f3c48371c40834436e432e61a393d92a.ppt
- Количество слайдов: 67
Biology 4900 Biocomputing
Chapter 10 Protein Analysis and Proteomics
Composition of living organisms 5 major components Proteins Nucleic acids Lipids (fats) Water Carbohydrates Pevsner, Bioinformatics and Functional Genomics, 2009
Roles of DNA and Proteins • If we think of constructing an organism like building a house, DNA would be the blueprint and proteins would be most of the construction materials • Protein functions include: – – – Structural roles (e. g. , actin in the cytoskeleton) Enzyme catalysts (e. g. , trypsin, a serine protease) Intra- and intercellular transporters Molecular signaling Cellular regulation (e. g. , Nrf 2) Pevsner, Bioinformatics and Functional Genomics, 2009
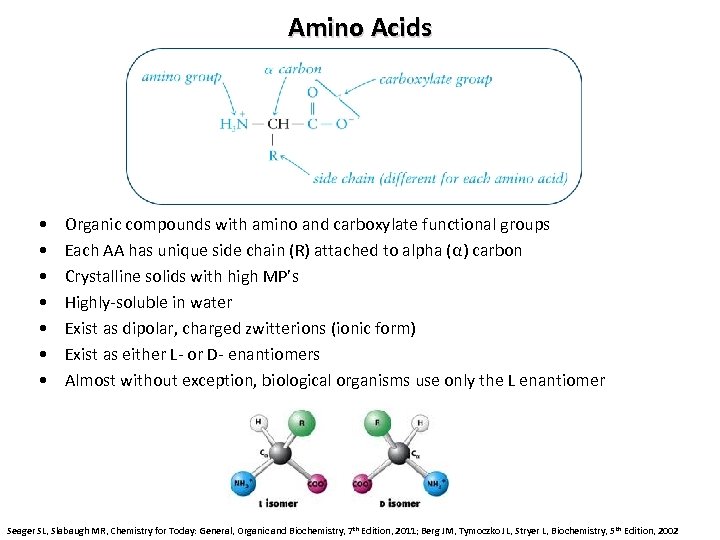
Amino Acids • • Organic compounds with amino and carboxylate functional groups Each AA has unique side chain (R) attached to alpha (α) carbon Crystalline solids with high MP’s Highly-soluble in water Exist as dipolar, charged zwitterions (ionic form) Exist as either L- or D- enantiomers Almost without exception, biological organisms use only the L enantiomer Seager SL, Slabaugh MR, Chemistry for Today: General, Organic and Biochemistry, 7 th Edition, 2011; Berg JM, Tymoczko JL, Stryer L, Biochemistry, 5 th Edition, 2002
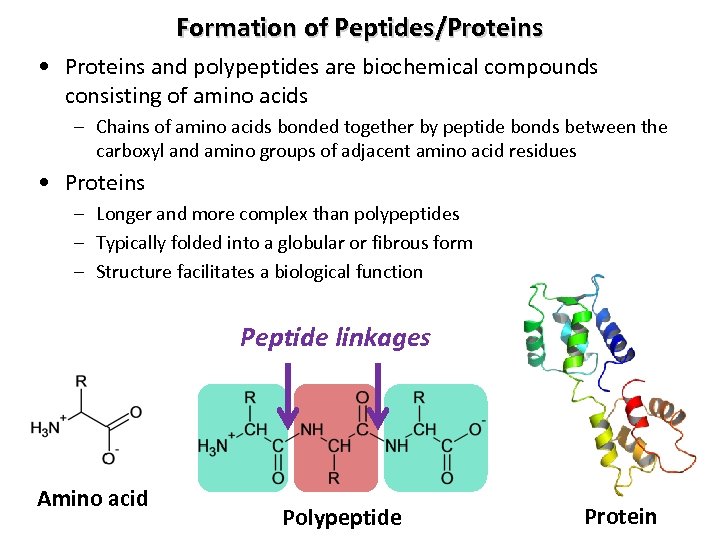
Formation of Peptides/Proteins • Proteins and polypeptides are biochemical compounds consisting of amino acids – Chains of amino acids bonded together by peptide bonds between the carboxyl and amino groups of adjacent amino acid residues • Proteins – Longer and more complex than polypeptides – Typically folded into a globular or fibrous form – Structure facilitates a biological function Peptide linkages Amino acid Polypeptide Protein
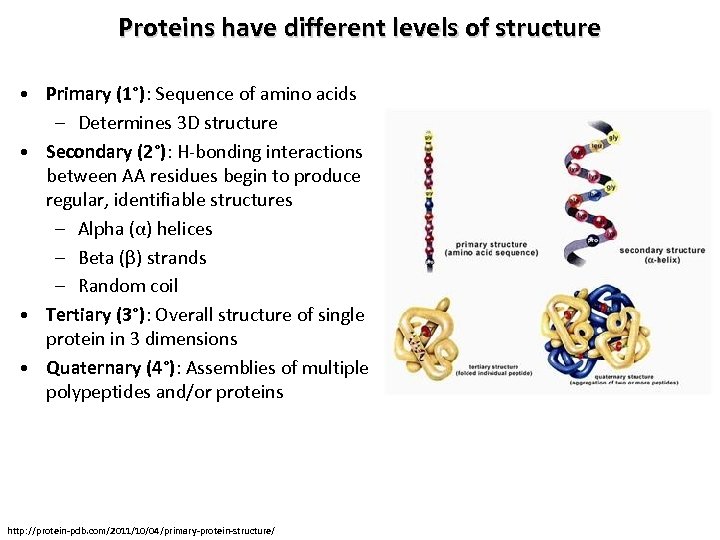
Proteins have different levels of structure • Primary (1°): Sequence of amino acids – Determines 3 D structure • Secondary (2°): H-bonding interactions between AA residues begin to produce regular, identifiable structures – Alpha (α) helices – Beta (β) strands – Random coil • Tertiary (3°): Overall structure of single protein in 3 dimensions • Quaternary (4°): Assemblies of multiple polypeptides and/or proteins http: //protein-pdb. com/2011/10/04/primary-protein-structure/
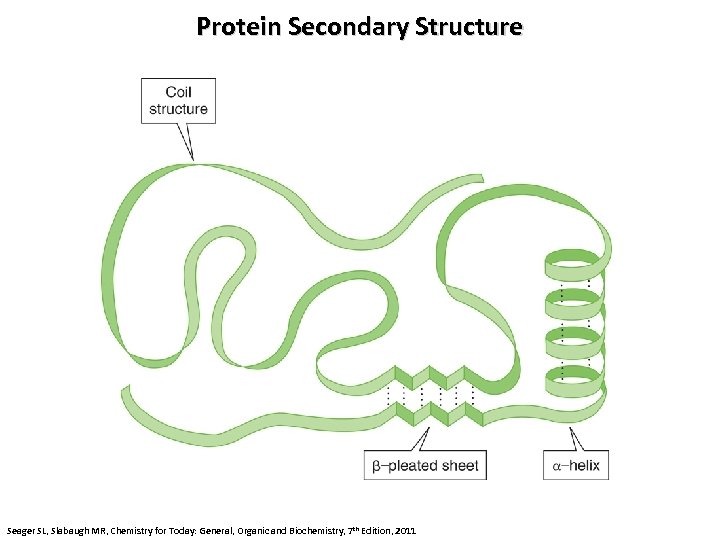
Protein Secondary Structure Seager SL, Slabaugh MR, Chemistry for Today: General, Organic and Biochemistry, 7 th Edition, 2011
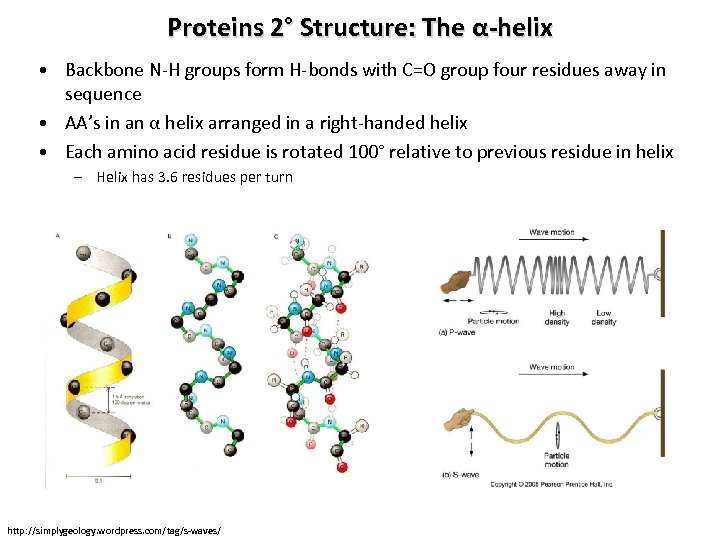
Proteins 2° Structure: The α-helix • Backbone N-H groups form H-bonds with C=O group four residues away in sequence • AA’s in an α helix arranged in a right-handed helix • Each amino acid residue is rotated 100° relative to previous residue in helix – Helix has 3. 6 residues per turn http: //simplygeology. wordpress. com/tag/s-waves/
