b42a24f86c319284ea3464da36d13017.ppt
- Количество слайдов: 30
Bioinformatics B 90901099 劉兆昕
Outline • • • What is Bioinformatics? Applications of Bioinformatics DNA Databases Protein Databases FASTA BLAST 2
What is Bioinformatics? • Computational biology. • This word has not a clear definition. • It involves the analysis and interpretation of data and the development of algorithms and statistics. • The term was coined to encompass computer applications in biological sciences but is now used to mean rather different things, from artificial intelligence and robotics to genome analysis. • The term was originally applied to the computational manipulation and analysis of biological sequence data (DNA and/or protein), but now tends also to be used to embrace the manipulation and analysis of 3 D structural data. 3
What is Bioinformatics? • Bioinformatics is the application of computational techniques to the management and analysis of biological information. 4
Applications of Bioinformatics • • Sequencing of the Human Genome Annotation of the Genome Protein Interactions Find Proteins that are Better Suited for Treating with Drugs • Development of New Drugs • Optimization of Known Therapies on Individuals 5

Sequencing of the Human Genome 6
DNA Databases • Gen. Bank (USA) • EMBL (European Molecular Biology Laboratory) • The DNA Database of Japan 7

Gen. Bank 8
EMBL 9

DDBJ 10

A Sample DNA Database Entry ID TRBG 361 standard; m. RNA; PLN; 1859 BP. XX AC X 56734; S 46826; XX SV X 56734. 1 XX DT 12 -SEP-1991 (Rel. 29, Created) DT 15 -MAR-1999 (Rel. 59, Last updated, Version 9) XX DE Trifolium repens m. RNA for non-cyanogenic beta-glucosidase XX KW beta-glucosidase. XX OS Trifolium repens (white clover) OC Eukaryota; Viridiplantae; Streptophyta; Embryophyta; Tracheophyta; OC Spermatophyta; Magnoliophyta; eudicotyledons; core eudicots; rosids; OC eurosids I; Fabales; Fabaceae; Papilionoideae; Trifolium. XX RN [5] RP 1 -1859 RX MEDLINE; 91322517. RX PUBMED; 1907511. RA Oxtoby E. , Dunn M. A. , Pancoro A. , Hughes M. A. ; RT "Nucleotide and derived amino acid sequence of the cyanogenic RT beta-glucosidase (linamarase) from white clover (Trifolium repens L. ). "; RL Plant Mol. Biol. 17(2): 209 -219(1991). XX RN [6] RP 1 -1859 RA Hughes M. A. ; RT ; RL Submitted (19 -NOV-1990) to the EMBL/Gen. Bank/DDBJ databases. RL M. A. Hughes, UNIVERSITY OF NEWCASTLE UPON TYNE, MEDICAL SCHOOL, NEW CASTLE RL UPON TYNE, NE 2 4 HH, UK XX FH Key Location/Qualifiers FH FT source 1. . 1859 FT /db_xref="taxon: 3899" FT /mol_type="m. RNA" FT /organism="Trifolium repens" FT /tissue_type="leaves" FT /clone_lib="lambda gt 10" FT /clone="TRE 361" FT CDS 14. . 1495 FT /db_xref="GOA: P 26204" FT /db_xref="HSSP: P 26205" FT /db_xref="Uni. Prot/Swiss-Prot: P 26204" FT /note="non-cyanogenic" FT /EC_number="3. 2. 1. 21" FT /product="beta-glucosidase" FT /protein_id="CAA 40058. 1" FT /translation="MDFIVAIFALFVISSFTITSTNAVEASTLLDIGNLSRSSFPRGFI FT FGAGSSAYQFEGAVNEGGRGPSIWDTFTHKYPEKIRDGSNADITVDQYHRYKEDVGIMK FT DQNMDSYRFSISWPRILPKGKLSGGINHEGIKYYNNLINELLANGIQPFVTLFHWDLPQ FT VLEDEYGGFLNSGVINDFRDYTDLCFKEFGDRVRYWSTLNEPWVFSNSGYALGTNAPGR FT CSASNVAKPGDSGTGPYIVTHNQILAHAEAVHVYKTKYQAYQKGKIGITLVSNWLMPLD FT DNSIPDIKAAERSLDFQFGLFMEQLTTGDYSKSMRRIVKNRLPKFSKFESSLVNGSFDF FT IGINYYSSSYISNAPSHGNAKPSYSTNPMTNISFEKHGIPLGPRAASIWIYVYPYMFIQ FT EDFEIFCYILKINITILQFSITENGMNEFNDATLPVEEALLNTYRIDYYYRHLYYIRSA FT IRAGSNVKGFYAWSFLDCNEWFAGFTVRFGLNFVD" FT m. RNA 1. . 1859 FT /evidence=EXPERIMENTAL XX SQ Sequence 1859 BP; 609 A; 314 C; 355 G; 581 T; 0 other; aaacca aatatggatt ttattgtagc catatttgct ctgtta ttagctcatt 60 cacaattact tccacaaatg cagttgaagc ttctactctt cttgacatag gtaacctgag 120 tcggagcagt tttcctcgtg gcttcatctt tggtgctgga tcttcagcat accaatttga 180 aggtgcagta aacgaaggcg gtagaggacc aagtatttgg gataccttca cccataaata 240 tccagaaaaa ataagggatg gaagcaatgc agacatcacg gttgaccaat atcaccgcta 300 caaggaagat gttgggatta tgaaggatca aaatatggat tcgtatagat tctcaatctc 360 ttggccaaga atactcccaa agggaaagtt gagcggaggc ataaatcacg aaggaatcaa 420 atattacaac aaccttatca acgaactatt ggctaacggt atacaaccat ttgtaactct 480 ttttcattgg gatcttcccc aagtcttaga agatgagtat ggtggtttct taaactccgg 540 tgtaataaat gattttcgag actatacgga tctttgcttc aaggaatttg gagatagagt 600 gaggtattgg agtactctaa atgagccatg ggtgtttagc aattctggat atgcactagg 660 aacaaatgca ccaggtcgat gttcggcctc caacgtggcc aagcctggtg attctggaac 720 aggaccttat atagttacac acaatcaaat tcttgctcat gcagaagctg tacatgtgta 780 taagactaaa taccaggcat atcaaaaggg aaagataggc ataacgttgg tatctaactg 840 gttaatgcca cttgatgata atagcatacc agatataaag gctgccgaga gatcacttga 900 cttccaattt ggattgttta tggaacaatt aacaacagga gattattcta agagcatgcg 960 gcgtatagtt aaaaaccgat tacctaagtt ctcaaaattc gaatcaagcc tagtgaatgg 1020 ttcatttgat tttattggta taaactatta ctcttctagt tatattagca atgccccttc 1080 acatggcaat gccaaaccca gttactcaac aaatcctatg accaatattt catttgaaaa 1140 acatgggata cccttaggtc caagggctgc ttcaatttgg atatatgttt atccatatat 1200 gtttatccaa gaggacttcg agatcttttg ttacatatta aaaata taacaatcct 1260 gcaattttca atcactgaaa atggtatgaattcaac gatgcaacac ttccagtaga 1320 agaagctctt ttgaatactt acagaattga ttactattac cgtcacttat actacattcg 1380 ttctgcaatc agggct caaatgtgaa gggtttttac gcatggtcat ttttggactg 1440 taatgg tttgcaggct ttactgttcg ttttggatta aactttgtag attagaaaga 1500 tggattaaaa aggtacccta agctttctgc ccaatggtac aagaactttc tcaaaagaaa 1560 ctagta ttattaaaag aactttgtag tagattacag tacatcgttt gaagttgagt 1620 tggtgcacct aattaaataa aagaggttac tcttaacata tttttaggcc attcgttgtg 1680 aagttgttag gctgttattt ctattatact atgttgtagt aataagtgca ttgttgtacc 1740 agaagctatg atcataacta taggttgatc cttcatgtat cagtttgatg ttgagaatac 1800 tttgaattaa aagtcttttt ttattttttt aaaaaaaaaa 1859 // 11

