f34db32556f9ff5103367fb078ebf0d2.ppt
- Количество слайдов: 32
Bioinformatics in the 90’s Ø Ø Origins : data storage needs related to the sequencing effort. . . …but storage was hardly enough : additional needs: è è è è Assembly, comparison and annotation of sequences Prediction of genes Reconstruction of evolutionary trees Modelisation & prediction of 3 D structures. . . IT : on-line databases and software tools Science : modeling, computational representations, algorithms Février 2002 – Journée PréARC « Algèbres de processus et processus biologiques »
The post-genomic phase transition 1. 2. Availability of complete genome sequences High-throughput experimental techniques yield new types of results : 1. 2. 3. 4. 5. 6. 7. 3. SNP (Single Nucleotide Polymorphisms) m. RNA expression levels (“DNA chips”) Systematic determination of 3 D structure Protein expression levels Protein- protein interactions Systematic mutagenesis. . . New needs & opportunities : è è è Processing and analysis of each type of data Integration of heterogeneous data Reconstruction and simulation of cellular mechanisms… Février 2002 – Journée PréARC « Algèbres de processus et processus biologiques »

Corporate Information Founded: December 1997 Headquarters and laboratories: Central Paris Employees: 60 as of end 2001 Intellectual Property : 57 patents on technology, interactions and targets Equity raised: c. 30 million Ownership: Advent (B), Alafi (US), Apax (F), Auriga (F), IMH(D), Health Cap (S), Lombard-Odier (CH), Medicis (D), Rendex (B) Février 2002 – Journée PréARC « Algèbres de processus et processus biologiques »

Hybrigenics’ business strategy Ø Own drug discovery programs è è Ø in the fields of infectious diseases, cancer and metabolic disorders the resulting novel validated targets being exploited for the Company’s own product pipeline Collaboration and licensing agreements with biopharmaceutical companies è è in any disease field for out-licensing Février 2002 – Journée PréARC « Algèbres de processus et processus biologiques »