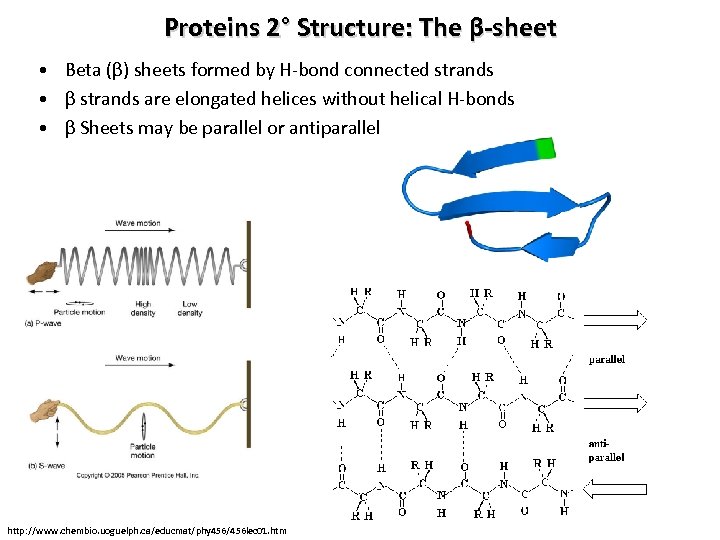
Proteins 2° Structure: The β-sheet • Beta (β) sheets formed by H-bond connected strands • β strands are elongated helices without helical H-bonds • β Sheets may be parallel or antiparallel http: //www. chembio. uoguelph. ca/educmat/phy 456/456 lec 01. htm
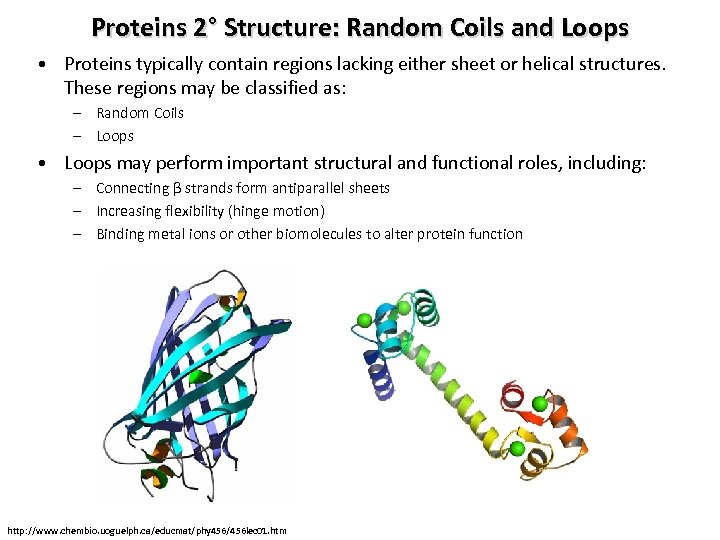
Proteins 2° Structure: Random Coils and Loops • Proteins typically contain regions lacking either sheet or helical structures. These regions may be classified as: – Random Coils – Loops • Loops may perform important structural and functional roles, including: – Connecting β strands form antiparallel sheets – Increasing flexibility (hinge motion) – Binding metal ions or other biomolecules to alter protein function http: //www. chembio. uoguelph. ca/educmat/phy 456/456 lec 01. htm
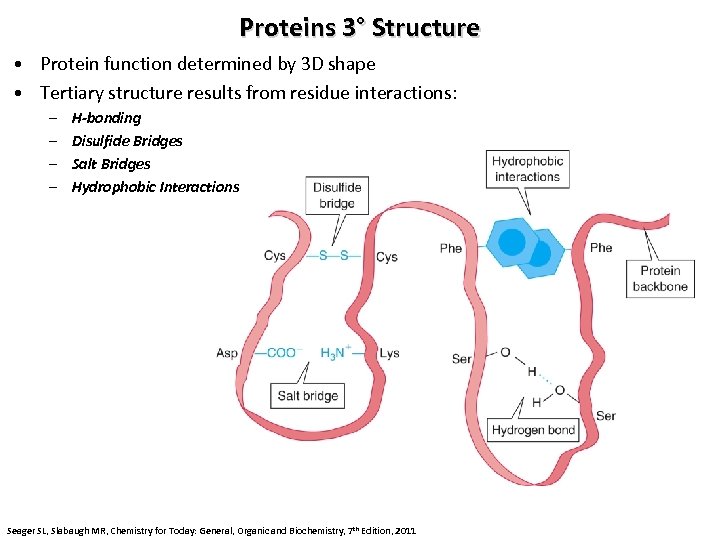
Proteins 3° Structure • Protein function determined by 3 D shape • Tertiary structure results from residue interactions: – – H-bonding Disulfide Bridges Salt Bridges Hydrophobic Interactions Seager SL, Slabaugh MR, Chemistry for Today: General, Organic and Biochemistry, 7 th Edition, 2011
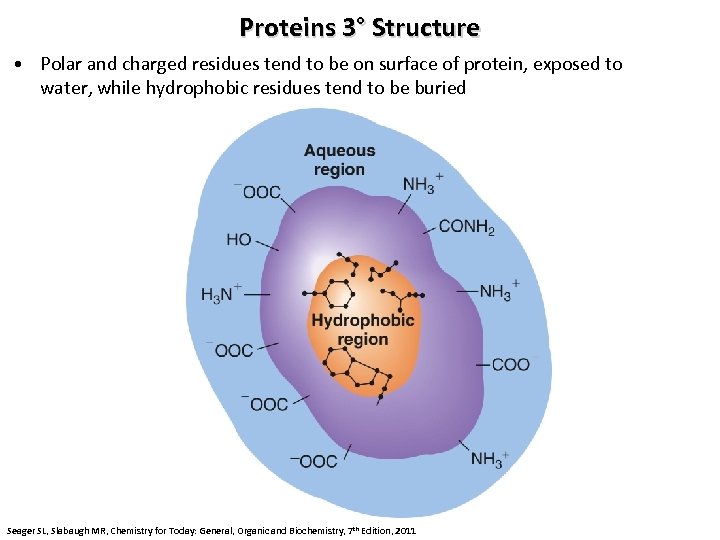
Proteins 3° Structure • Polar and charged residues tend to be on surface of protein, exposed to water, while hydrophobic residues tend to be buried Seager SL, Slabaugh MR, Chemistry for Today: General, Organic and Biochemistry, 7 th Edition, 2011
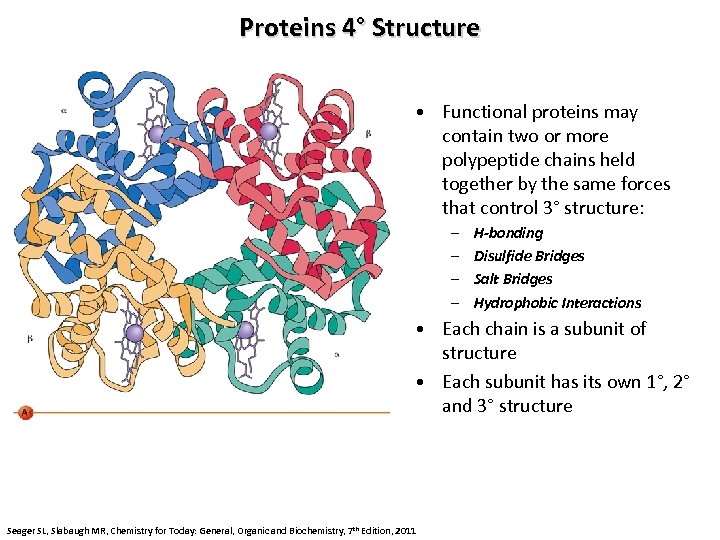
Proteins 4° Structure • Functional proteins may contain two or more polypeptide chains held together by the same forces that control 3° structure: – – H-bonding Disulfide Bridges Salt Bridges Hydrophobic Interactions • Each chain is a subunit of structure • Each subunit has its own 1°, 2° and 3° structure Seager SL, Slabaugh MR, Chemistry for Today: General, Organic and Biochemistry, 7 th Edition, 2011
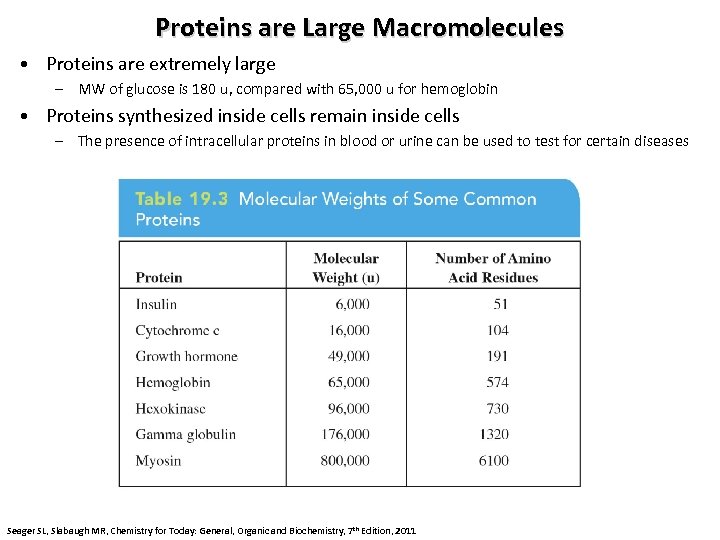
Proteins are Large Macromolecules • Proteins are extremely large – MW of glucose is 180 u, compared with 65, 000 u for hemoglobin • Proteins synthesized inside cells remain inside cells – The presence of intracellular proteins in blood or urine can be used to test for certain diseases Seager SL, Slabaugh MR, Chemistry for Today: General, Organic and Biochemistry, 7 th Edition, 2011
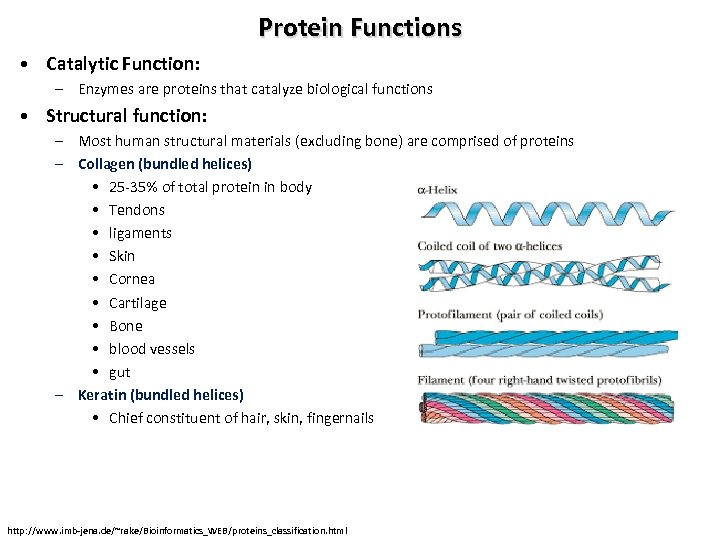
Protein Functions • Catalytic Function: – Enzymes are proteins that catalyze biological functions • Structural function: – Most human structural materials (excluding bone) are comprised of proteins – Collagen (bundled helices) • 25 -35% of total protein in body • Tendons • ligaments • Skin • Cornea • Cartilage • Bone • blood vessels • gut – Keratin (bundled helices) • Chief constituent of hair, skin, fingernails http: //www. imb-jena. de/~rake/Bioinformatics_WEB/proteins_classification. html
