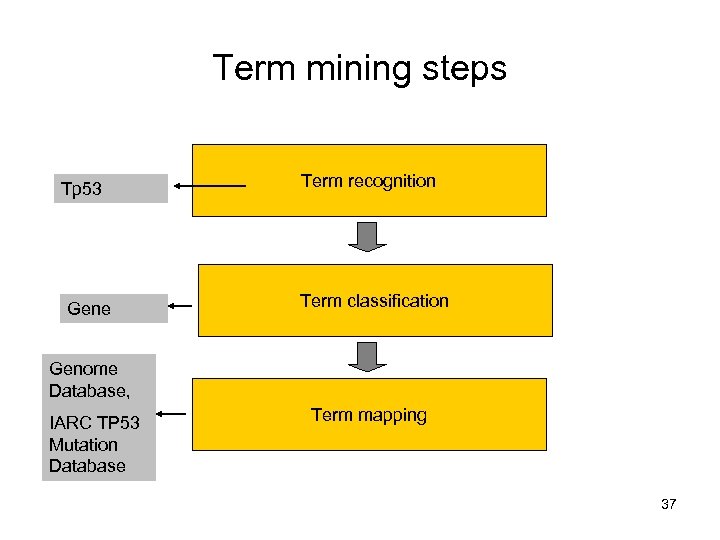
Term mining steps Tp 53 Gene Term recognition Term classification Genome Database, IARC TP 53 Mutation Database Term mapping 37
Term recognition techniques • ATR extracts terms (variants) from a collection of document • Distinguishes terms vs non-terms • In NER the steps of recognition and classification are merged, a classified terminological instance is a named entity • The tasks of ATR and NER share techniques but their ultimate goals are different – ATR for resource building, lexica & ontologies – NER first step of IE, text mining 38

Overview papers 1. S. Ananiadou & G. Nenadic (2006) Automatic Terminology Management in Biomedicine, Text Mining for Biology and Biomedicine, pp. 67 - 97. 2. M. Krauthammer & G. Nenadic (2004) Term identification in the biomedical literature, JBI 37 (2004) 512 -526 3. J. C. Park & J. Kim (2006) Named Entity Recognition, Text Mining for Biology and Biomedicine, pp. 121 -142 Detailed bibliography in Bio-Text Mining 1. BLIMPhttp: //blimp. cs. queensu. ca/ 2. http: //www. ccs. neu. edu/home/futrelle/bionlp/ Book on Bio. Text Mining 1. S. Ananiadou & J. Mc. Naught (eds) (2006) Text Mining for Biology and Biomedicine, Artech House. Other Bio-Text Mining tutorials Kevin Cohen (NAACL 2007 tutorial) U. Colorado 39
Biomedical IE/IR Systems • i. HOP – http: //www. ihop-net. org/Uni. Pub/i. HOP/ • EBIMed – http: //www. ebi. ac. uk/Rebholz-srv/ebimed/index. jsp • Go. Pub. Med – http: //www. gopubmed. org/ • Pub. Finder – http: //www. glycosciences. de/tools/Pub. Finder • Textpresso – http: //www. textpresso. org/ 40
Acronyms • Very productive type of term variation • Acronym variation (synonymy) – NF kappa B/ NF k. B / nuclear factor kappa B • Acronym ambiguity (polysemy) even in controlled vocabularies GR glucocorticoid receptor glutathione reductase 41
Acronym recognition • Swartz, A. & Hearst, M. (2003) A simple algorithm for identifying abbreviation definitions in biomedical text, PSB 2003, 8, 451 -462 • Adar, E. (2004) Sa. RAD: a simple and robust abbreviation dictionary, Bioinformatics, 20(4) 527 -533 • Chang, J. T. & Schutze, H. (2006) Abbreviations in biomedical text, Text Mining for Biology and Biomedicine, pp. 99 -119, Artech • Tsuruoka, Y. , Ananiadou, S. & Tsujii, J. (2005) A Machine learning approach to automatic acronym generation, ISMB, Bio. Link SIG, 25 -31 • Okazaki, N. & S. Ananiadou (2006) Acronym recognition based on term identification, Bioinformatics 42
The importance of acronym recognition • Acronyms are among the most productive type of term variation – 64, 242 new acronyms are introduced in 2004 [Chang and Schütze 06] • Acronyms are used more frequently than full terms – 5, 477 documents could be retrieved by using the acronym JNK while only 3, 773 documents could be retrieved by using its full term, c-jun N-terminal kinase [Wren et al. 05] • No rules or exact patterns for the creation of acronyms from their full form 43

Recognition • Extracting pairs of short and long forms <acronym, long form> – Distinguishing acronyms from parenthetical expressions – Search for parentheses in text; single or more words; e. g. Ab (antibody) – Limit context around ( ); limit number of words according to number of letters in acronym 44
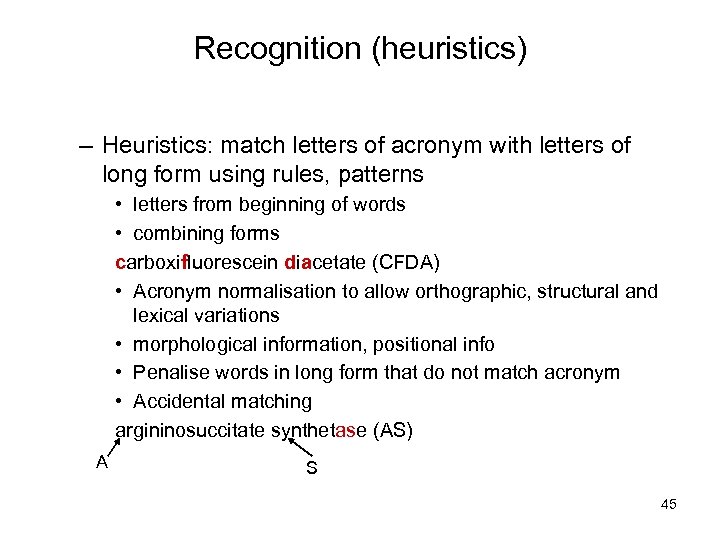
Recognition (heuristics) – Heuristics: match letters of acronym with letters of long form using rules, patterns • letters from beginning of words • combining forms carboxifluorescein diacetate (CFDA) • Acronym normalisation to allow orthographic, structural and lexical variations • morphological information, positional info • Penalise words in long form that do not match acronym • Accidental matching argininosuccitate synthetase (AS) A S 45

