07d23c2ca22b764082f360c206a997a5.ppt
- Количество слайдов: 28

BIological Netw. Ork Manager Cytoscape plugin Andrei Zinovyev Institut Curie/INSERM/Ecole de Mines, UMR 900 “Computational Systems Biology of Cancer” http: //bioinfo. curie. fr/sysbio ICSB Web-services - 28 August 2008
ICSB Web-services - 28 August 2008
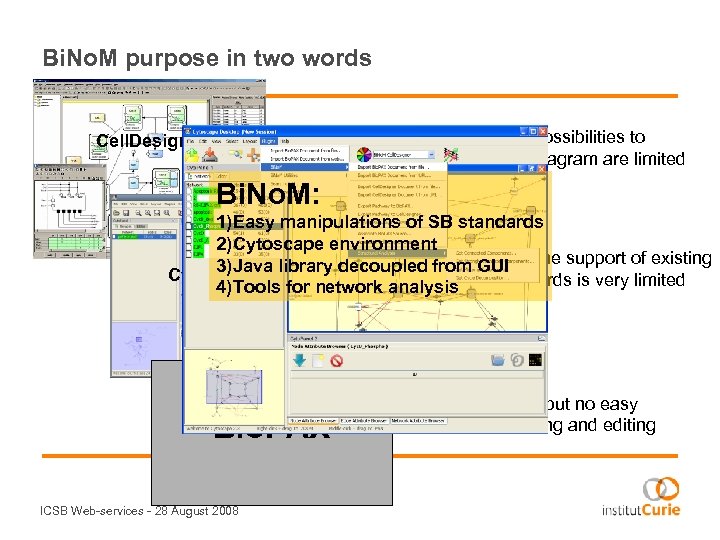
Bi. No. M purpose in two words …very user friendly, but the possibilities to analyze and manipulate the diagram are limited Cell. Designer Bi. No. M: 1)Easy manipulations of SB standards 2)Cytoscape environment …very interactive, 3)Java library decoupled from GUIbut the support of existing Cytoscape Systems biology standards is very limited 4)Tools for network analysis Bio. PAX ICSB Web-services - 28 August 2008 …very useful, but no easy tools for creating and editing