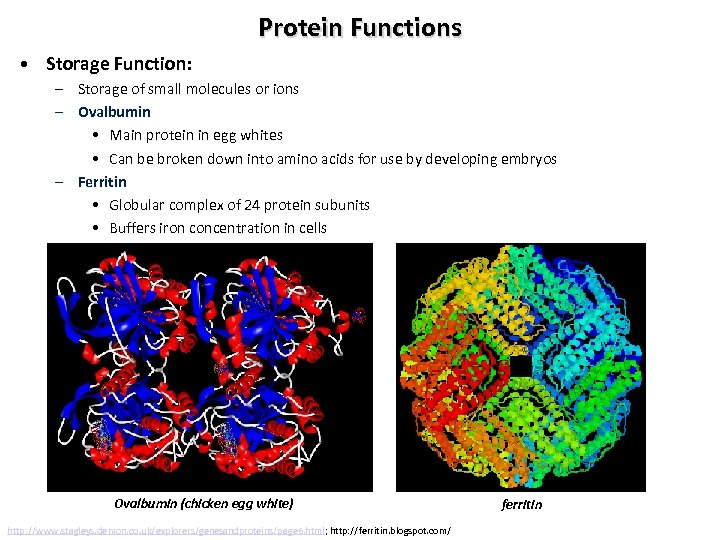
Protein Functions • Storage Function: – Storage of small molecules or ions – Ovalbumin • Main protein in egg whites • Can be broken down into amino acids for use by developing embryos – Ferritin • Globular complex of 24 protein subunits • Buffers iron concentration in cells Ovalbumin (chicken egg white) http: //www. stagleys. demon. co. uk/explorers/genesandproteins/page 6. html; http: //ferritin. blogspot. com/ ferritin
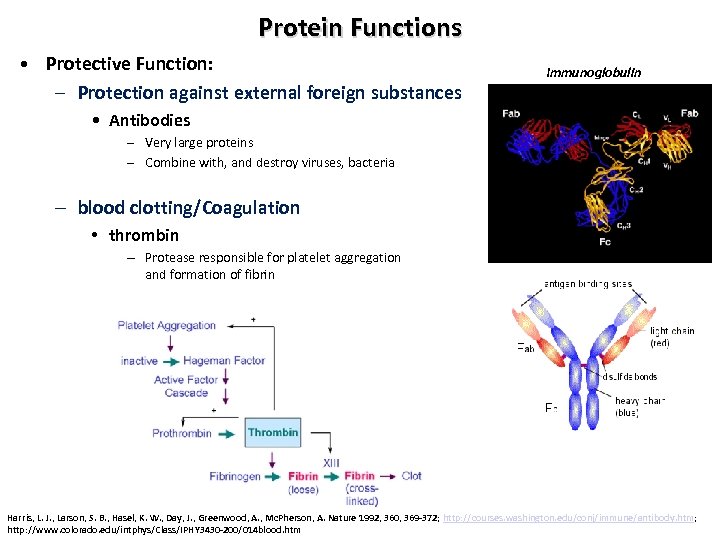
Protein Functions • Protective Function: – Protection against external foreign substances Immunoglobulin • Antibodies – Very large proteins – Combine with, and destroy viruses, bacteria – blood clotting/Coagulation • thrombin – Protease responsible for platelet aggregation and formation of fibrin Harris, L. J. , Larson, S. B. , Hasel, K. W. , Day, J. , Greenwood, A. , Mc. Pherson, A. Nature 1992, 360, 369 -372; http: //courses. washington. edu/conj/immune/antibody. htm; http: //www. colorado. edu/intphys/Class/IPHY 3430 -200/014 blood. htm
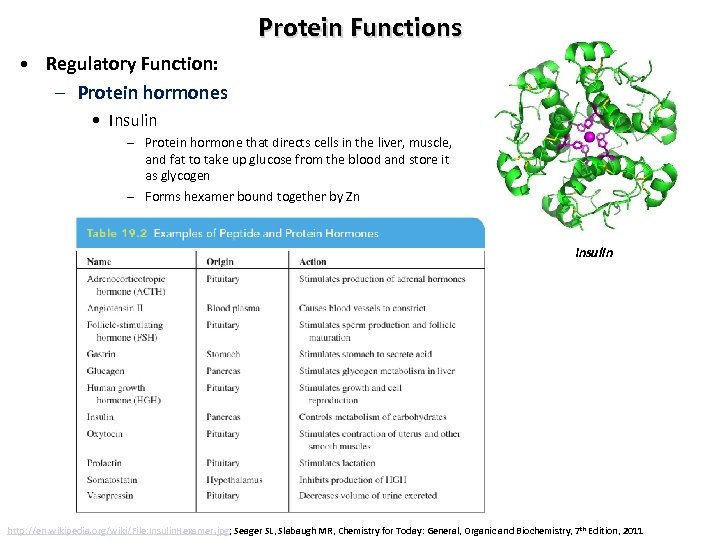
Protein Functions • Regulatory Function: – Protein hormones • Insulin – Protein hormone that directs cells in the liver, muscle, and fat to take up glucose from the blood and store it as glycogen – Forms hexamer bound together by Zn Insulin http: //en. wikipedia. org/wiki/File: Insulin. Hexamer. jpg; Seager SL, Slabaugh MR, Chemistry for Today: General, Organic and Biochemistry, 7 th Edition, 2011
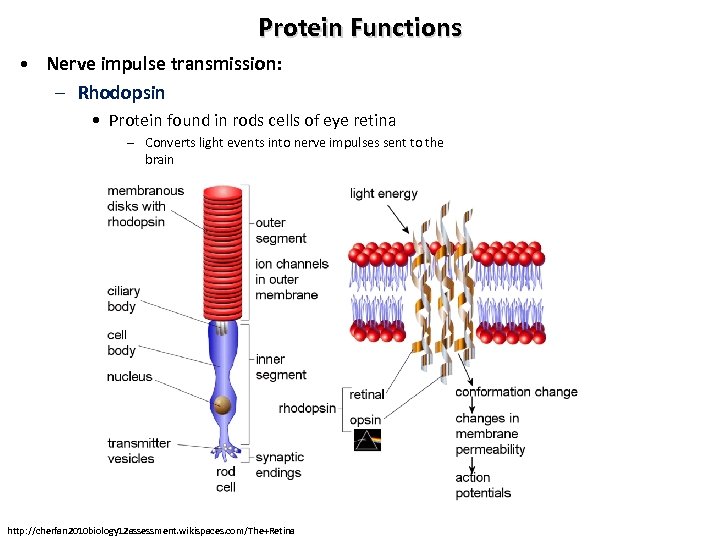
Protein Functions • Nerve impulse transmission: – Rhodopsin • Protein found in rods cells of eye retina – Converts light events into nerve impulses sent to the brain http: //cherfan 2010 biology 12 assessment. wikispaces. com/The+Retina
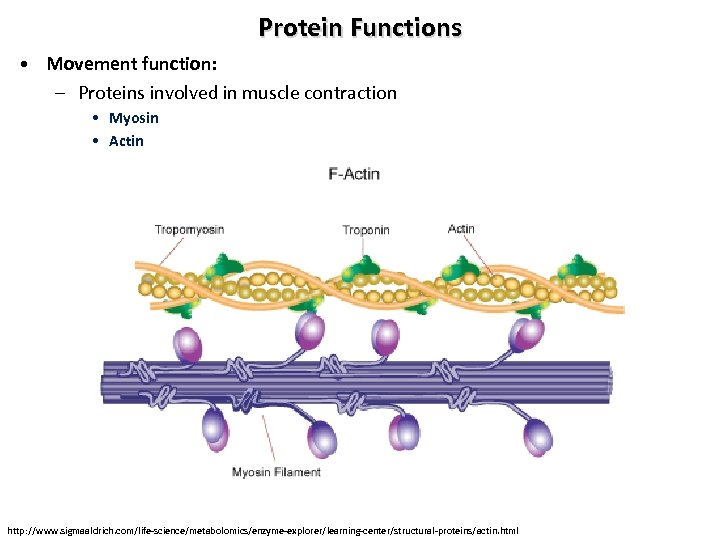
Protein Functions • Movement function: – Proteins involved in muscle contraction • Myosin • Actin http: //www. sigmaaldrich. com/life-science/metabolomics/enzyme-explorer/learning-center/structural-proteins/actin. html
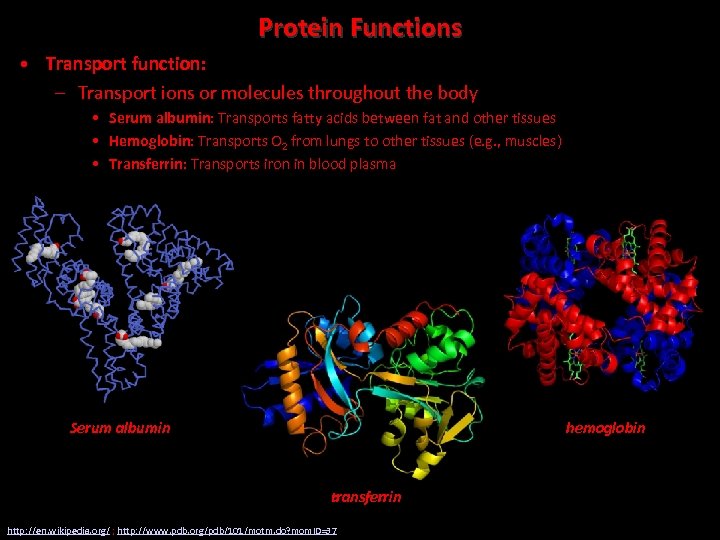
Protein Functions • Transport function: – Transport ions or molecules throughout the body • Serum albumin: Transports fatty acids between fat and other tissues • Hemoglobin: Transports O 2 from lungs to other tissues (e. g. , muscles) • Transferrin: Transports iron in blood plasma Serum albumin hemoglobin transferrin http: //en. wikipedia. org/ ; http: //www. pdb. org/pdb/101/motm. do? mom. ID=37
Protein Databases – NCBI Ref. Seq – Uni. Prot/Swiss-Prot Tr. EMBL (merged with PIR) (http: //www. ebi. ac. uk/uniprot/) – Ensembl (http: //useast. ensembl. org/index. html) – Protein Data. Bank Some of these DB’s have been consolidated over the years. Efforts are being made to develop community standards for reporting protein data HUPO