Letter matching – Alignment: find all matches between letters of acronyms and their long forms and calculate likelihood (Chang & Schütze) • Solves problem of acronyms containing letters not occurring in LF • Choose best alignment based on features, e. g. position of letter etc. • Finding optimal weight for each feature challenge http: //abbreviation. stanford. edu/ 46
Acronym Recognition Okazaki, N. , Ananiadou, S. (2006) Building an abbreviation dictionary using a term recognition approach. Bioinformatics. S. Ananiadou Na. CTe. M 47
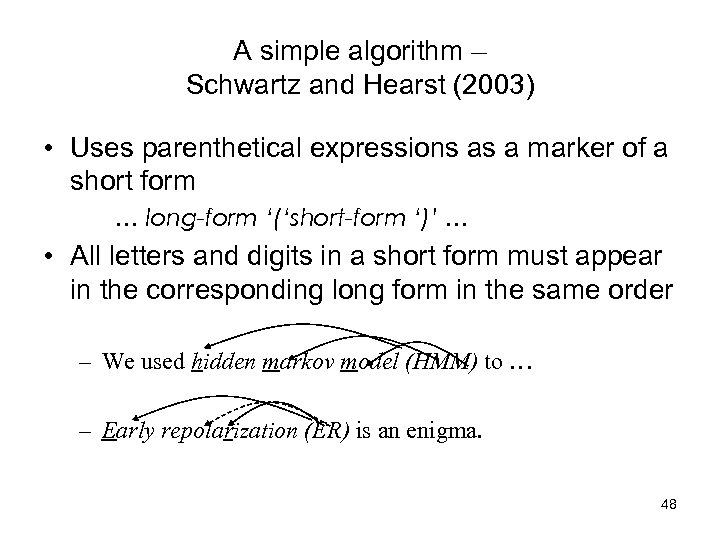
A simple algorithm – Schwartz and Hearst (2003) • Uses parenthetical expressions as a marker of a short form … long-form ‘(‘short-form ‘)’ … • All letters and digits in a short form must appear in the corresponding long form in the same order – We used hidden markov model (HMM) to … – Early repolarization (ER) is an enigma. 48

