f5f6494e5213f24d03521cc66577a09f.ppt
- Количество слайдов: 24

Bioinformatics Resources and Tools on the Web: A Primer Joel H. Graber Center for Advanced Biotechnology Boston University

Outline • Introduction: What is bioinformatics? • The basics – The five sites that all biologists should know • Some examples – Using the tools in a somewhat less-than-naïve manner • Questions/comments are welcome at all points • Much of this material comes from the Boston University course: BF 527 Bioinformatic Applications (http: //matrix. bu. edu/BF 527/)
What is bioinformatics?
Examples of Bioinformatics • Database interfaces – Genbank/EMBL/DDBJ, Medline, Swiss. Prot, PDB, … • Sequence alignment – BLAST, FASTA • Multiple sequence alignment – Clustal, Mult. Alin, Di. Align • Gene finding – Genscan, Genome. Scan, Gene. Mark, GRAIL • Protein Domain analysis and identification – pfam, BLOCKS, Pro. Dom, • Pattern Identification/Characterization – Gibbs Sampler, Align. ACE, MEME • Protein Folding prediction – Predict. Protein, Swiss. Modeler

Things to know and remember about using web server-based tools • You are using someone else’s computer • You are (probably) getting a reduced set of options or capacity • Servers are great for sporadic or proof-ofprinciple work, but for intensive work, the software should be obtained and run locally

Five websites that all biologists should know • NCBI (The National Center for Biotechnology Information; – http: //www. ncbi. nlm. nih. gov/ • EBI (The European Bioinformatics Institute) – http: //www. ebi. ac. uk/ • The Canadian Bioinformatics Resource – http: //www. cbr. nrc. ca/ • Swiss. Prot/Ex. PASy (Swiss Bioinformatics Resource) – http: //expasy. cbr. nrc. ca/sprot/ • PDB (The Protein Databank) – http: //www. rcsb. org/PDB/

NCBI (http: //www. ncbi. nlm. nih. gov/) • Entrez interface to databases – Medline/OMIM – Genbank/Genpept/Structures • BLAST server(s) – Five-plus flavors of blast • Draft Human Genome • Much, much more…

EBI (http: //www. ebi. ac. uk/) • SRS database interface – EMBL, Swiss. Prot, and many more • Many server-based tools – Clustal. W, DALI, …

Swiss. Prot (http: //expasy. cbr. nrc. ca/sprot/) • Curation!!! – Error rate in the information is greatly reduced in comparison to most other databases. • Extensive cross-linking to other data sources • Swiss. Prot is the ‘gold-standard’ by which other databases can be measured, and is the best place to start if you have a specific protein to investigate

A few more resources to be aware of • Human Genome Working Draft – http: //genome. ucsc. edu/ • TIGR (The Institute for Genomics Research) – http: //www. tigr. org/ • Celera – http: //www. celera. com/ • (Model) Organism specific information: – – – Yeast: http: //genome-www. stanford. edu/Saccharomyces/ Arabidopis: http: //www. tair. org/ Mouse: http: //www. jax. org/ Fruitfly: http: //www. fruitfly. org/ Nematode: http: //www. wormbase. org/ • Nucleic Acids Research Database Issue – http: //nar. oupjournals. org/ (First issue every year)
Example 1: Searching a new genome for a specific protein • Specific problem: We want to find the closest match in C. elegans of D. melanogaster protein NTF 1, a transcription factor • First- understanding the different forms of blast
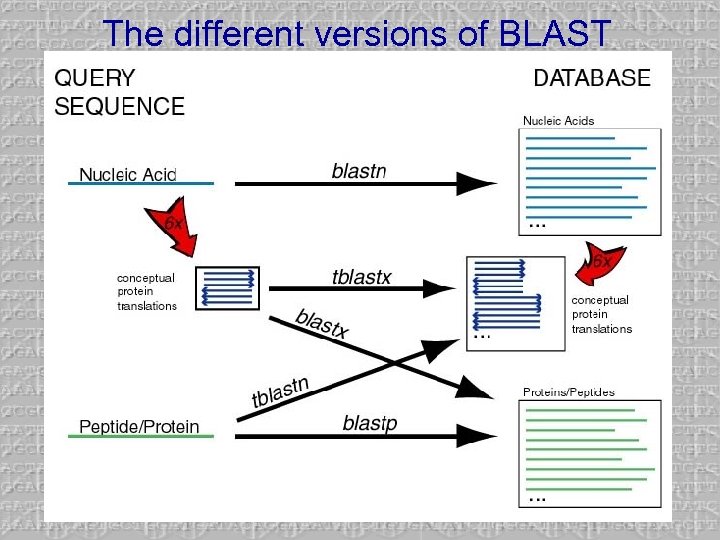
The different versions of BLAST
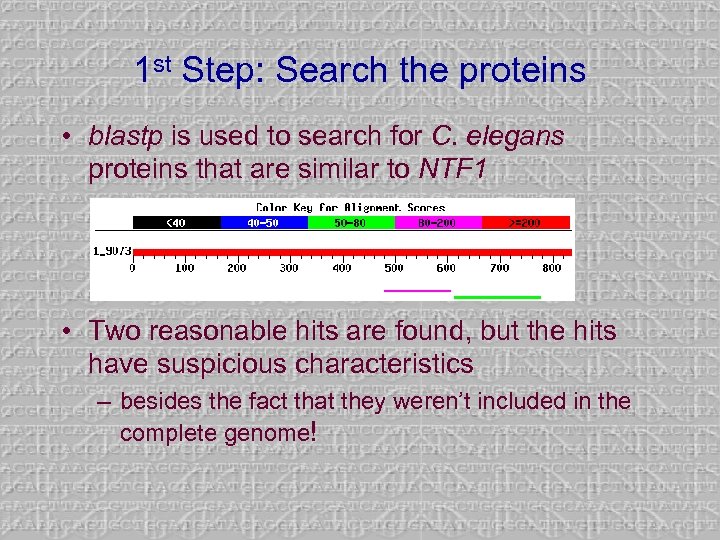
1 st Step: Search the proteins • blastp is used to search for C. elegans proteins that are similar to NTF 1 • Two reasonable hits are found, but the hits have suspicious characteristics – besides the fact that they weren’t included in the complete genome!
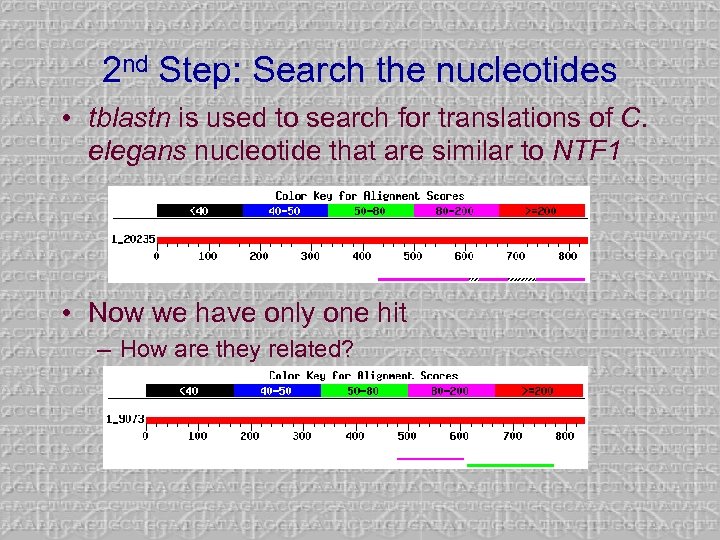
2 nd Step: Search the nucleotides • tblastn is used to search for translations of C. elegans nucleotide that are similar to NTF 1 • Now we have only one hit – How are they related?
Conclusion: Incorrect gene prediction/annotation • The two predicted proteins have essentially identical annotation • The protein-protein alignments are disjoint and consecutive on the protein • The protein-nucleotide alignment includes both protein-protein alignments in the proper order • Why/how does this happen?