The Human Proteome Organisation (HUPO) Proteomics Standards Initiative (PSI) http: //www. psidev. info/ HUPO organized into working groups that focus on different aspects of protein research • • • Gel Electrophoresis Mass Spectrometry Molecular Interactions Protein Modifications Proteomics Informatics Sample Processing Goals: Defining standards for proteomic data representation to facilitate the comparison, exchange, and verification of data • Controlled vocabularies • MIAPE: Minimum information about a proteomics experiment
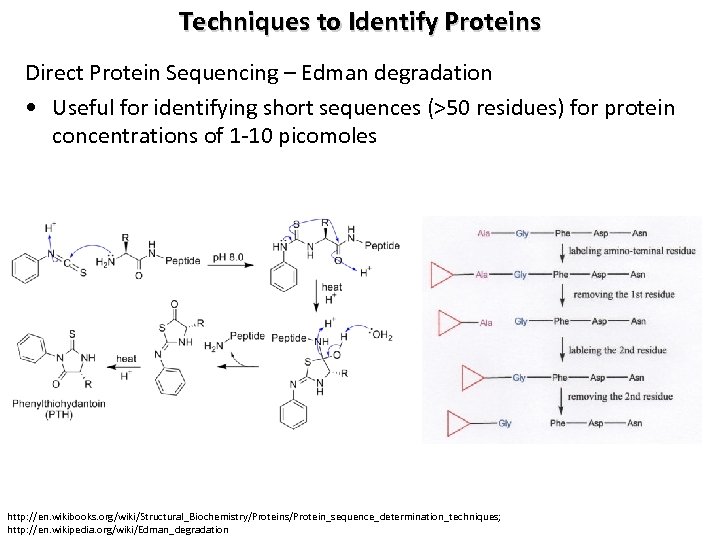
Techniques to Identify Proteins Direct Protein Sequencing – Edman degradation • Useful for identifying short sequences (>50 residues) for protein concentrations of 1 -10 picomoles http: //en. wikibooks. org/wiki/Structural_Biochemistry/Proteins/Protein_sequence_determination_techniques; http: //en. wikipedia. org/wiki/Edman_degradation
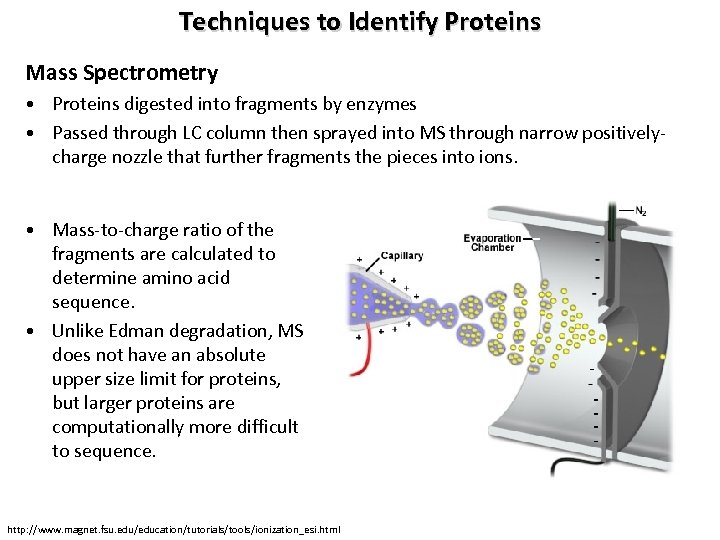
Techniques to Identify Proteins Mass Spectrometry • Proteins digested into fragments by enzymes • Passed through LC column then sprayed into MS through narrow positivelycharge nozzle that further fragments the pieces into ions. • Mass-to-charge ratio of the fragments are calculated to determine amino acid sequence. • Unlike Edman degradation, MS does not have an absolute upper size limit for proteins, but larger proteins are computationally more difficult to sequence. http: //www. magnet. fsu. edu/education/tutorials/tools/ionization_esi. html
Outline: Protein analysis and proteomics Perspectives on Individual proteins Perspective 1: Protein families (domains and motifs) Perspective 2: Physical properties (3 D structure) Perspective 3: Localization Perspective 4: Function

Perspective 1: Protein domains and motifs Page 389
Definitions Signature: a protein category such as a domain or motif Domain: a region of a protein that can adopt a 3 D structure (a fold) Examples: – zinc finger domain – immunoglobulin domain Family: a group of proteins that share a domain Motif (or fingerprint): A short, conserved region of a protein; typically 10 to 20 contiguous amino acid residues Pevsner, Bioinformatics and Functional Genomics, 2009
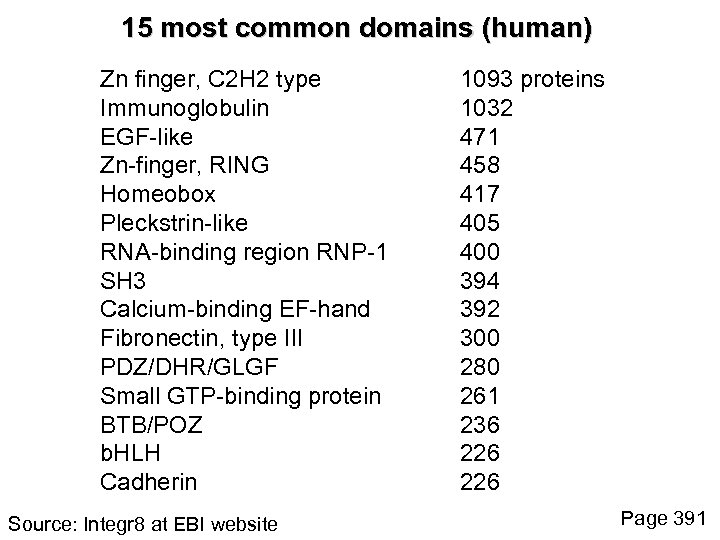
15 most common domains (human) Zn finger, C 2 H 2 type Immunoglobulin EGF-like Zn-finger, RING Homeobox Pleckstrin-like RNA-binding region RNP-1 SH 3 Calcium-binding EF-hand Fibronectin, type III PDZ/DHR/GLGF Small GTP-binding protein BTB/POZ b. HLH Cadherin Source: Integr 8 at EBI website 1093 proteins 1032 471 458 417 405 400 394 392 300 280 261 236 226 Page 391
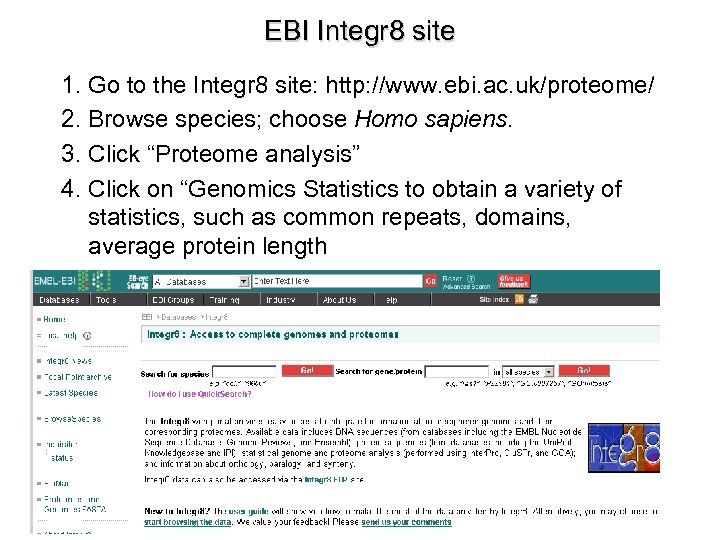