Problems of letter-matching approach • Highly dependent on the expressions in the target text – o acquired immuno deficiency syndrome (AIDS) – x acquired syndrome (AIDS) – x a patient with human immunodeficiency syndrome (AIDS) – ? magnetic resonance imaging unit (MRI) – ! beta 2 adrenergic receptor (ADRB 2) – ! gamma interferon (IFN-GAMMA) (These examples are obtained from actual MEDLINE abstracts) • Naive with respect to term variations 49
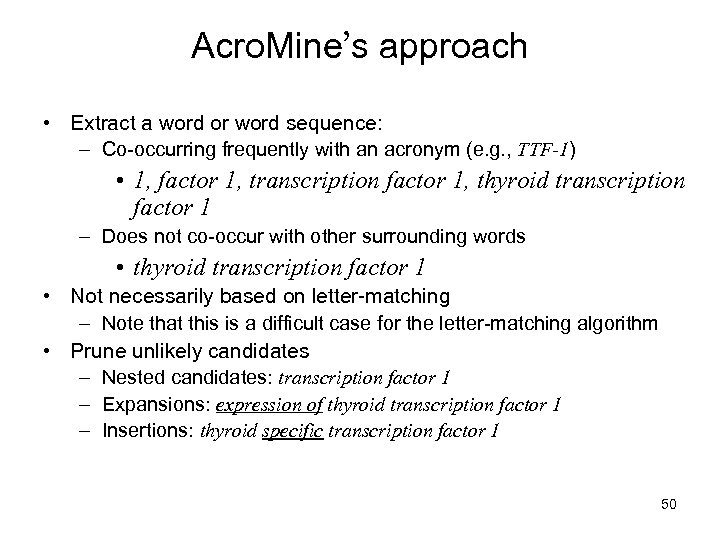
Acro. Mine’s approach • Extract a word or word sequence: – Co-occurring frequently with an acronym (e. g. , TTF-1) • 1, factor 1, transcription factor 1, thyroid transcription factor 1 – Does not co-occur with other surrounding words • thyroid transcription factor 1 • Not necessarily based on letter-matching – Note that this is a difficult case for the letter-matching algorithm • Prune unlikely candidates – Nested candidates: transcription factor 1 – Expansions: expression of thyroid transcription factor 1 – Insertions: thyroid specific transcription factor 1 50
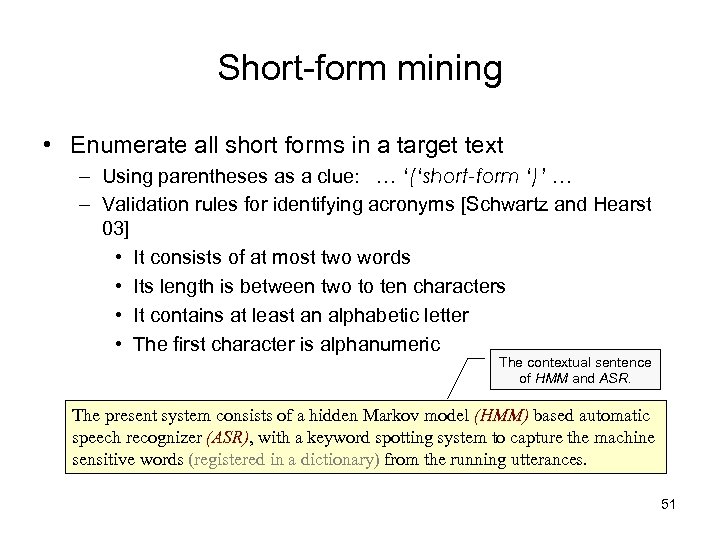
Short-form mining • Enumerate all short forms in a target text – Using parentheses as a clue: … ‘(‘short-form ‘)’ … – Validation rules for identifying acronyms [Schwartz and Hearst 03] • It consists of at most two words • Its length is between two to ten characters • It contains at least an alphabetic letter • The first character is alphanumeric The contextual sentence of HMM and ASR. The present system consists of a hidden Markov model (HMM) based automatic speech recognizer (ASR), with a keyword spotting system to capture the machine sensitive words (registered in a dictionary) from the running utterances. 51