Hybrigenics’ discovery programs Cancer Proteins involved in basic cellular functions Proteins involved in apoptosis Proteins involved in cell cycle regulation Metabolic disorders / Obesity Proteins involved in adipogenesis Anti-infectious diseases Antibacterial Essential proteins of the pathogens HIV, HCV : protein-protein interactions between the host cell and the pathogen Février 2002 – Journée PréARC « Algèbres de processus et processus biologiques »
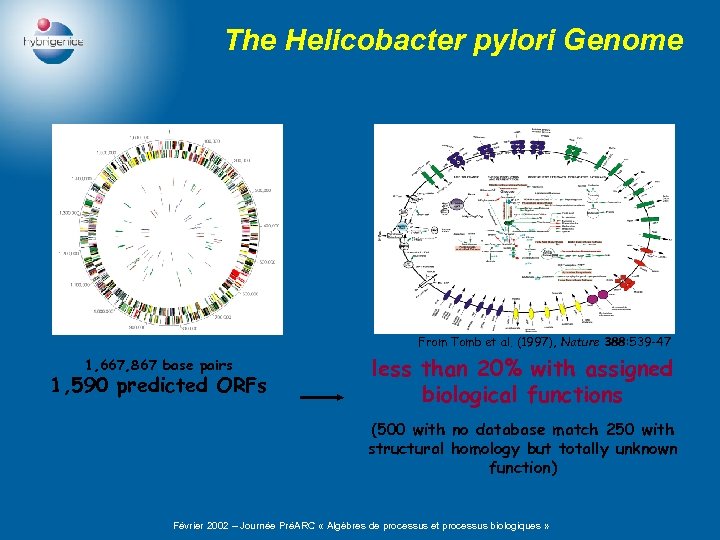
The Helicobacter pylori Genome From Tomb et al. (1997), Nature 388: 539 -47 1, 667, 867 base pairs 1, 590 predicted ORFs less than 20% with assigned biological functions (500 with no database match 250 with structural homology but totally unknown function) Février 2002 – Journée PréARC « Algèbres de processus et processus biologiques »
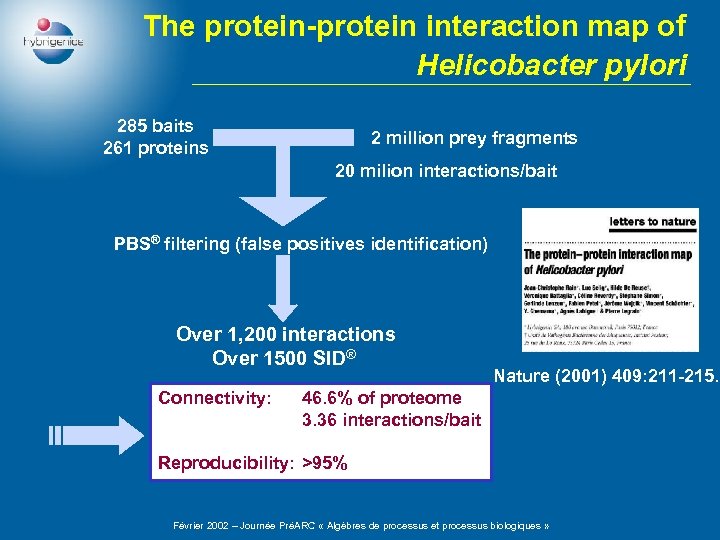
The protein-protein interaction map of Helicobacter pylori 285 baits 261 proteins 2 million prey fragments 20 milion interactions/bait PBS® filtering (false positives identification) Over 1, 200 interactions Over 1500 SID® Connectivity: Nature (2001) 409: 211 -215. 46. 6% of proteome 3. 36 interactions/bait Reproducibility: >95% Février 2002 – Journée PréARC « Algèbres de processus et processus biologiques »
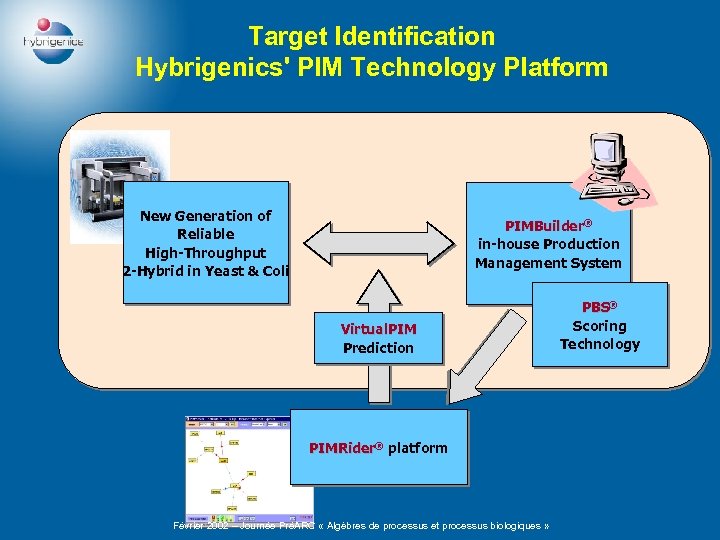
Target Identification Hybrigenics' PIM Technology Platform New Generation of Reliable High-Throughput 2 -Hybrid in Yeast & Coli PIMBuilder® in-house Production Management System Virtual. PIM Prediction PIMRider® platform Février 2002 – Journée PréARC « Algèbres de processus et processus biologiques » PBS® Scoring Technology
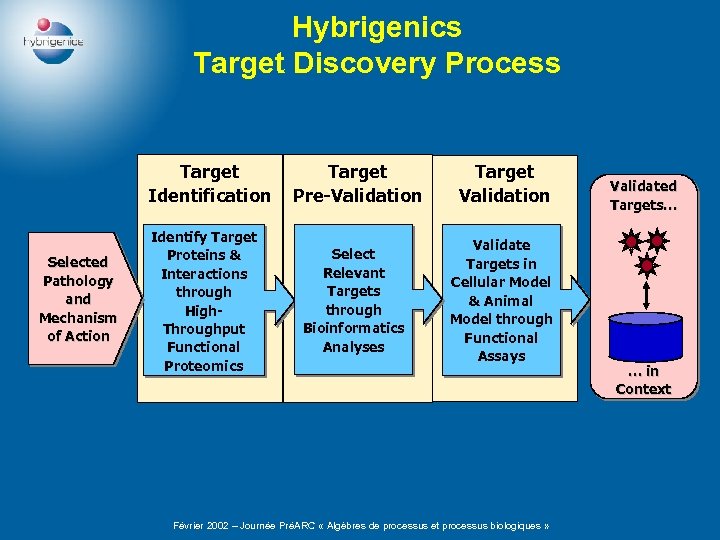
Hybrigenics Target Discovery Process Target Identification Selected Pathology and Mechanism of Action Target Pre-Validation Target Validation Identify Target Proteins & Interactions through High. Throughput Functional Proteomics Select Relevant Targets through Bioinformatics Analyses Validate Targets in Cellular Model & Animal Model through Functional Assays Février 2002 – Journée PréARC « Algèbres de processus et processus biologiques » Validated Targets… … in Context