EBI Integr 8 site 1. Go to the Integr 8 site: http: //www. ebi. ac. uk/proteome/ 2. Browse species; choose Homo sapiens. 3. Click “Proteome analysis” 4. Click on “Genomics Statistics to obtain a variety of statistics, such as common repeats, domains, average protein length
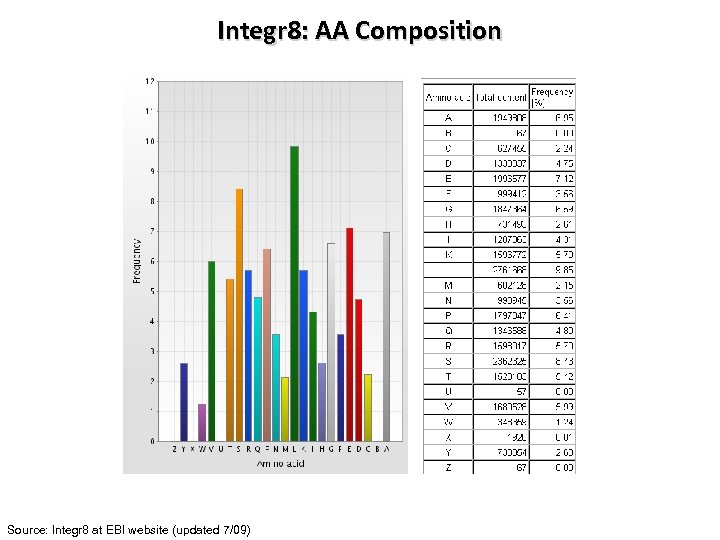
Integr 8: AA Composition Source: Integr 8 at EBI website (updated 7/09)
![Analysis of full-length proteins [fragments excluded] Avg protein length : 412 +/- 548 amino Analysis of full-length proteins [fragments excluded] Avg protein length : 412 +/- 548 amino](https://present5.com/presentation/f3c48371c40834436e432e61a393d92a/image-33.jpg)
Analysis of full-length proteins [fragments excluded] Avg protein length : 412 +/- 548 amino acid residues Size range: 4 - 34942 amino acid residues Source: Integr 8 at EBI website (updated 7/09)
Definitions of a domain According to Inter. Pro at EBI (http: //www. ebi. ac. uk/interpro/): A domain is an independent structural unit, found alone or in conjunction with other domains or repeats. Domains are evolutionarily related. According to SMART (http: //smart. embl-heidelberg. de): A domain is a conserved structural entity with distinctive secondary structure content and a hydrophobic core. Homologous domains with common functions usually show sequence similarities. Page 390
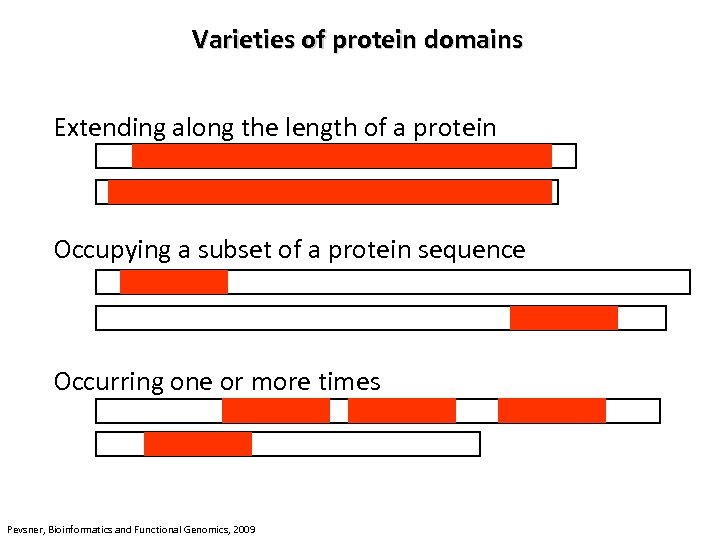
Varieties of protein domains Extending along the length of a protein Occupying a subset of a protein sequence Occurring one or more times Pevsner, Bioinformatics and Functional Genomics, 2009
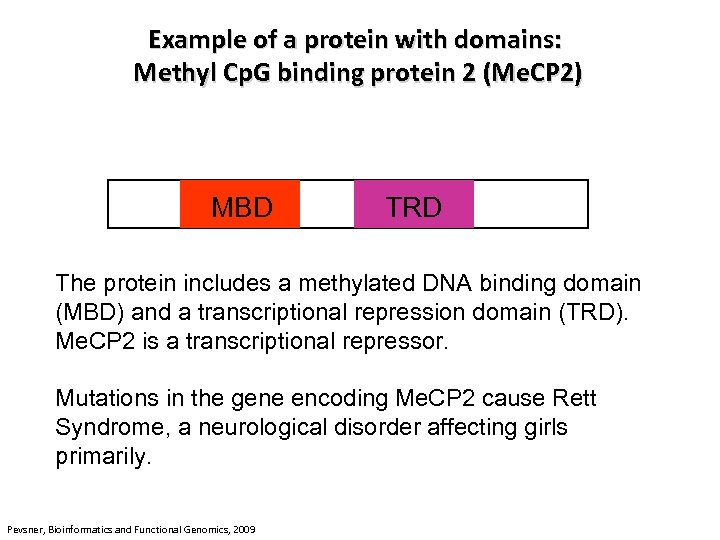
Example of a protein with domains: Methyl Cp. G binding protein 2 (Me. CP 2) MBD TRD The protein includes a methylated DNA binding domain (MBD) and a transcriptional repression domain (TRD). Me. CP 2 is a transcriptional repressor. Mutations in the gene encoding Me. CP 2 cause Rett Syndrome, a neurological disorder affecting girls primarily. Pevsner, Bioinformatics and Functional Genomics, 2009
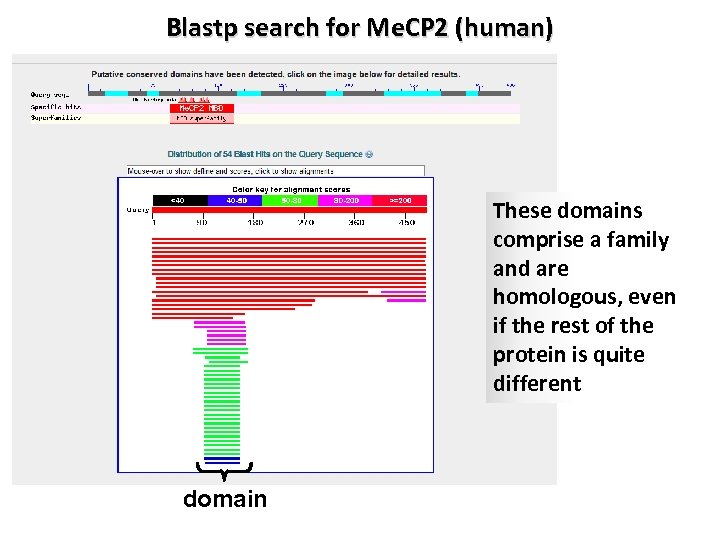
Blastp search for Me. CP 2 (human) These domains comprise a family and are homologous, even if the rest of the protein is quite different domain
Example of a multidomain protein: HIV-1 pol • Multi-domain proteins such as HIV-1 gag-pol are common • Pol (NP_789740), 995 amino acids long • Gag-Pol (NP_057849), 1435 amino acids • cleaved into three proteins with distinct activities: – -- aspartyl protease – -- reverse transcriptase – -- integrase • We will explore HIV-1 pol through Uni. Prot. Pevsner, Bioinformatics and Functional Genomics, 2009

www. uniprot. org Three protein databases merged to form Uni. Prot: • Swiss. Prot • Tr. EMBL (translated European Molecular Biology Lab) • Protein Information Resource (PIR) You can search for information on your favorite protein there; a BLAST server is provided. Pevsner, Bioinformatics and Functional Genomics, 2009