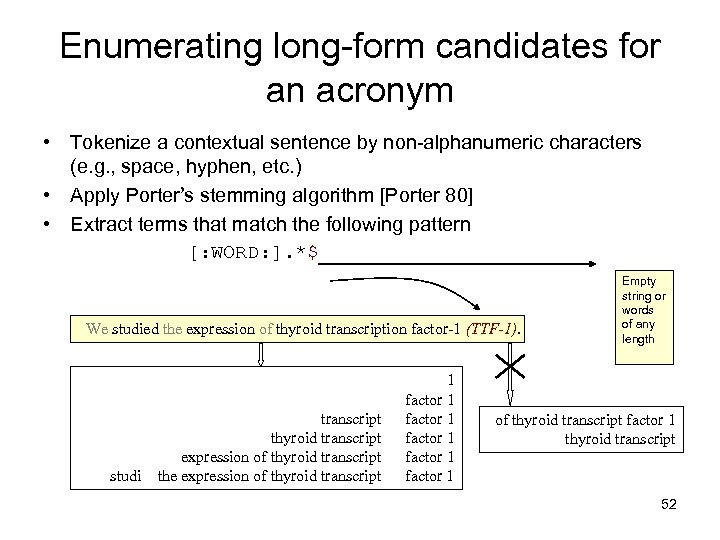
Enumerating long-form candidates for an acronym • Tokenize a contextual sentence by non-alphanumeric characters (e. g. , space, hyphen, etc. ) • Apply Porter’s stemming algorithm [Porter 80] • Extract terms that match the following pattern [: WORD: ]. *$ We studied the expression of thyroid transcription factor-1 (TTF-1). studi transcript thyroid transcript expression of thyroid transcript the expression of thyroid transcript 1 factor 1 factor 1 Empty string or words of any length of thyroid transcript factor 1 thyroid transcript 52
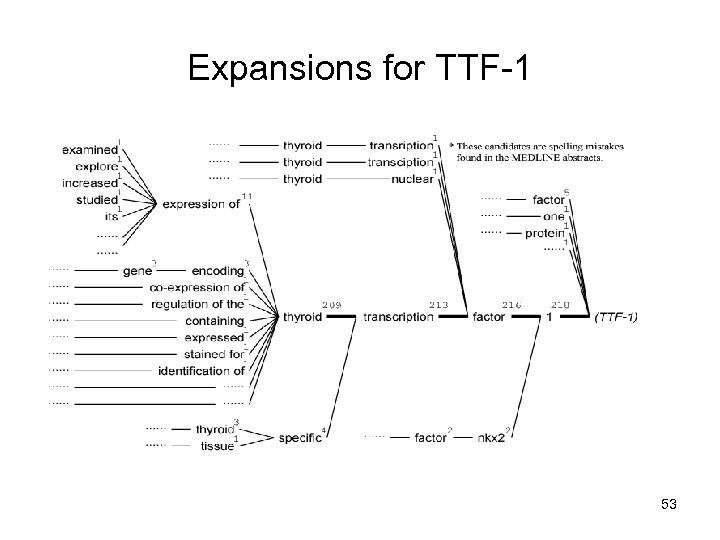
Expansions for TTF-1 53
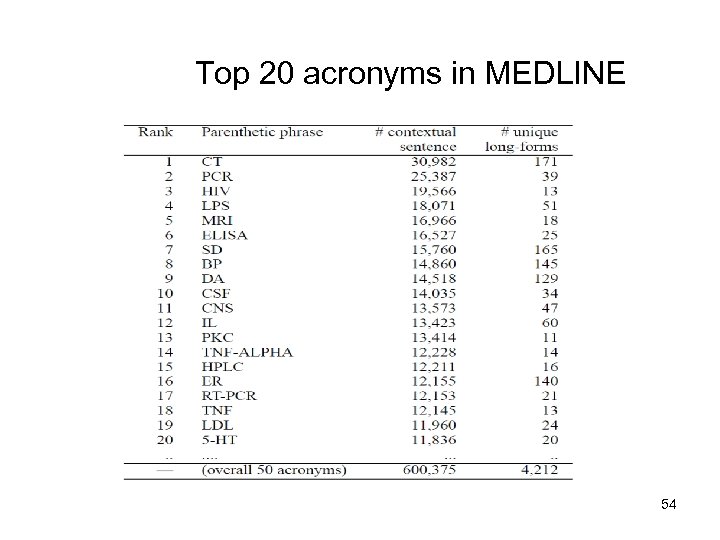
Top 20 acronyms in MEDLINE 54
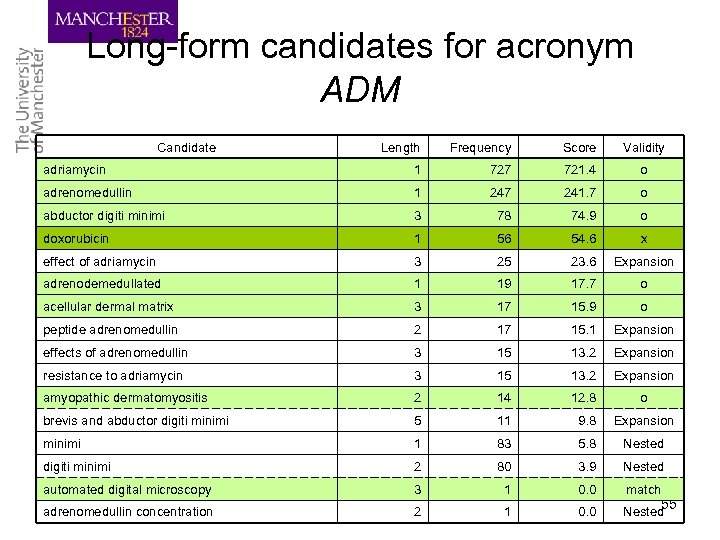
Long-form candidates for acronym ADM Candidate Length Frequency Score Validity adriamycin 1 727 721. 4 o adrenomedullin 1 247 241. 7 o abductor digiti minimi 3 78 74. 9 o doxorubicin 1 56 54. 6 x effect of adriamycin 3 25 23. 6 Expansion adrenodemedullated 1 19 17. 7 o acellular dermal matrix 3 17 15. 9 o peptide adrenomedullin 2 17 15. 1 Expansion effects of adrenomedullin 3 15 13. 2 Expansion resistance to adriamycin 3 15 13. 2 Expansion amyopathic dermatomyositis 2 14 12. 8 o brevis and abductor digiti minimi 5 11 9. 8 Expansion minimi 1 83 5. 8 Nested digiti minimi 2 80 3. 9 Nested automated digital microscopy 3 1 0. 0 match adrenomedullin concentration 2 1 0. 0 Nested 55