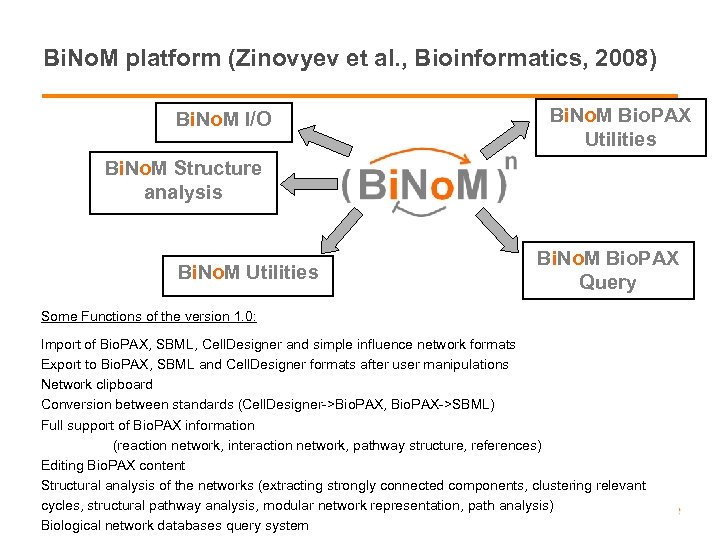
Bi. No. M platform (Zinovyev et al. , Bioinformatics, 2008) Bi. No. M I/O Bi. No. M Bio. PAX Utilities Bi. No. M Structure analysis Bi. No. M Utilities Bi. No. M Bio. PAX Query Some Functions of the version 1. 0: Import of Bio. PAX, SBML, Cell. Designer and simple influence network formats Export to Bio. PAX, SBML and Cell. Designer formats after user manipulations Network clipboard Conversion between standards (Cell. Designer->Bio. PAX, Bio. PAX->SBML) Full support of Bio. PAX information (reaction network, interaction network, pathway structure, references) Editing Bio. PAX