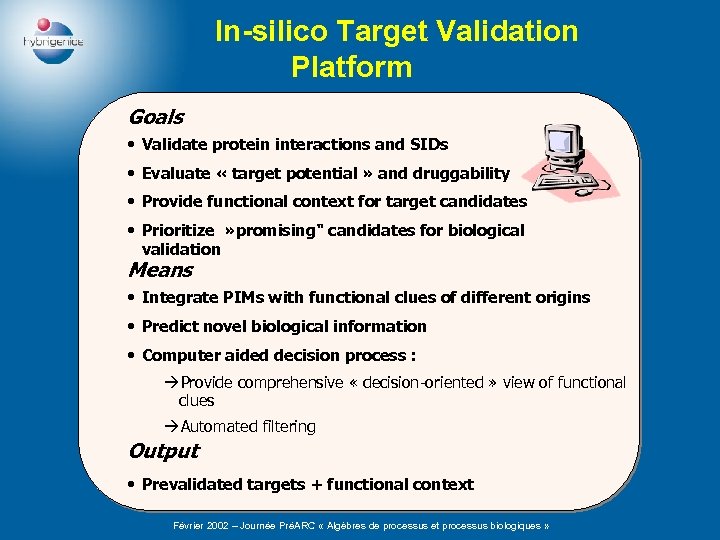
In-silico Target Validation Platform Goals • Validate protein interactions and SIDs • Evaluate « target potential » and druggability • Provide functional context for target candidates • Prioritize » promising" candidates for biological validation Means • Integrate PIMs with functional clues of different origins • Predict novel biological information • Computer aided decision process : àProvide comprehensive « decision-oriented » view of functional clues àAutomated filtering Output • Prevalidated targets + functional context Février 2002 – Journée PréARC « Algèbres de processus et processus biologiques »
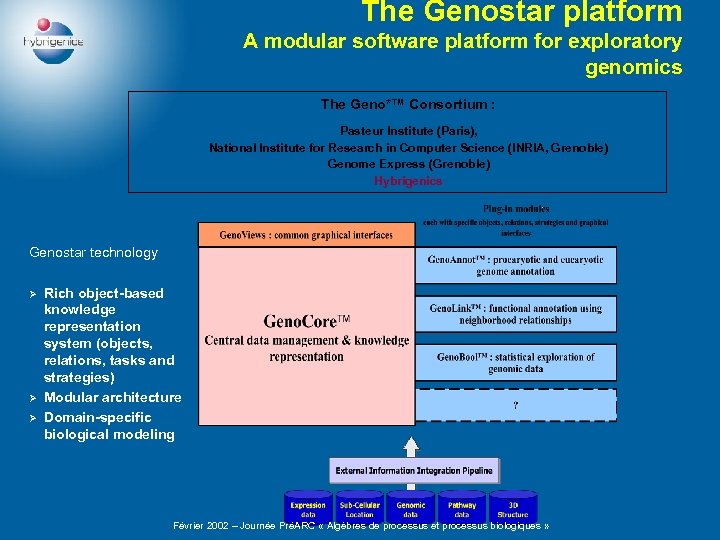
The Genostar platform A modular software platform for exploratory genomics The Geno*™ Consortium : Pasteur Institute (Paris), National Institute for Research in Computer Science (INRIA, Grenoble) Genome Express (Grenoble) Hybrigenics Genostar technology Ø Ø Ø Rich object-based knowledge representation system (objects, relations, tasks and strategies) Modular architecture Domain-specific biological modeling Février 2002 – Journée PréARC « Algèbres de processus et processus biologiques »
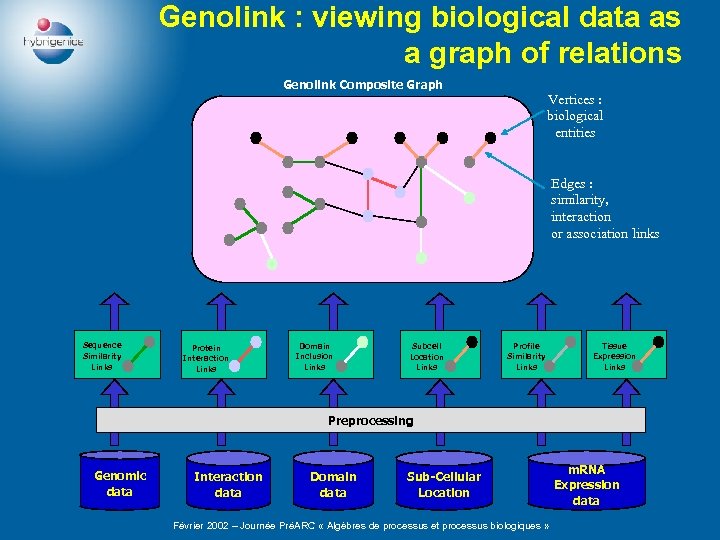
Genolink : viewing biological data as a graph of relations Genolink Composite Graph Vertices : biological entities Edges : similarity, interaction or association links Sequence Similarity Links Protein Interaction Links Domain Inclusion Links Subcell Location Links Profile Similarity Links Tissue Expression Links Preprocessing Genomic data Interaction data Domain data Sub-Cellular Location Février 2002 – Journée PréARC « Algèbres de processus et processus biologiques » m. RNA Expression data
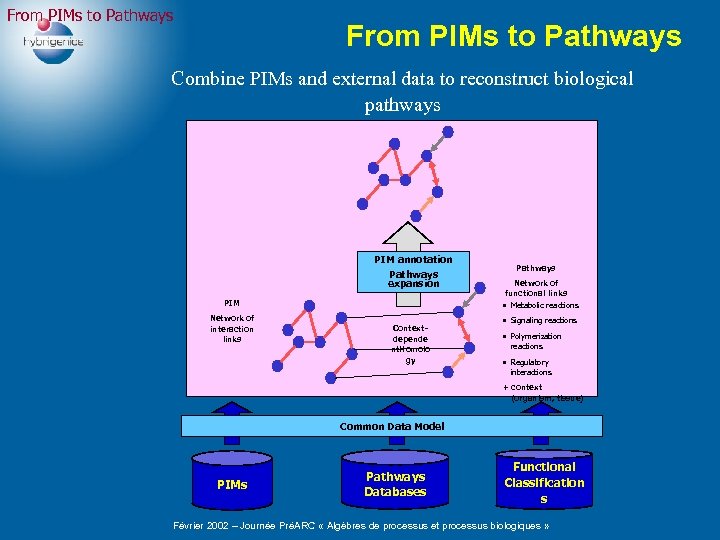
From PIMs to Pathways Combine PIMs and external data to reconstruct biological pathways PIM annotation Pathways expansion PIM Network of interaction links Contextdepende nt. Homolo gy Pathways Network of functional links • Metabolic reactions • Signaling reactions • Polymerization reactions • Regulatory interactions + context (organism, tissue) Common Data Model PIMs Pathways Databases Functional Classification s Février 2002 – Journée PréARC « Algèbres de processus et processus biologiques »