Long-form extraction • Long-form candidates are sorted with their scores in a descending order • A long-form candidate is considered valid if: – It has a score greater than 2. 0 – The words in the long form can be rearranged so that all alphanumeric letters appear in the same order as the short form – It is not nested or expansion of the previously chosen long forms 56
http: //www. nactem. ac. uk/software/acromine/
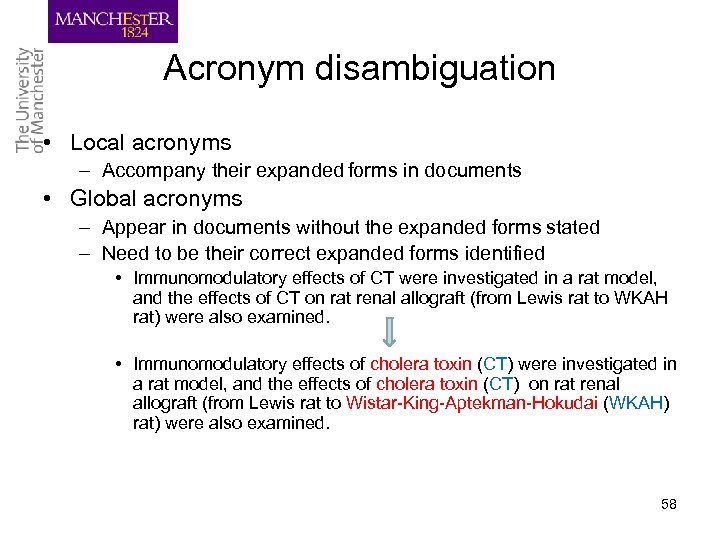
Acronym disambiguation • Local acronyms – Accompany their expanded forms in documents • Global acronyms – Appear in documents without the expanded forms stated – Need to be their correct expanded forms identified • Immunomodulatory effects of CT were investigated in a rat model, and the effects of CT on rat renal allograft (from Lewis rat to WKAH rat) were also examined. • Immunomodulatory effects of cholera toxin (CT) were investigated in a rat model, and the effects of cholera toxin (CT) on rat renal allograft (from Lewis rat to Wistar-King-Aptekman-Hokudai (WKAH) rat) were also examined. 58
Acronym disambiguation Sample text: Considerations in the identification of functional RNA structural elements in genomic alignments (Tomas Babak et al) http: //www. biomedcentral. com/1471 -2105/8/33
Term structuring • term clustering (linking semantically similar terms) and term classification (assigning terms to classes from a pre -defined classification scheme) • Hypothesis: similar terms tend to appear in similar contexts (patterns) • combining various sources of similarity: – – lexical syntactic contextual Ontological (using external resources) 60
Term structuring • Based on term similarities – choice of features: – domain specific – linguistic ontology text • ontology-based similarity • textual similarity – internal features – contextual features 61