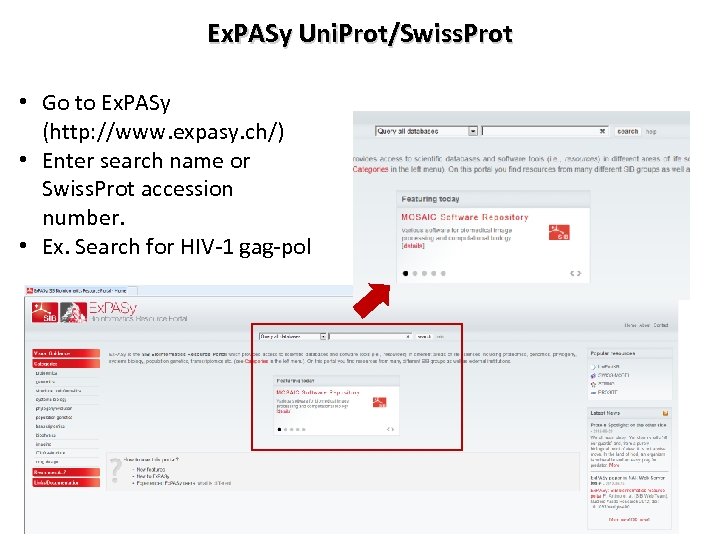
Ex. PASy Uni. Prot/Swiss. Prot • Go to Ex. PASy (http: //www. expasy. ch/) • Enter search name or Swiss. Prot accession number. • Ex. Search for HIV-1 gag-pol
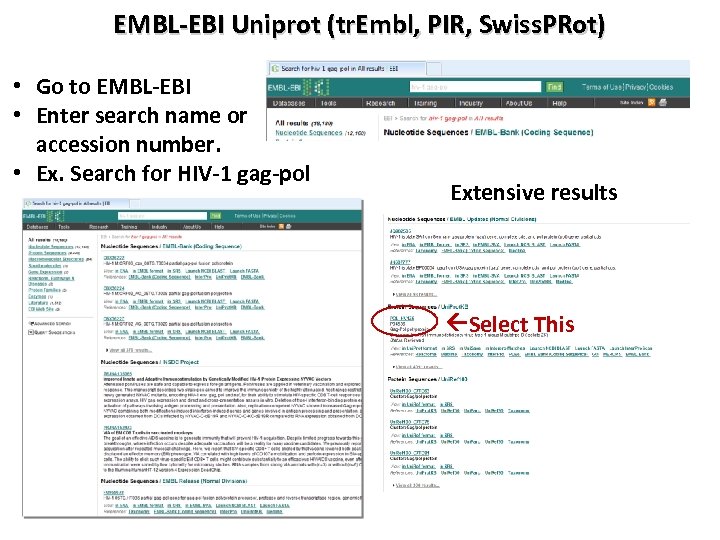
EMBL-EBI Uniprot (tr. Embl, PIR, Swiss. PRot) • Go to EMBL-EBI • Enter search name or accession number. • Ex. Search for HIV-1 gag-pol Extensive results Select This
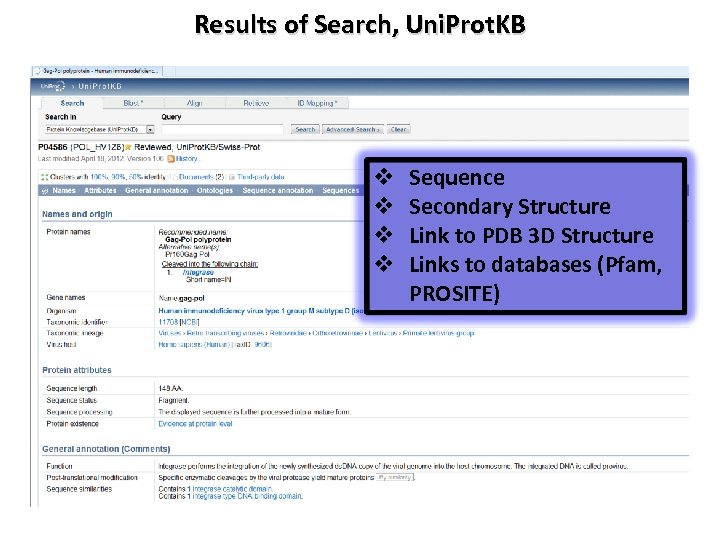
Results of Search, Uni. Prot. KB v v Sequence Secondary Structure Link to PDB 3 D Structure Links to databases (Pfam, PROSITE)
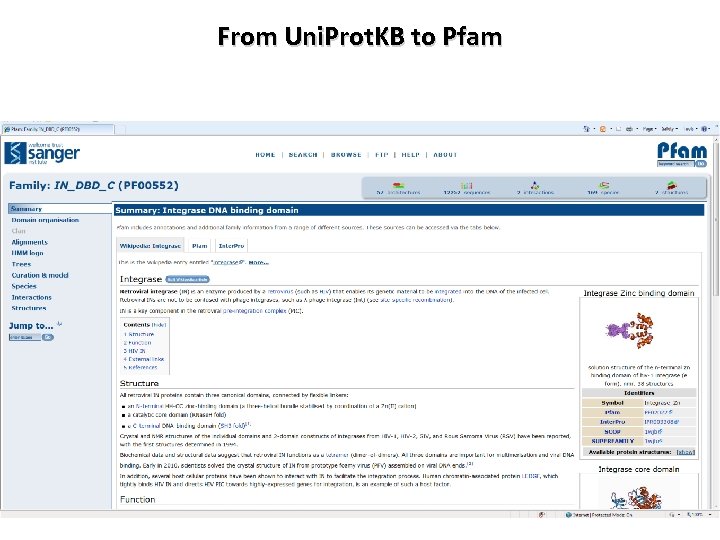
From Uni. Prot. KB to Pfam
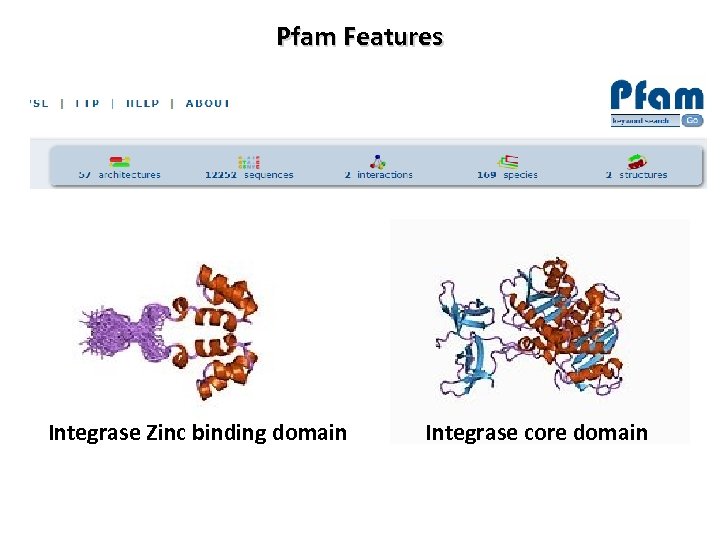
Pfam Features Integrase Zinc binding domain Integrase core domain
Pfam Features: Domains Select This
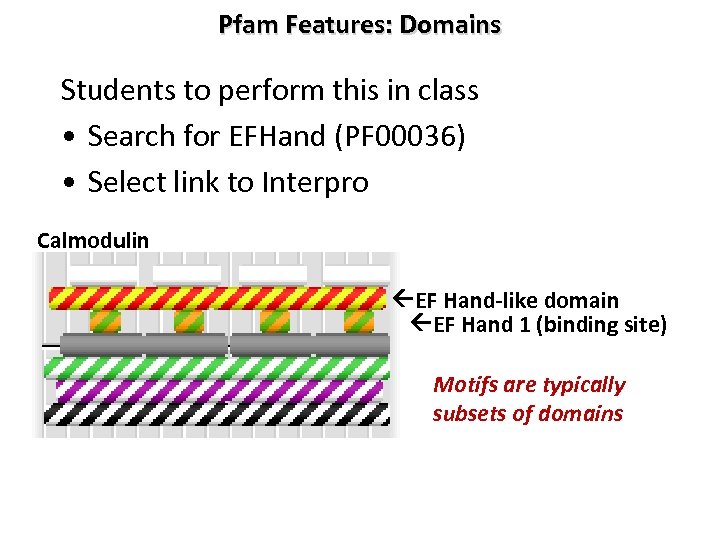
Pfam Features: Domains Students to perform this in class • Search for EFHand (PF 00036) • Select link to Interpro Calmodulin EF Hand-like domain EF Hand 1 (binding site) Motifs are typically subsets of domains
Definition of a motif • Motif (or fingerprint): A short, conserved region of a protein (10 to 20 amino acids). • Simple motifs include (but are not limited to): – transmembrane domains – phosphorylation sites – calcium-binding sites • These do not imply homology when found in a group of proteins. • PROSITE (www. expasy. org/prosite) is a dictionary of motifs. • In PROSITE, a pattern is a qualitative motif description (a protein either matches a pattern, or not). • In contrast, a profile is a quantitative motif description. We will encounter profiles in Pfam, Pro. Dom, SMART, and other databases. Pevsner, Bioinformatics and Functional Genomics, 2009
Calcium-binding protein sequence patterns

Perspective 2: Physical properties of proteins
Physical properties of proteins Many websites are available for the analysis of individual proteins. Ex. PASy is an excellent resource. The accuracy of these programs varies. Predictions based on primary amino acid sequence (such as molecular weight prediction) are likely to be more trustworthy. For many other properties (such as posttranslational modification of proteins by specific sugars), experimental evidence may be required rather than prediction algorithms. Pevsner, Bioinformatics and Functional Genomics, 2009
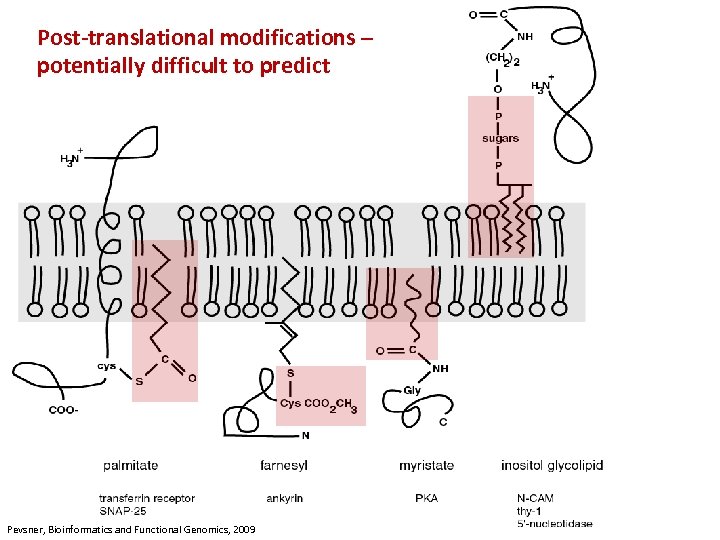
Post-translational modifications – potentially difficult to predict Pevsner, Bioinformatics and Functional Genomics, 2009
Calculate Protein MW/p. I • p. I: The p. H at which a particular molecule or surface carries no net electrical charge • p. I and MW can be calculated from a sequence using Protein Calculator (http: //www. scripps. edu/~cdputnam/protcalc. html) • Lets look at some examples!