content Structural analysis of the networks (extracting strongly connected components, clustering relevant cycles, structural 28 August 2008 ICSB Web-services -pathway analysis, modular network representation, path analysis) Biological network databases query system
Typical scenarios 1) Import Cell. Designer diagram, manipulate, convert to Bio. PAX 2) Import Cell. Designer diagram, put colors and save it back 3) Import Cell. Designer diagram, analyze, decompose into modules, create a modular view of the diagram 4) Import Bio. PAX file, extract a part, export to SBML, create a model 5) Import Bio. PAX file, edit, save it back 6) Index huge Bio. PAX file (whole Reactome), make a query, save result as small Bio. PAX file 7) Create a new Bio. PAX file from simple text file etc. ICSB Web-services - 28 August 2008
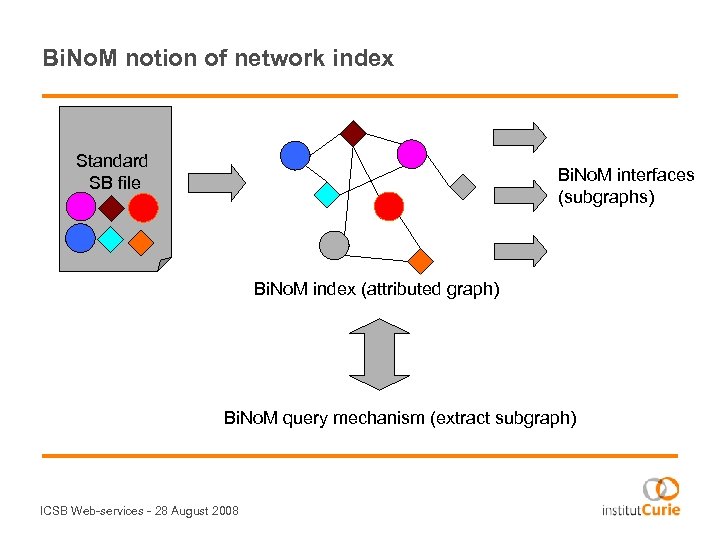
Bi. No. M notion of network index Standard SB file Bi. No. M interfaces (subgraphs) Bi. No. M index (attributed graph) Bi. No. M query mechanism (extract subgraph) ICSB Web-services - 28 August 2008
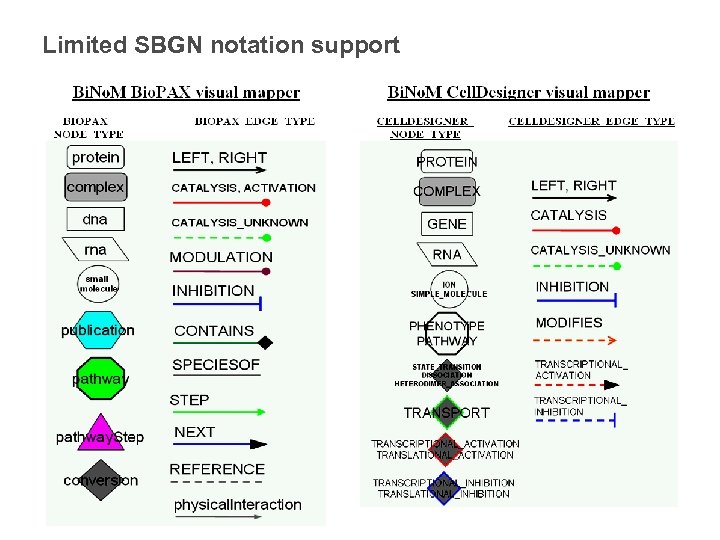
Limited SBGN notation support
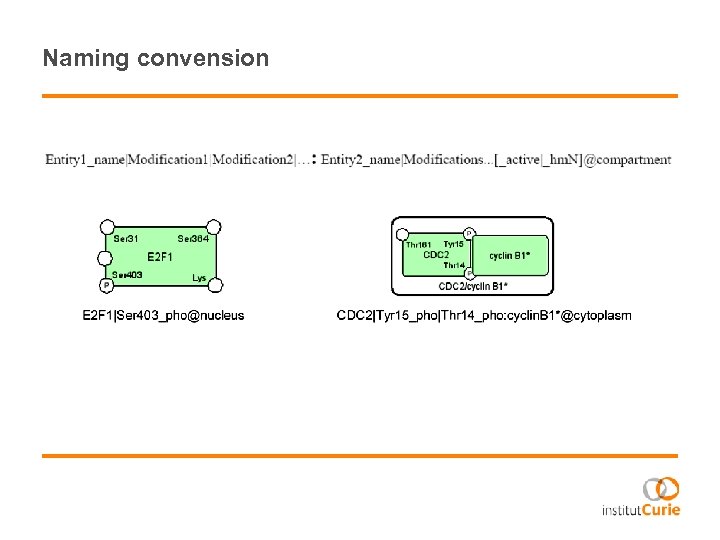
Naming convension
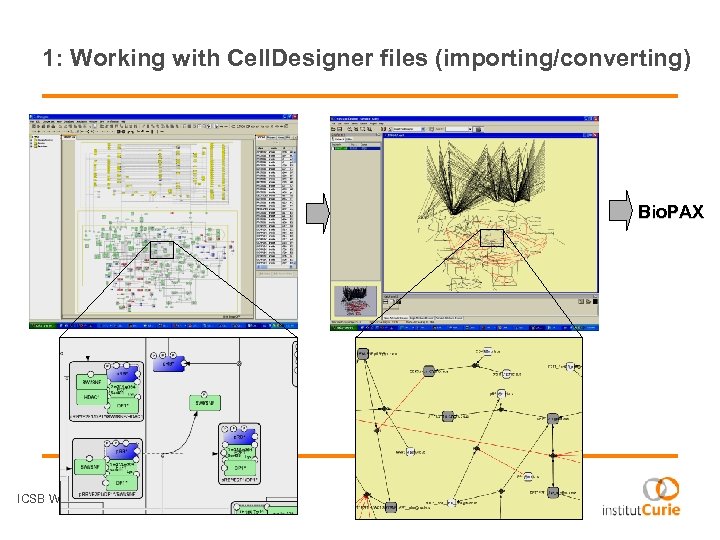
1: Working with Cell. Designer files (importing/converting) Bio. PAX ICSB Web-services - 28 August 2008
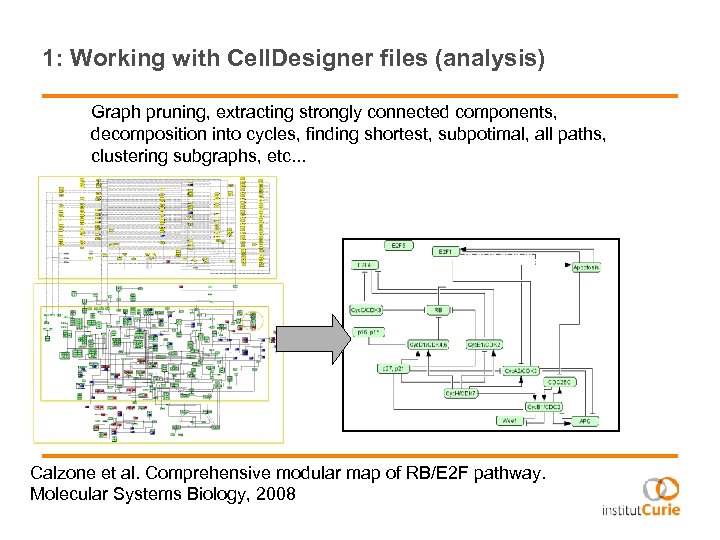