Using ontologies • two terms should match if they are: – identified as variants – siblings in the is-a hierarchy – in the is-a or part-whole relation • the distance between the corresponding nodes in the ontology should be transformed into the matching score ► I. Spasic presentation MIE Tutorial http: //www. nactem. ac. uk/ 62
Using text • number of neologisms: terms are not in the ontologies • Use of text based techniques to calculate similarities • edit distance (ED) – the minimal number (or cost) of changes needed to transform one string into the other • edit operations: insertion deletion replacement transposition. . . a-c. . . abc. . . a-c. . . adc. . . acb. . . • use of dynamic programming 63
Pattern-matching IE – Usual limitations with non inclusion of semantic processing – Large amount of surface grammatical structures = too many patterns (Zipf’s law) – Cannot explore syntactic generalisations (active, passive voice) – Systems extract phrases or entire sentences with matched patterns; restricted usefulness for subsequent mining 64
Pattern-matching systems (1) Ø Bio. IE uses patterns to extract sentences, protein families, structures, functions. . Ø Presents user with relevant information, improvement from classic IR Ø Bio. RAT uses “deeper” analysis, tagging, apply RE over POS tags, stemming, gazetter categories etc Ø Templates apply to extract matching phrases, primitive filters (verbs are not proteins, etc) 65
Pattern matching systems (2) Ø RLIMS-P (Hu) protein phosphorylation by looking for enzymes, substrates, sites assigned to agent, theme, site roles of phosphorylation relations Ø Pos tagger, trained on newswire, chunking, semantic typing of chunks, identification of relations using pattern-matching rules Ø Semantic typing of NPs: using combination of clue words, suffixes, acronyms etc Ø Semantically typed sentences matched with rules Ø Patterns target sentences containing phosphorylate 66

Full parsing approaches • Link Grammar applied for protein-protein interactions; general English grammar adapted to bio-text • Link Grammar finds all possible linkages according to its grammar • Number of analyses reduced by random sampling, heuristics, processing constraints relaxed – 10, 000 results permitted per sentence – 60% of protein interactions extracted – Problems: missing possessive markers & determiners, coordination of compound noun modifiers 67

Full parsing IE (2) • Not all parsing strategies suitable for bio-text mining • Text type, abstracts, “ungrammaticality” related with sublanguage characteristics? • Ambiguity and full parsing; fragmentary phrases (titles, headings, text in table cells, etc) • CADERIGE project used Link grammar but on shallow parsing mode • Kim & Park (Bio. IE) use combinatorial categorial grammar, annotated with GO concepts, extract general biological interactions • 1, 300 patterns applied to find instances of patterns with keywords 68
Full parsing (3) • Keywords indicate basic biological interactions • Patterns find potential arguments of the interaction keywords (verbs or nominalisations) – Validated arguments mapped into GO concepts – Difficult to generalise interaction keyword patterns • Bio. IE’s syntactic parsing performance improved after adding subcategorisation frames on verbal interaction keywords 69
Full parsing (4) – 1. 2. 3. 4. 5. Daraselia(2004) use full parsing and domain specific filter to extract protein interactions All syntactic analyses discovered using CFG and variant of LFG Each alternative parse mapped to its corresponding semantic representation Output= set of semantic trees, lexemes linked by relations indicating thematic or attributive roles Apply custom-built, frame based ontology to filter representations of each sentence Preference mechanism controls construction of frame tree, high precision, low recall (21%) 70

Sublanguage-driven IE (1) • Language of a special community (e. g. biology) • Particular set of constraints re GL • Constraints operate at all linguistic levels – – Special vocabulary (terms) Specialised term formation rules Sublanguage syntactic patterns Sublanguage semantics • These constraints give rise to the informational structure of the domain (Z. Harris) • See JBI 35(4) Special Issue on Sublanguage 71
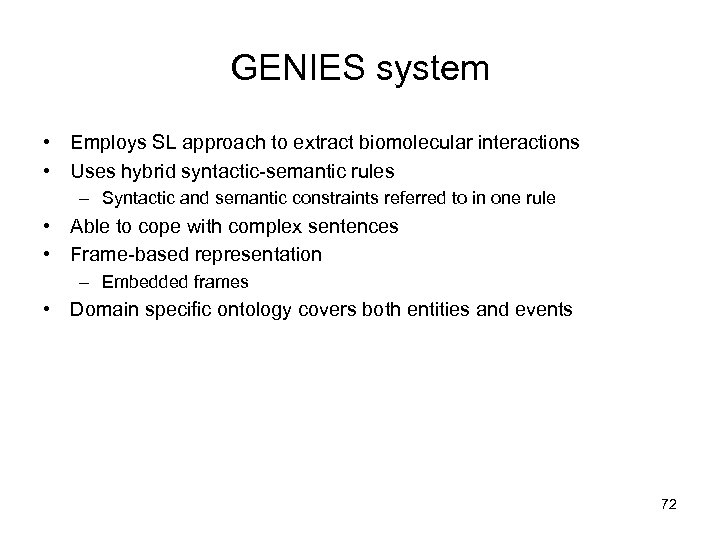
GENIES system • Employs SL approach to extract biomolecular interactions • Uses hybrid syntactic-semantic rules – Syntactic and semantic constraints referred to in one rule • Able to cope with complex sentences • Frame-based representation – Embedded frames • Domain specific ontology covers both entities and events 72

GENIES system • Default strategy: full parsing – Robust due to sublanguage constraints – Much ambiguity excluded • If full parse fails, partial parsing invoked – Maintains good level of recall • Precision: 96%, Recall: 63% 73