The Bio. Pathways Consortium Ø Mission : è Ø Goals : è Ø è è è Ø Forum open to interested participants (academics, pharmas, biotechs, software vendors) Achievements : è Ø Scientific community buildup, standards recommendation, public outreach, industryacademia collaboration support, coordination with other groups Means : è Ø Foster development of pathways informatics & systems biology Launched June 2000 by 3 rd Millennium (Boston) and Hybrigenics (Paris) 1 st Meeting at ISMB 2000 -> Work Groups 2 nd Meeting at PSB 2001 -> First results on evaluation of pathways representations 3 rd Meeting – Satellite Meeting of ISMB 2001, Copenhagen -> Focus on ontologies and pathways reconstruction (>150 attendants), new workgroups Several sponsors (pharmas, biotechs, IT companies) Over 200 participants from academia & industry Février 2002 – Journée PréARC « Algèbres de processus et processus biologiques »

Annotation fonctionnelle Ø Ø Objectif : assigner une/des “fonction(s)” à un gène ou à une protéine de séquence connue Méthodes traditionnelles: è è Ø Résultats expérimentaux Variations sur le thème : propagation d’annotations d’origine expérimentale via similitude de séquences Fonction ? è è è Locale et précise (Ex : la protéine P est un enzyme catalysant la réaction R) Globale et vague : appartenance à un processus biologique de haut niveau (Ex : P intervient dans la dégradation du glucose) Ce qui est propagé : mots clefs, nœud d’un arbre de classification fonctionnelle Février 2002 – Journée PréARC « Algèbres de processus et processus biologiques »
An effort toward consensus : Gene Ontology The Gene Ontology Consortium (2000) Nature Genet. 25: 25 -29 Février 2002 – Journée PréARC « Algèbres de processus et processus biologiques »
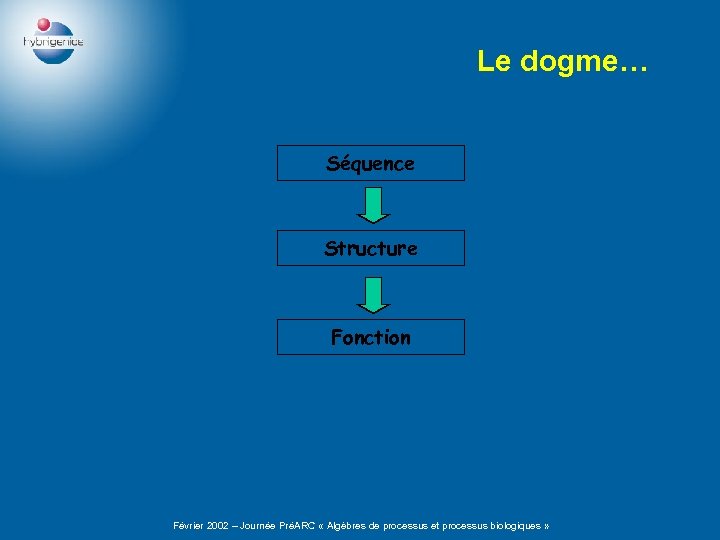
Le dogme… Séquence Structure Fonction Février 2002 – Journée PréARC « Algèbres de processus et processus biologiques »
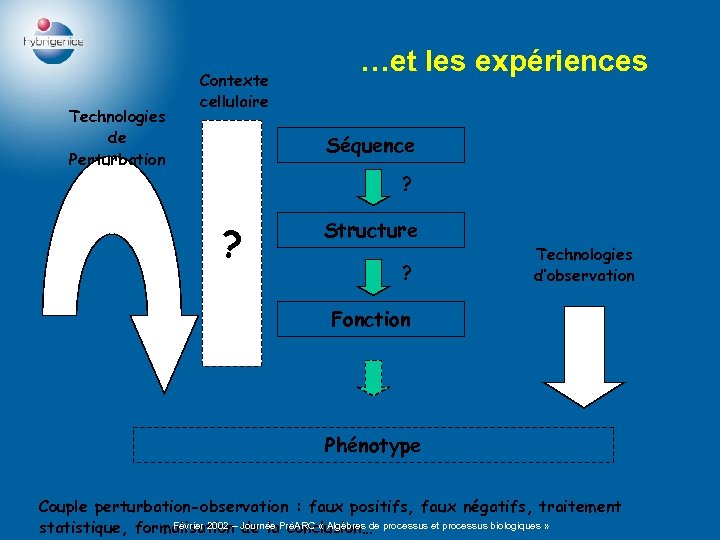