1: Working with Cell. Designer files (analysis) Graph pruning, extracting strongly connected components, decomposition into cycles, finding shortest, subpotimal, all paths, clustering subgraphs, etc. . . Calzone et al. Comprehensive modular map of RB/E 2 F pathway. Molecular Systems Biology, 2008
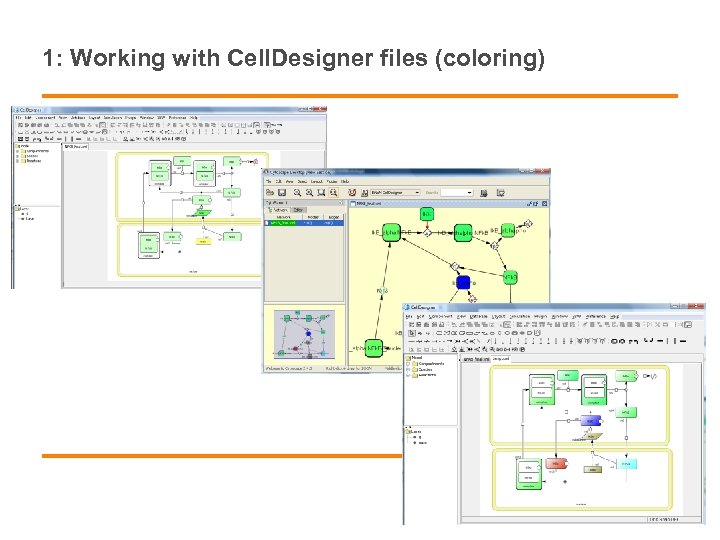
1: Working with Cell. Designer files (coloring)
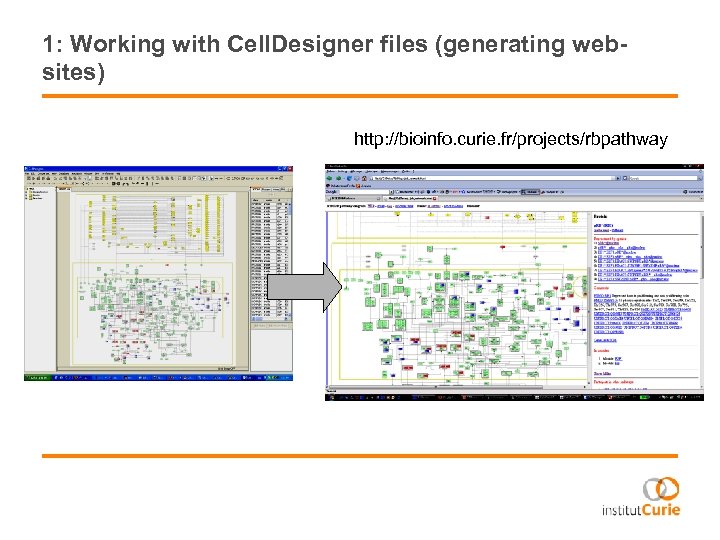
1: Working with Cell. Designer files (generating websites) http: //bioinfo. curie. fr/projects/rbpathway
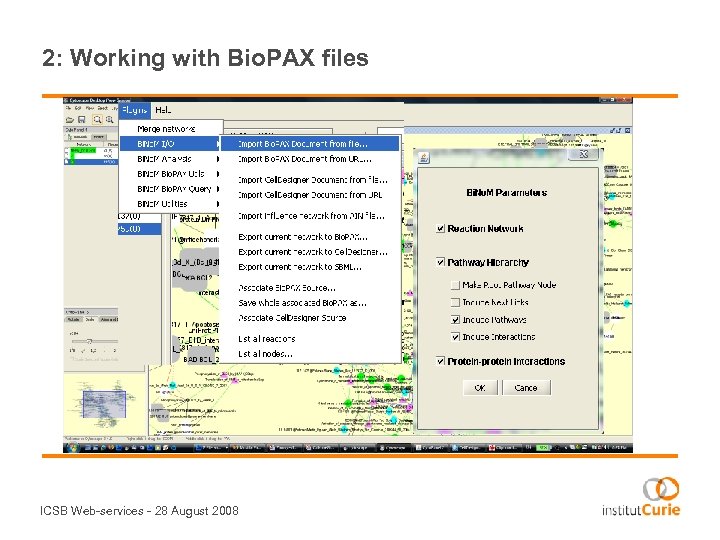
2: Working with Bio. PAX files ICSB Web-services - 28 August 2008
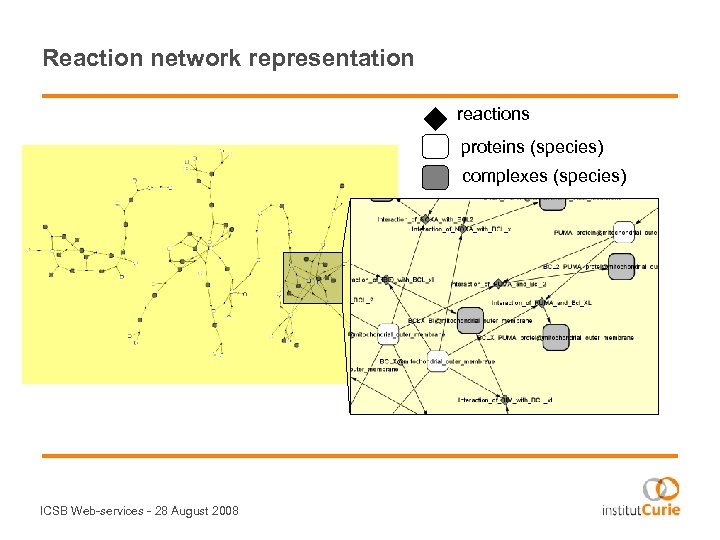
Reaction network representation reactions proteins (species) complexes (species) ICSB Web-services - 28 August 2008