Ontology-driven IE • Until recently most rule based IE have used neither linguistic lexica nor ontologies – Reliance on gazetteers – Small number of semantic categories • Gazetteer approach not well suited in bio. IE • Ontology based vs ontology driven – Passive use of ontologies, map discovered entity to concept – Active use, ontology guides and constrains analysis, fewer rules • Examples: PASTA, Gen. IE not SL • GENIES, SL and ontology driven 74
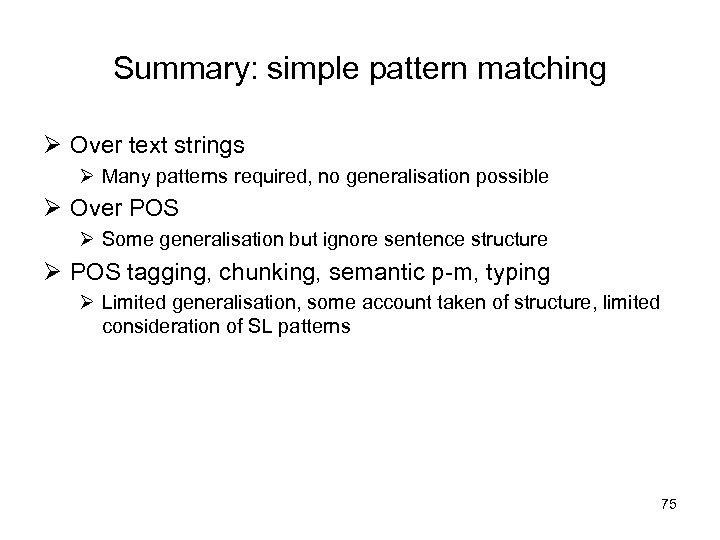
Summary: simple pattern matching Ø Over text strings Ø Many patterns required, no generalisation possible Ø Over POS Ø Some generalisation but ignore sentence structure Ø POS tagging, chunking, semantic p-m, typing Ø Limited generalisation, some account taken of structure, limited consideration of SL patterns 75

Summary: full parsing Ø Full parsing on its own, parsing done in combination with chunking, partial parsing, heuristics) to reduce ambiguity, filter out implausible readings Ø Ø GL theories not appropriate Difficult to specialise for biotext Many analyses per sentence Missing information due to sublanguage meaning 76

Summary: sublanguage approach Ø Ø Exploits a rich SL lexicon Describes SL verbs in detail Syntactic-semantic grammar Current systems would benefit from adopting ontologydriven approach 77

Ontology-driven Ø Uses event concept frames to guide processing Ø Integration of extracted information Ø Current systems would benefit from adopting also SL approach 78

Domain Adaptation from News to Biomedical?

What is domain adaptation? 80
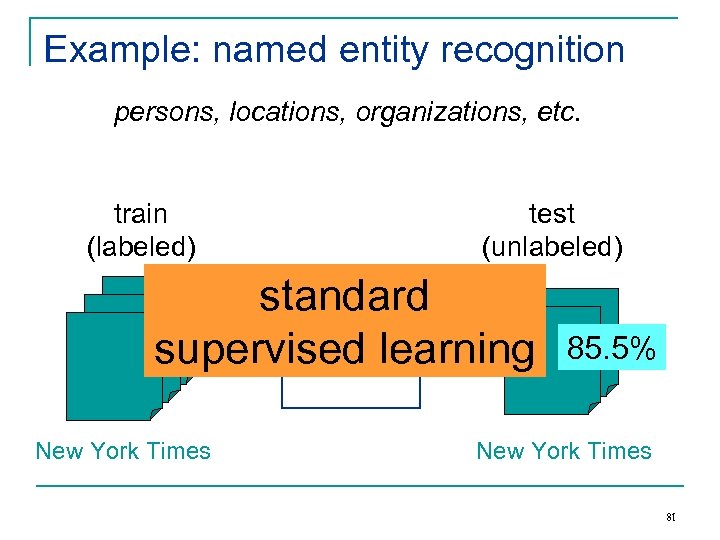
Example: named entity recognition persons, locations, organizations, etc. train (labeled) test (unlabeled) standard NER supervised learning Classifier New York Times 85. 5% New York Times 81
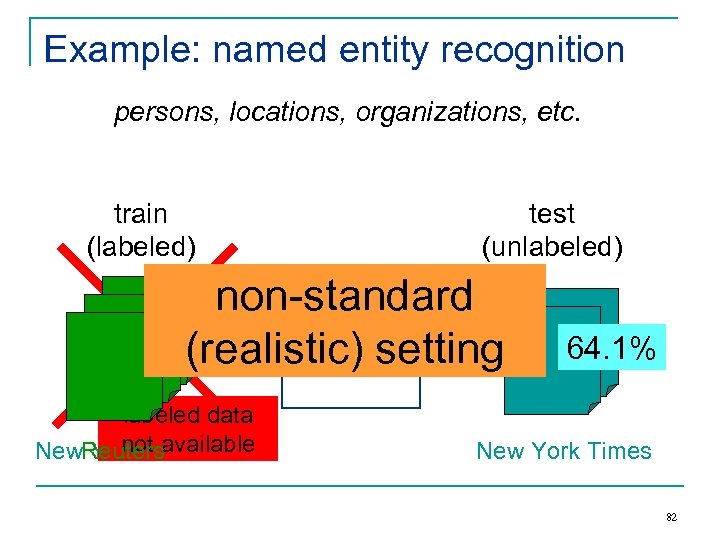
Example: named entity recognition persons, locations, organizations, etc. train (labeled) test (unlabeled) non-standard NER (realistic) setting Classifier labeled data not New. Reutersavailable York Times 64. 1% New York Times 82
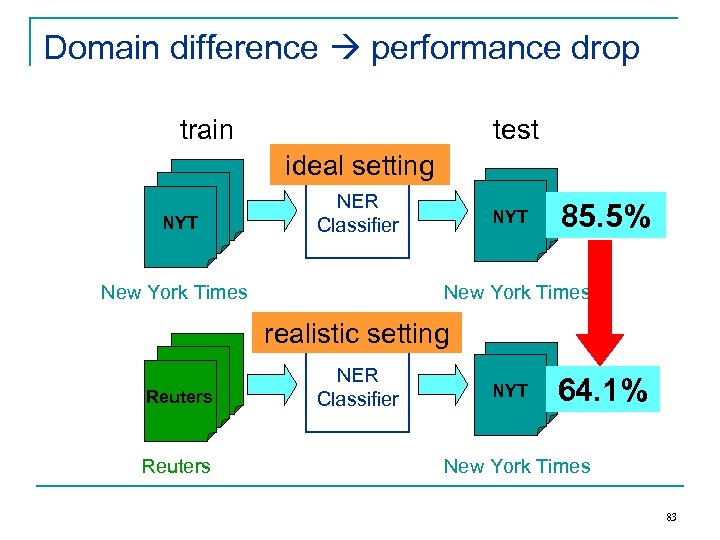
Domain difference performance drop train test ideal setting NYT NER Classifier New York Times NYT 85. 5% New York Times realistic setting Reuters NER Classifier NYT 64. 1% New York Times 83
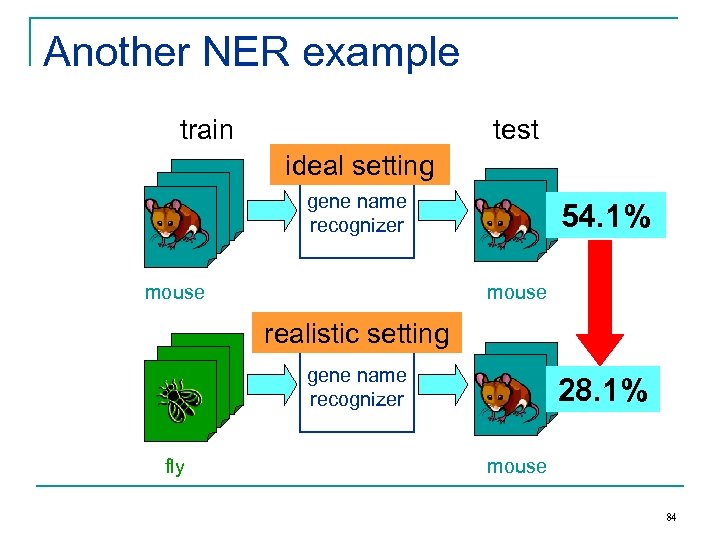
Another NER example train test ideal setting gene name recognizer mouse 54. 1% mouse realistic setting gene name recognizer fly 28. 1% mouse 84