Technologies de Perturbation Contexte cellulaire …et les expériences Séquence ? ? Structure ? Technologies d’observation Fonction Phénotype Couple perturbation-observation : faux positifs, faux négatifs, traitement Février 2002 – Journée PréARC « Algèbres de processus et processus biologiques » statistique, formalisation de la conclusion…
Integration of heterogeneous data Ø Joint use of functional clues from a variety of experimental approaches to : è è Ø Ø Validate the biological relevance of interactions Determine the function of proteins Validate targets in-silico Examples : • Interaction + expression • Interaction + 3 D structure • Location + expression • Phylogenetic profiles + domain fusion Recent problem, drug discovery efforts bottleneck Frontier for the bioinformatics community Technology : normalization, formats, ontologies Science : automate (some) biological reasoning ? Février 2002 – Journée PréARC « Algèbres de processus et processus biologiques »

Evaluating pathways representations Vincent Schächter, Hybrigenics, Paris Aviv Regev, Tel-Aviv University Bio. Pathways Formalisms Workgroup Février 2002 – Journée PréARC « Algèbres de processus et processus biologiques »
Evaluation scope : untangling the web. . . Ø Large body of literature, focusing on different biological phenomena and different theoretical issues Ø A typical article on pathways may include one or more of the following : è è A data-model, describing (a fraction of) the pathway “universe of discourse” A formalism, used to describe the data-model and to express algorithms / functions Description of algorithms based on characteristics of both the formalism and the data model Description of implementations of data-storage functionalities and/or of some of the above algorithms Février 2002 – Journée PréARC « Algèbres de processus et processus biologiques »