Final(? ) Check: Gene prediction • Genscan is the best available ab initio gene predictor – http: //genes. mit. edu/GENSCAN. html • Genscan’s prediction spans both protein alignments, reinforcing our conclusion of a bad prediction
Ab initio vs. similarity vs. hybrid models for gene finding • Ab initio: The gene looks like the average of many genes – Genscan, Gene. Mark, GRAIL… • Similarity: The gene looks like a specific known gene – Procrustes, … • Hybrid: A combination of both – Genomescan (http: //genes. mit. edu/genomescan/)
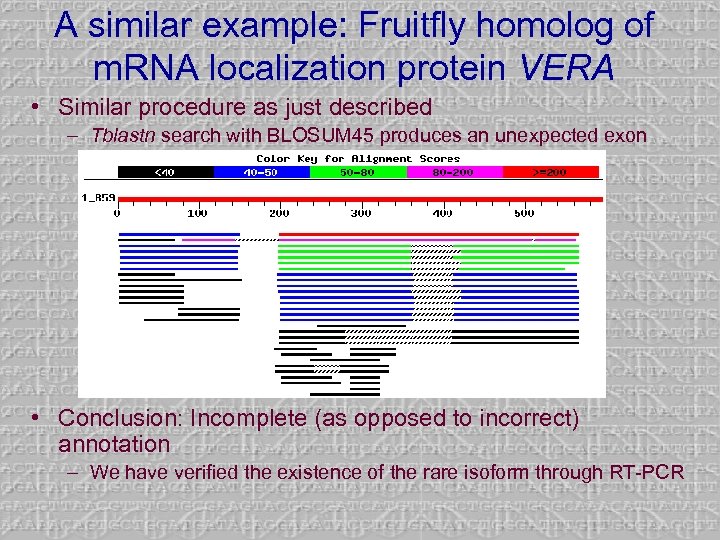
A similar example: Fruitfly homolog of m. RNA localization protein VERA • Similar procedure as just described – Tblastn search with BLOSUM 45 produces an unexpected exon • Conclusion: Incomplete (as opposed to incorrect) annotation – We have verified the existence of the rare isoform through RT-PCR

Another example: Find all genes with pdz domains • Multiple methods are possible • The ‘best’ method will depend on many things – How much do you know about the domain? – Do you know the exact extent of the domain? – How many examples do you expect to find?
Some possible methods if the domain is a known domain: • Swiss. Prot – text search capabilities – good annotation of known domains – crosslinks to other databases (domains) • Databases of known domains: – BLOCKS (http: //blocks. fhcrc. org/) – Pfam (http: //pfam. wustl. edu/) – Others (Pro. Dom, Pro. Site, DOMO, …)

Determination of the nature of conservation in a domain • For new domains, multiple alignment is your best option – Global: clustalw – Local: Di. Align – Hidden Markov Model: HMMER • For known domains, this work has largely been done for you – BLOCKS – Pfam

If you have a protein, and want to search it to known domains • Search/Analysis tools – Pfam – BLOCKS – Predict. Protein (http: //cubic. bioc. columbia. edu/predictprotein. html)
Different representations of conserved domains • BLOCKS – Gapless regions – Often multiple blocks for one domain • PFAM – Statistical model, based on HMM – Since gaps are allowed, most domains have only one pfam model

Conclusions • We have only touched small parts of the elephant • Trial and error (intelligently) is often your best tool • Keep up with the main five sites, and you’ll have a pretty good idea of what is happening and available
f5f6494e5213f24d03521cc66577a09f.ppt