Other examples n Spam filtering: q n Sentiment analysis of product reviews q q n n Public email collection personal inboxes Digital cameras cell phones Movies books Can we do better than standard supervised learning? Domain adaptation: to design learning methods that are aware of the training and test domain difference. 85

How do we solve the problem in general? 86
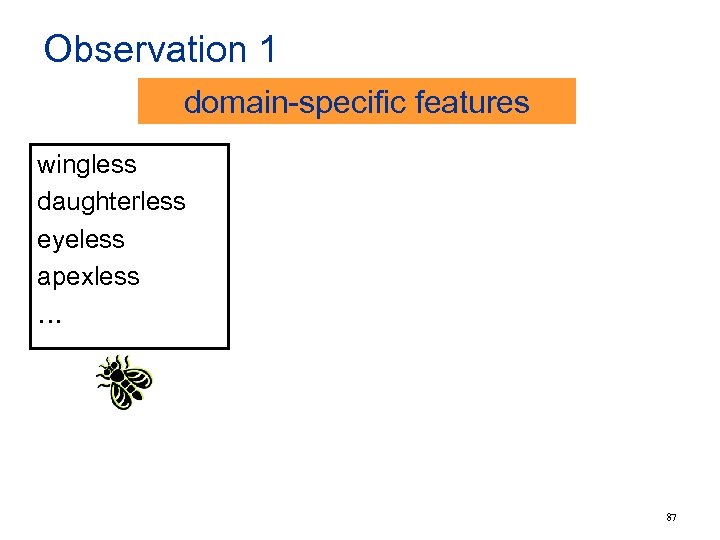
Observation 1 domain-specific features wingless daughterless eyeless apexless … 87
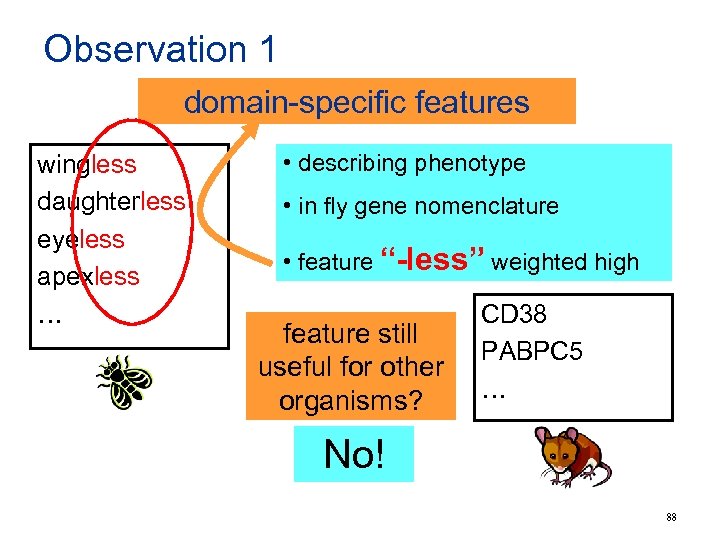
Observation 1 domain-specific features wingless daughterless eyeless apexless … • describing phenotype • in fly gene nomenclature • feature “-less” weighted high feature still useful for other organisms? CD 38 PABPC 5 … No! 88
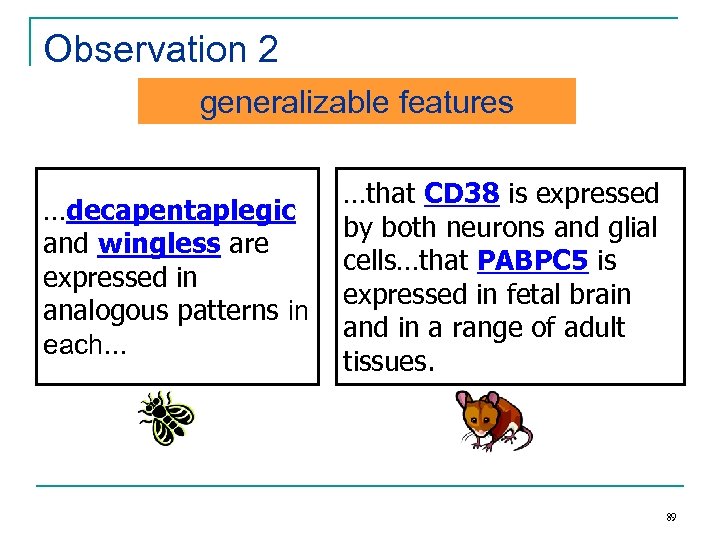
Observation 2 generalizable features …decapentaplegic and wingless are expressed in analogous patterns in each… …that CD 38 is expressed by both neurons and glial cells…that PABPC 5 is expressed in fetal brain and in a range of adult tissues. 89
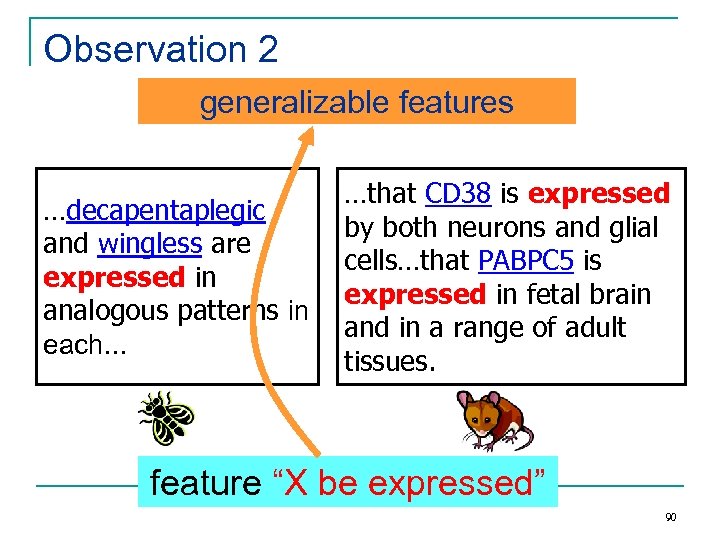
Observation 2 generalizable features …decapentaplegic and wingless are expressed in analogous patterns in each… …that CD 38 is expressed by both neurons and glial cells…that PABPC 5 is expressed in fetal brain and in a range of adult tissues. feature “X be expressed” 90
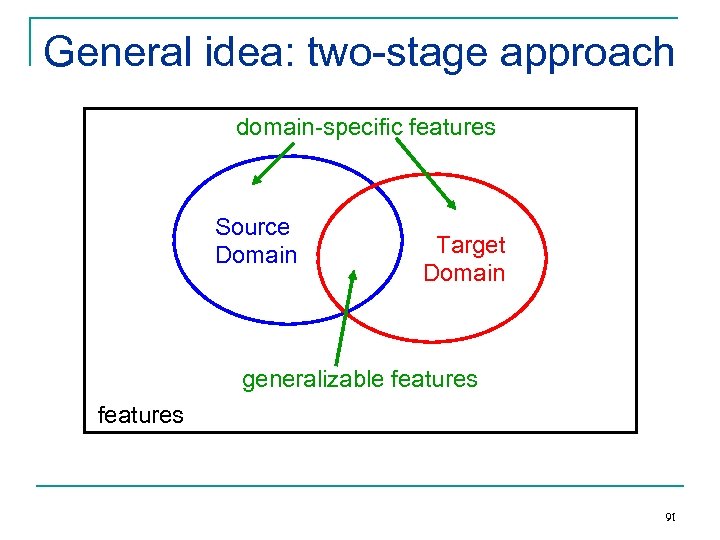
General idea: two-stage approach domain-specific features Source Domain Target Domain generalizable features 91
Goal Source Domain Target Domain features 92
Regular classification Source Domain Target Domain features 93
Generalization: to emphasize generalizable features in the trained model Source Domain features Target Domain Stage 1 94
Adaptation: to pick up domain-specific features for the target domain Source Domain features Target Domain Stage 2 95
Regular semi-supervised learning Source Domain Target Domain features 96
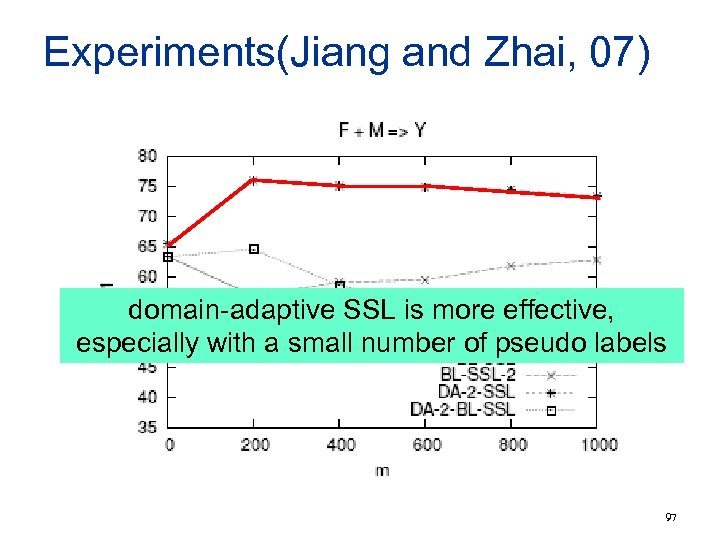
Experiments(Jiang and Zhai, 07) domain-adaptive SSL is more effective, especially with a small number of pseudo labels 97
623d77f469a01f2e07fbc2888f32498b.ppt