Excerpt from target evaluation list : “non DE” formalisms è è è è Petri nets (basic, hybrid, self-modifying, time-dependent, hierarchical, mobile) Process algebra (basic and stochastic pi-calculus) Markup languages (Cell. ML and SBML) Biocalculus Regulatory grammars (Collado-Vides) Semiotes (Kazic) Statecharts (Kam, Holcombe) Boolean networks (basic, multi-level) Hierarchical networks (Bodnar) Neural networks (Mjolsness) Molecular graph reaction networks (Mc. Caskill) Molecular interaction maps (Kohn) Electrical circuits (Keane) Février 2002 – Journée PréARC « Algèbres de processus et processus biologiques »

Quelques exemples de représentations “discrètes” Ø Modèles orienté-objet : è è Ø Réseaux booléens : è è Ø Simulation qualitative, reconstruction à partir de données d ’expression Appliqué aux « réseaux de régulation » Réseaux de Petri : è è è Ø Requêtes sur tous types de réseaux Reconstruction, mais problème de l ’information incomplète Simulation qualitative plus fine, analyse formelle du comportement Appliqué aux réseaux de régulation Application possible aux réseaux métaboliques et signalisation avec extensions (selfmodifying PN, Hybrid PN…) Algèbres de processus : è è Simulation, analyse formelle, reconstruction Appliqué aux réseaux de signalisation et de régulation (métabolisme avec extension stochastiques Février 2002 – Journée PréARC « Algèbres de processus et processus biologiques »
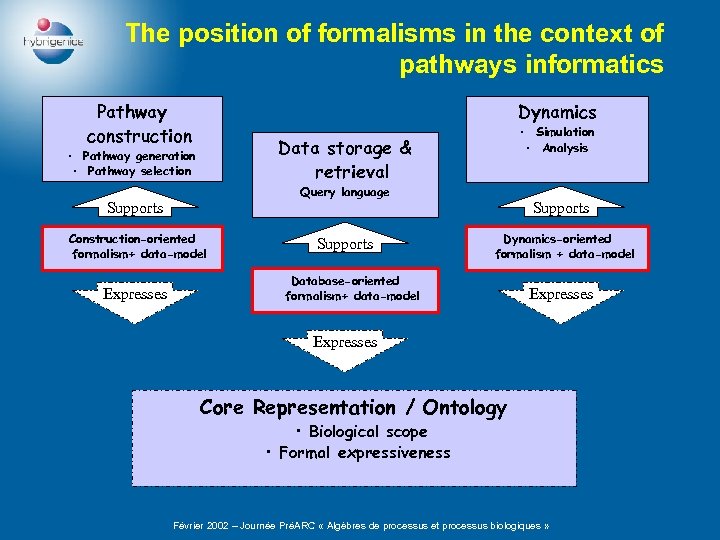
The position of formalisms in the context of pathways informatics Pathway construction Dynamics Data storage & retrieval • Pathway generation • Pathway selection Query language Supports Construction-oriented formalism+ data-model Expresses • Simulation • Analysis Supports Dynamics-oriented formalism + data-model Database-oriented formalism+ data-model Expresses Core Representation / Ontology • Biological scope • Formal expressiveness Février 2002 – Journée PréARC « Algèbres de processus et processus biologiques »
Evaluate and compare : a modular approach Ø Ø Ø Evaluate expressiveness/ease of use of representation relatively to specific goals/functionalities Compare representations in the categories for which they were designed Reduce each category to a set of evaluation items that can be rated and compared as objectively as possible Février 2002 – Journée PréARC « Algèbres de processus et processus biologiques »