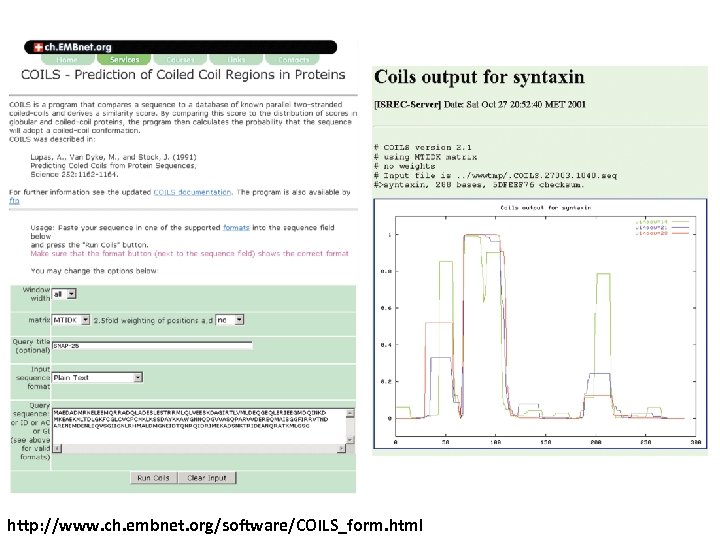
http: //www. ch. embnet. org/software/COILS_form. html
Protein secondary structure is determined by the amino acid side chains. Myoglobin is an example of a protein having many a-helices. These are formed by amino acid stretches 4 -40 residues in length. Thioredoxin from E. coli is an example of a protein with many b sheets, formed from b strands composed of 5 -10 residues. They are arranged in parallel or antiparallel orientations. Pevsner, Bioinformatics and Functional Genomics, 2009
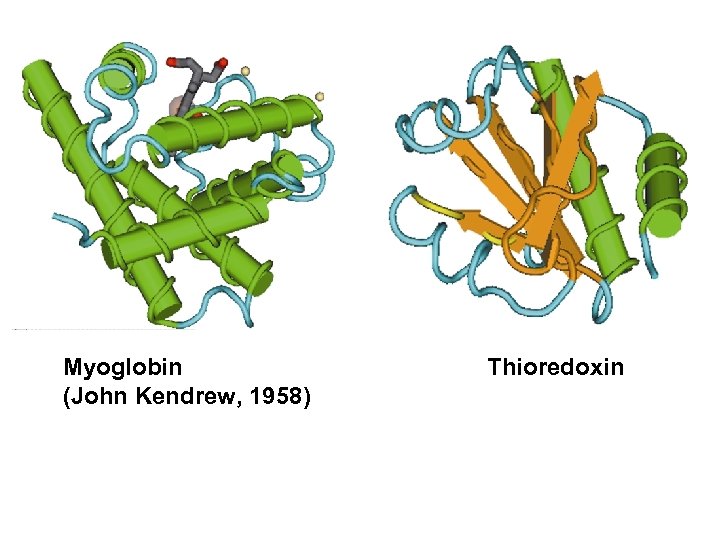
Myoglobin (John Kendrew, 1958) Thioredoxin
Secondary structure prediction • Chou and Fasman (1974) developed an algorithm based on the frequencies of amino acids found in a helices, bsheets, and turns. – Proline: occurs at turns, but not in a helices. • GOR (Garnier, Osguthorpe, Robson): related algorithm • Modern algorithms: use multiple sequence alignments and achieve higher success rate (about 70 -75%) Pevsner, Bioinformatics and Functional Genomics, 2009 Web servers: GOR 4 Jpred NNPREDICT PHD Predator Predict. Protein PSIPRED SAM-T 99 sec PDBSum
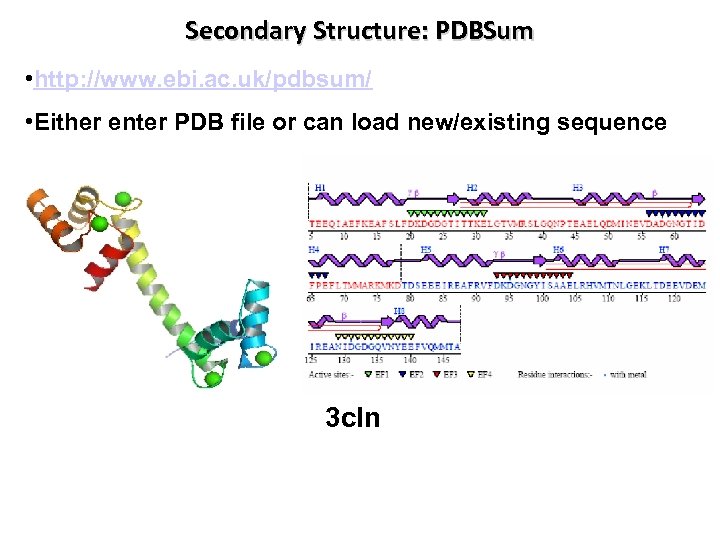
Secondary Structure: PDBSum • http: //www. ebi. ac. uk/pdbsum/ • Either enter PDB file or can load new/existing sequence 3 cln
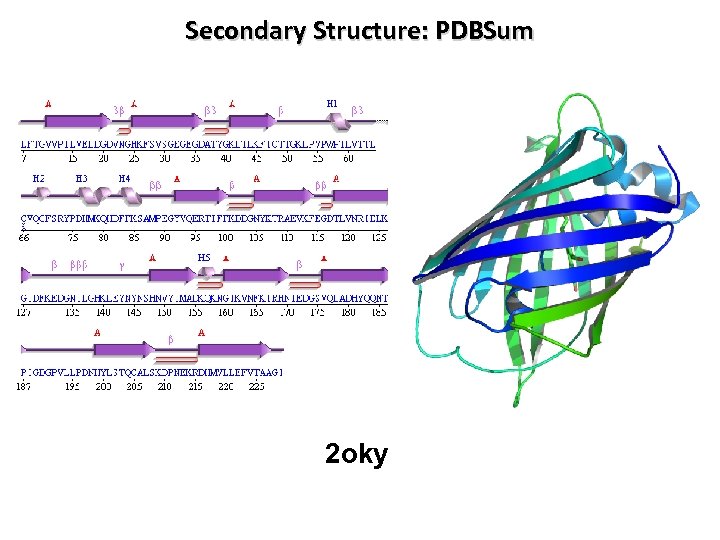
Secondary Structure: PDBSum 2 oky
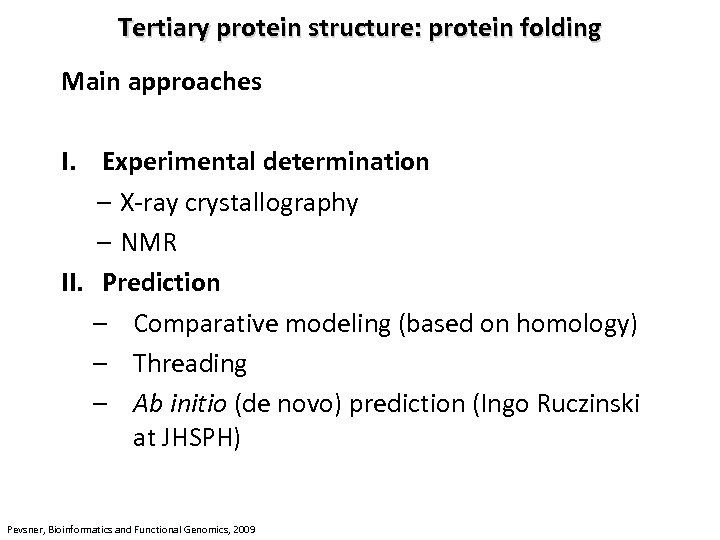
Tertiary protein structure: protein folding Main approaches I. Experimental determination – X-ray crystallography – NMR II. Prediction – Comparative modeling (based on homology) – Threading – Ab initio (de novo) prediction (Ingo Ruczinski at JHSPH) Pevsner, Bioinformatics and Functional Genomics, 2009
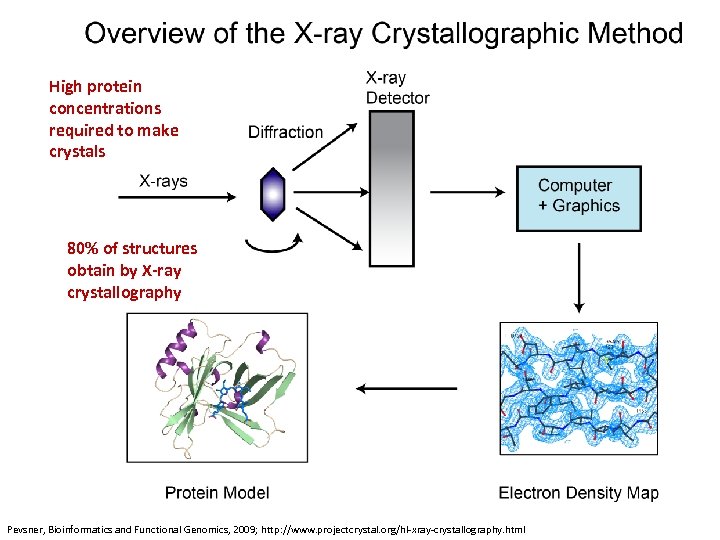
High protein concentrations required to make crystals 80% of structures obtain by X-ray crystallography Pevsner, Bioinformatics and Functional Genomics, 2009; http: //www. projectcrystal. org/hl-xray-crystallography. html
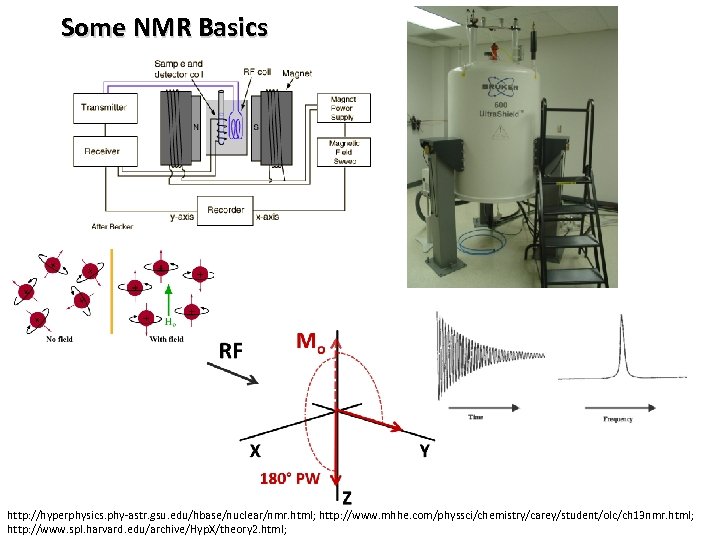
Some NMR Basics http: //hyperphysics. phy-astr. gsu. edu/hbase/nuclear/nmr. html; http: //www. mhhe. com/physsci/chemistry/carey/student/olc/ch 13 nmr. html; http: //www. spl. harvard. edu/archive/Hyp. X/theory 2. html;