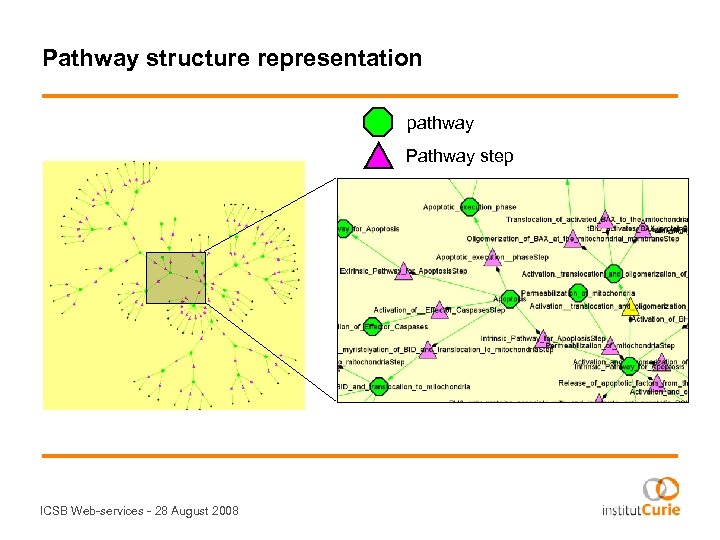
Pathway structure representation pathway Pathway step ICSB Web-services - 28 August 2008
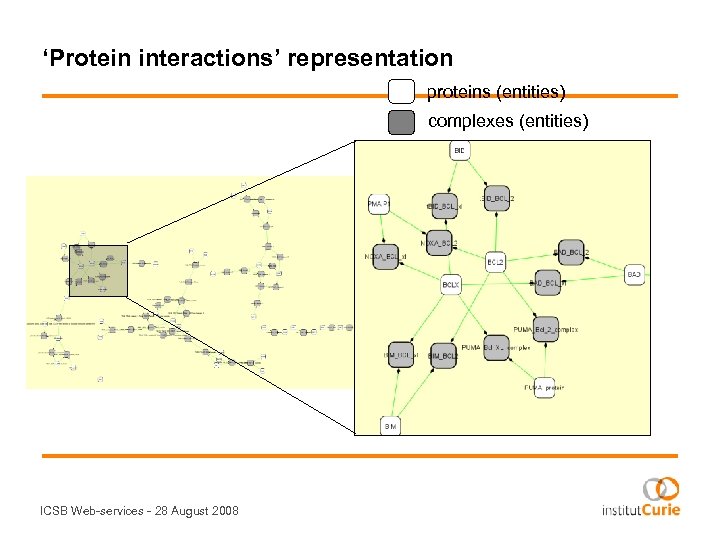
‘Protein interactions’ representation proteins (entities) complexes (entities) ICSB Web-services - 28 August 2008
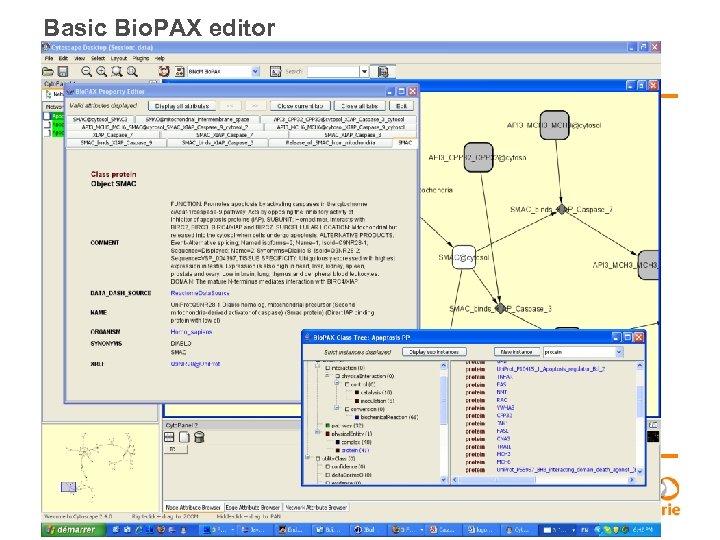
Basic Bio. PAX editor
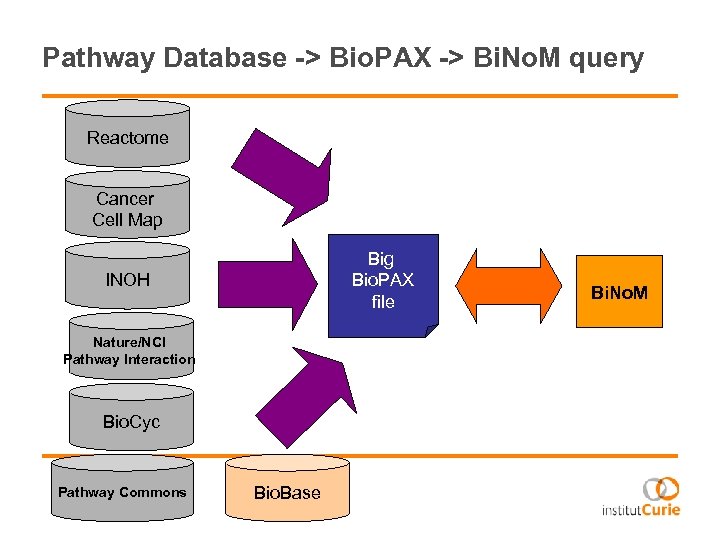
Pathway Database -> Bio. PAX -> Bi. No. M query Reactome Cancer Cell Map Big Bio. PAX file INOH Nature/NCI Pathway Interaction Bio. Cyc Pathway Commons Bio. Base Bi. No. M
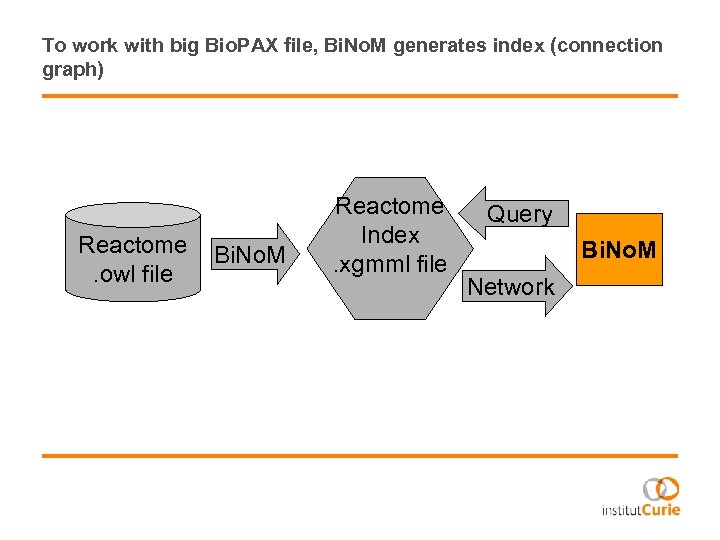
To work with big Bio. PAX file, Bi. No. M generates index (connection graph) Reactome. owl file Bi. No. M Reactome Index. xgmml file Query Bi. No. M Network