Core representation / “Ontology” Ø Ø Conceptual structure of the universe of discourse (abstract and concrete entities, relations, hierarchies. . . ) Constrains scope of phenomena that can be described, and thus queried, analyzed, reconstructed, and queried. Often implicit in a given pathway representation : need to extract. . . Possible evaluation schemes : 1. Compare “features” of ontology 2. Expressiveness benchmark : set of biological “situations” 3. Translation of data models into common formalisms + comparison How do you represent : “gene A inhibits gene B” in your data model ?
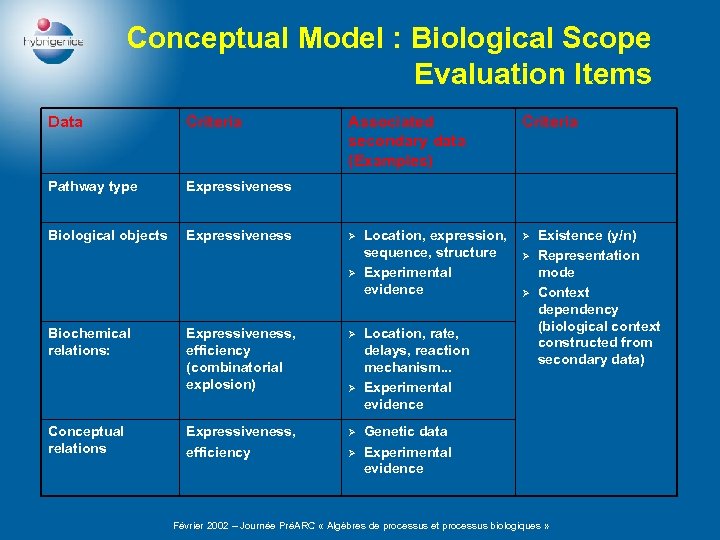
Conceptual Model : Biological Scope Evaluation Items Data Criteria Pathway type Expressiveness Biological objects Expressiveness Associated secondary data (Examples) Ø Ø Biochemical relations: Conceptual relations Expressiveness, efficiency (combinatorial explosion) Ø Expressiveness, efficiency Ø Ø Ø Location, expression, sequence, structure Experimental evidence Location, rate, delays, reaction mechanism. . . Experimental evidence Criteria Ø Ø Ø Existence (y/n) Representation mode Context dependency (biological context constructed from secondary data) Genetic data Experimental evidence Février 2002 – Journée PréARC « Algèbres de processus et processus biologiques »
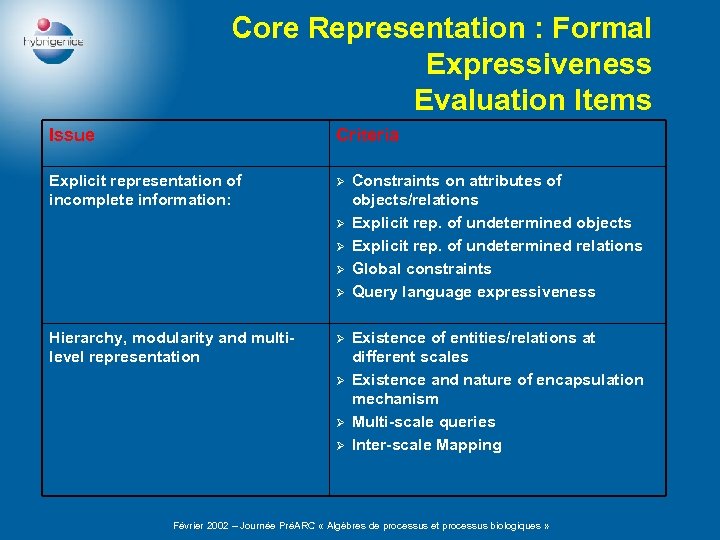
Core Representation : Formal Expressiveness Evaluation Items Issue Criteria Explicit representation of incomplete information: Ø Ø Ø Hierarchy, modularity and multilevel representation Ø Ø Constraints on attributes of objects/relations Explicit rep. of undetermined objects Explicit rep. of undetermined relations Global constraints Query language expressiveness Existence of entities/relations at different scales Existence and nature of encapsulation mechanism Multi-scale queries Inter-scale Mapping Février 2002 – Journée PréARC « Algèbres de processus et processus biologiques »