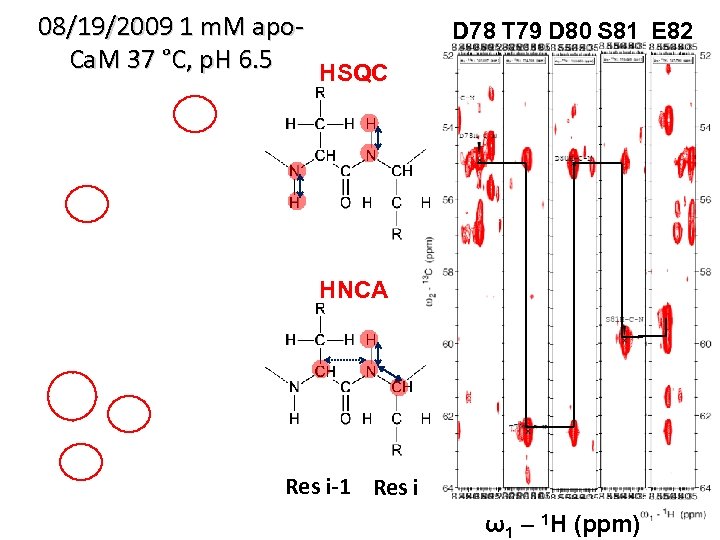
08/19/2009 1 m. M apo. Ca. M 37 °C, p. H 6. 5 HSQC D 78 T 79 D 80 S 81 E 82 HNCA Res i-1 Res i ω1 – 1 H (ppm)
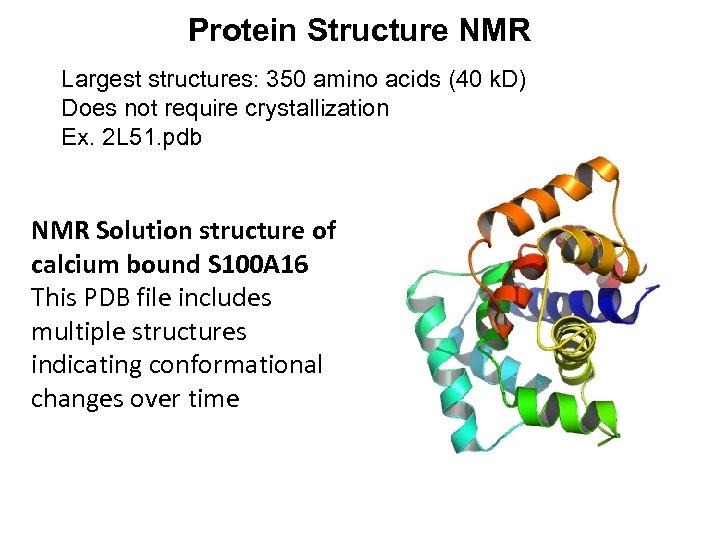
Protein Structure NMR Largest structures: 350 amino acids (40 k. D) Does not require crystallization Ex. 2 L 51. pdb NMR Solution structure of calcium bound S 100 A 16 This PDB file includes multiple structures indicating conformational changes over time
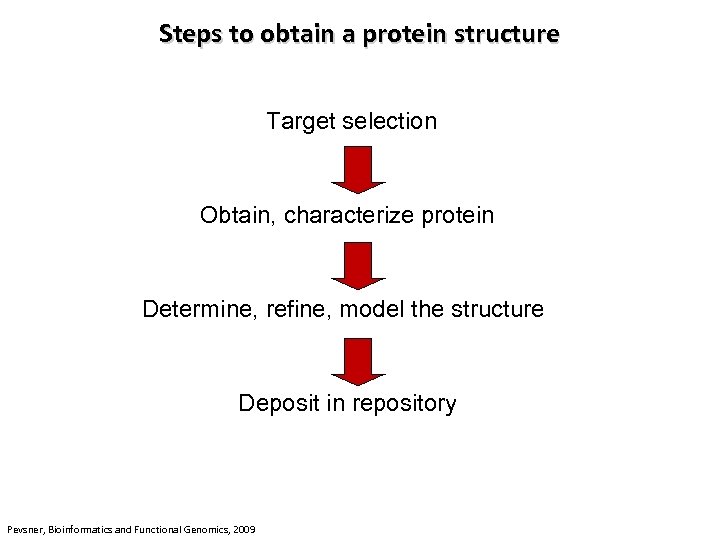
Steps to obtain a protein structure Target selection Obtain, characterize protein Determine, refine, model the structure Deposit in repository Pevsner, Bioinformatics and Functional Genomics, 2009
The Protein Data Bank (PDB) • Principal repository for protein structures (since 1971) • http: //www. rcsb. org/pdb/home. do
Protein Data Bank PDB Provides: • 3 D structural data • Fasta sequence • Citation Info (who solved it, related publications, etc. ) • experimental methods (X-Ray Diffraction, NMR) • resolution • classification (e. g. – metal transporter) • ligands, cofactors • Related PDB entries
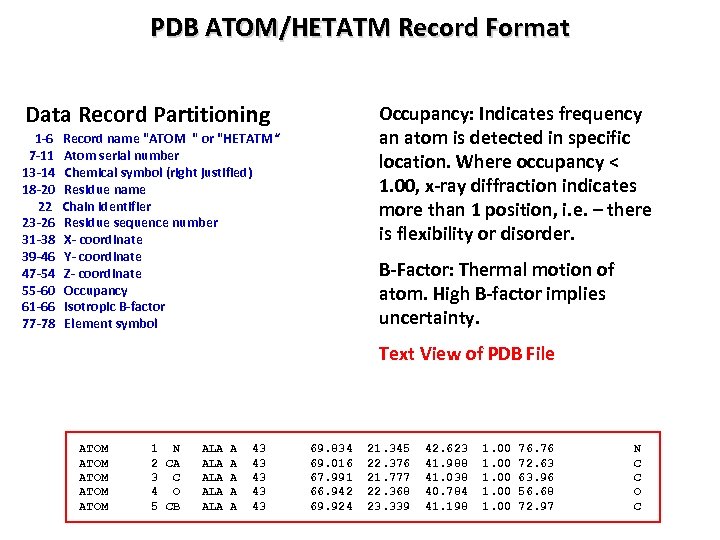
PDB ATOM/HETATM Record Format Data Record Partitioning 1 -6 7 -11 13 -14 18 -20 22 23 -26 31 -38 39 -46 47 -54 55 -60 61 -66 77 -78 Occupancy: Indicates frequency an atom is detected in specific location. Where occupancy < 1. 00, x-ray diffraction indicates more than 1 position, i. e. – there is flexibility or disorder. Record name "ATOM " or "HETATM“ Atom serial number Chemical symbol (right justified) Residue name Chain identifier Residue sequence number X- coordinate Y- coordinate Z- coordinate Occupancy Isotropic B-factor Element symbol B-Factor: Thermal motion of atom. High B-factor implies uncertainty. Text View of PDB File ATOM ATOM 1 N 2 CA 3 C 4 O 5 CB ALA ALA ALA A A 43 43 43 69. 834 69. 016 67. 991 66. 942 69. 924 21. 345 22. 376 21. 777 22. 368 23. 339 42. 623 41. 988 41. 038 40. 784 41. 198 1. 00 76. 76 72. 63 63. 96 56. 68 72. 97 N C C O C
f3c48371c40834436e432e61a393d92a.ppt