Example of query
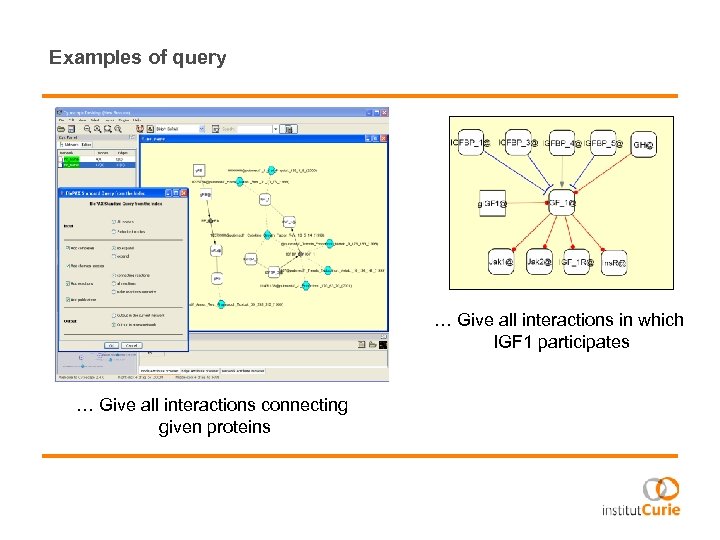
Examples of query … Give all interactions in which IGF 1 participates … Give all interactions connecting given proteins
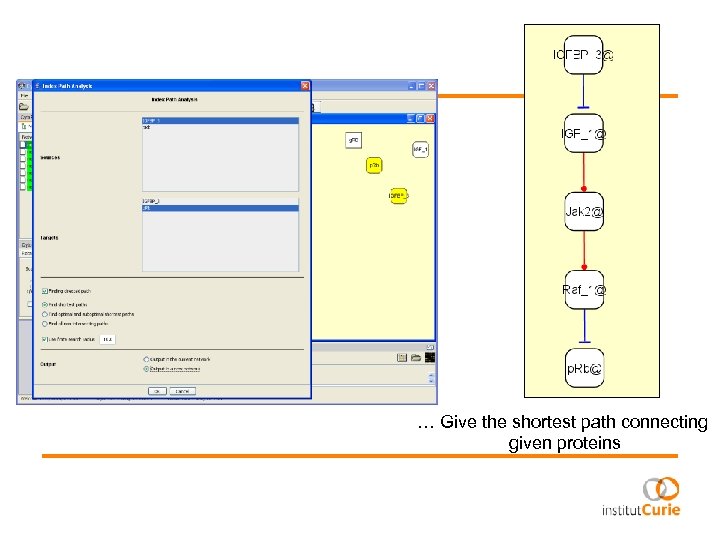
… Give the shortest path connecting given proteins
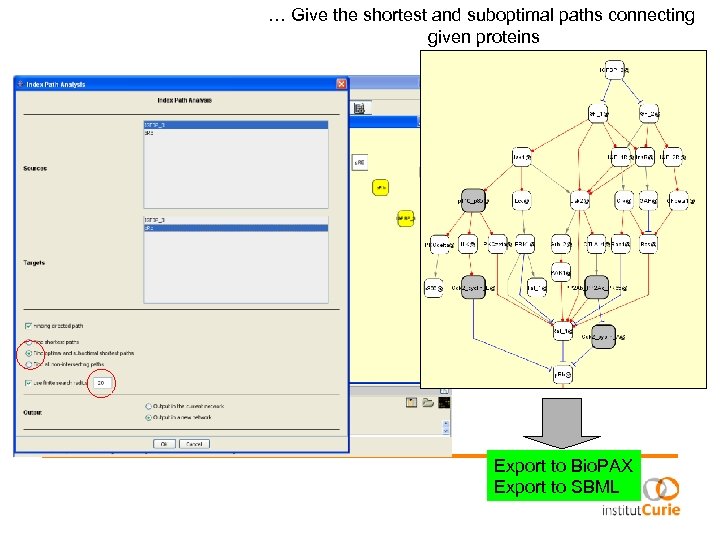
… Give the shortest and suboptimal paths connecting given proteins Export to Bio. PAX Export to SBML
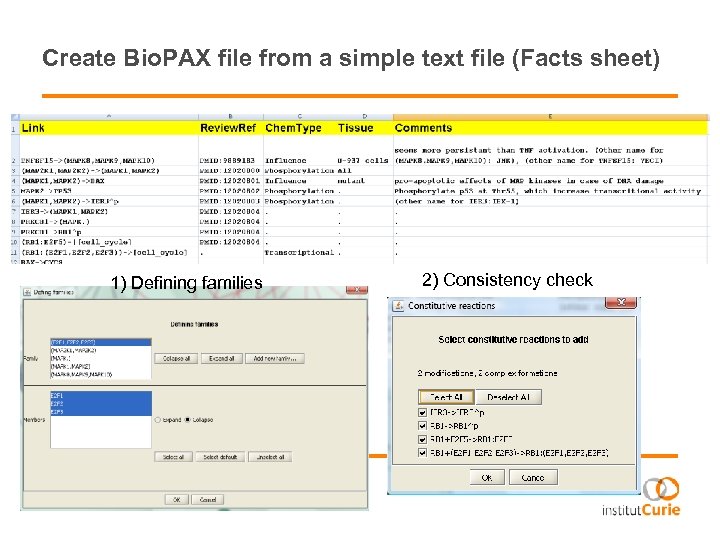
Create Bio. PAX file from a simple text file (Facts sheet) 1) Defining families 2) Consistency check
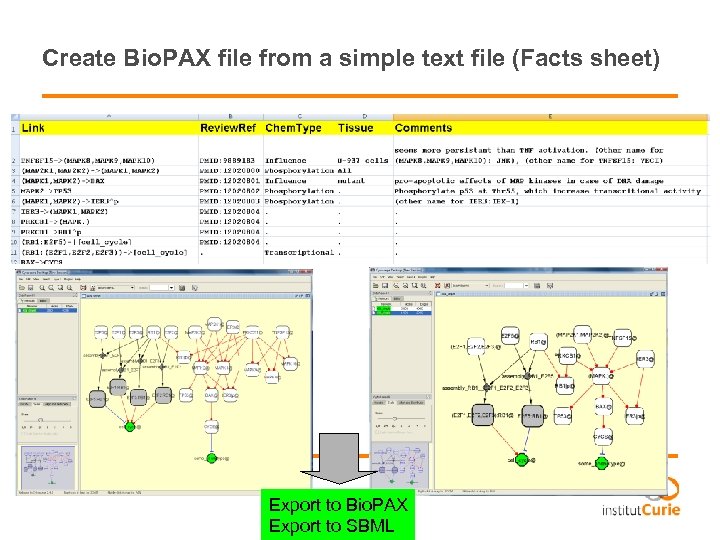
Create Bio. PAX file from a simple text file (Facts sheet) Export to Bio. PAX Export to SBML
Nearest future development 1) Bio. PAX 3. 0 support 2) Cell. Designer format 4. 0 support 3) Creating Bi. No. M plug-in for Cell. Designer 4. 0 4) New structural analysis algorithms ICSB Web-services - 28 August 2008
Install and Start Cytoscape+Bi. No. M Note: Bi. No. M is not yet compatible with the latest version of Cytoscape 2. 6. 0. These technical issues will be resolved very soon Detailed manual + Tutorial are available! Reference: Zinovyev et al, Bioinformatics 2008

Acknowledgements Eric Viara Laurence Calzone Gautier Stoll Emmanuel Barillot EU FP 6 project ESBIC-D (LSHG-CT-2005 -518192) High Council for Scientific and Technological Cooperation between France and Israel ICSB Web-services - 28 August 2008
07d23c2ca22b764082f360c206a997a5.ppt