Data Storage and Retrieval Storage and retrieval of data : “database-related” functionalities Ø Extremes : relational or OO models vs, e. g. , most simulationoriented formalisms. . . Ø A data-retrieval oriented formalism can be used « below » other formalisms Ø Query language : è è Retrieve information within a structured, homogeneous, compositional framework Shifting boundary with analysis and reconstruction algorithms Evaluation items / sub-categories · Robust database : implementation issue · Query language ease of use · Query language expressiveness · Limited by formalism and ontology expressiveness Février 2002 – Journée PréARC « Algèbres de processus et processus biologiques »

Pathway reconstruction Construction/prediction of pathways in given biological environment (organism, tissue, condition, location…) from a combination of : · experimental data · fully instantiated pathway information, · partially instantiated (or “incomplete”) pathway data, such as interaction data Special cases : reverse engineering, pathway inference Evaluation items / sub-categories : • Input data types • Pathway generation algorithm • Pathway selection algorithm : • Pathway “fitness” function • Pathway similarity/homology measure • Interactive validation ? Février 2002 – Journée PréARC « Algèbres de processus et processus biologiques »
Dynamics Study of network dynamics (regulatory networks, ST, MP) : Ø Simulation runs Ø Analysis of dynamic behavior Evaluation items / sub-categories : Ø Ø Ø States : nature, expressiveness, level of detail vs available data Evolution rules / Reaction model : rule, implementation Time : continuous/discrete, synchronous/asynchronous updates Space : continuous/discrete, topology, resolution Analysis : è è Scope : state reachability, liveness of transitions, substance flow. . . Formal methods available Comparative power Limited to steady state ? Février 2002 – Journée PréARC « Algèbres de processus et processus biologiques »
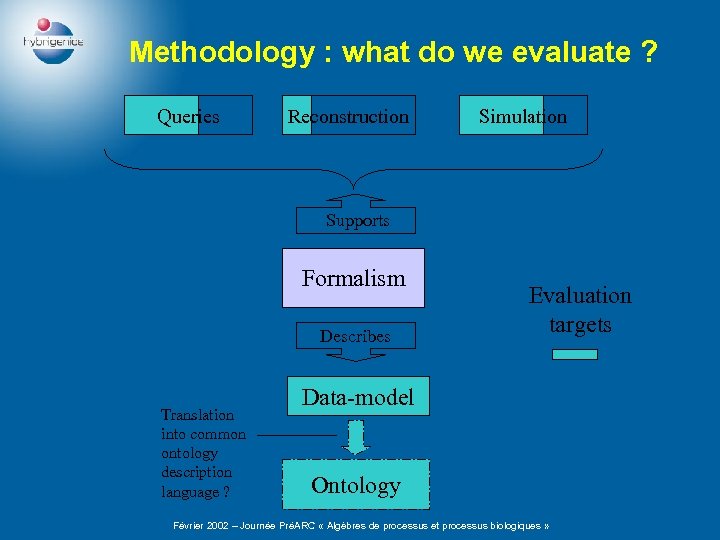
Methodology : what do we evaluate ? Queries Reconstruction Simulation Supports Formalism Describes Translation into common ontology description language ? Evaluation targets Data-model Ontology Février 2002 – Journée PréARC « Algèbres de processus et processus biologiques »
f34db32556f9ff5103367fb078ebf0d2.ppt