The Human Genome Project • Begun 1990 • Coordinated by the U. S. Department of Energy and the National Institutes of Health • Originally expected completion in 2005 (15 yrs), already completed 2003 (13 yrs). 12

The Human Genome Project 13

The Human Genome Project • Project goals were to – identify all the approximately 20, 000 -25, 000 genes in human DNA, – determine the sequences of the 3 billion chemical base pairs that make up human DNA, – store this information in databases, – improve tools for data analysis, – transfer related technologies to the private sector, and – address the ethical, legal, and social issues (ELSI) that may arise from the project 14
Protein Interactions 15
Protein Databases • PIR (Protein Information Resource) • Swiss-Prot • Uni. Prot (Universal Protein Resource) 16

PIR 17
Swiss-Prot 18
Uni. Prot 19
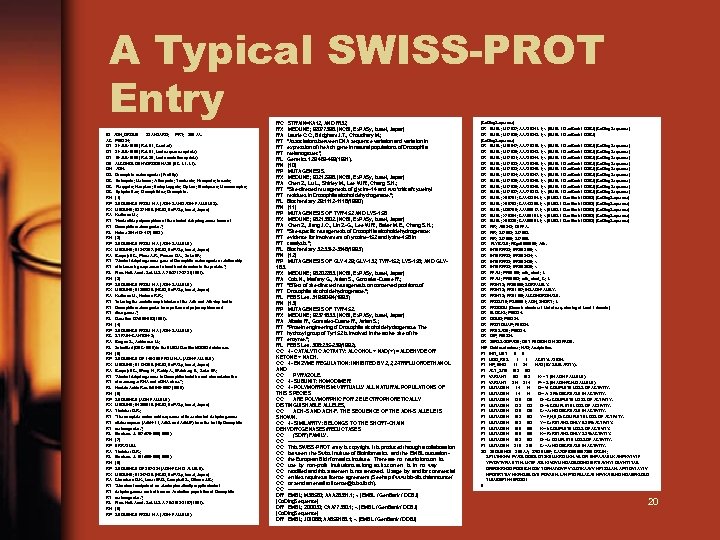
A Typical SWISS-PROT Entry ID AC DT DT DT DE GN OS OC OC OC RN RP RX RA RT RT RL RN RP RX RA RT RT RT RL RN RP RC RA RL RN RP RX RA RT RT RT RL RN RP ADH_DROME STANDARD; PRT; 255 AA. P 00334; 21 -JUL-1986 ( Rel. 01, Created) 21 -JUL-1986 ( Rel. 01, Last sequence update) 15 -JUL-1998 ( Rel. 36, Last annotation update) ALCOHOL DEHYDROGENASE (EC 1. 1). ADH. Drosophila melanogaster (Fruit fly). Eukaryota; Metazoa; Arthropoda; Tracheata; Hexapoda; Insecta; Pterygota; Neoptera; Endopterygota; Diptera; Brachycera; Muscomorpha; Ephydroidea; Drosophilidae; Drosophila. [1] SEQUENCE FROM N. A. (ADH-S AND ADH-F ALLELES). MEDLINE; 83271489. [NCBI, Ex. PASy, Israel, Japan] Kreitman M. ; "Nucleotide polymorphism at the alcohol dehydrogenase locus of Drosophila melanogaster. "; Nature 304: 412 -417(1983). [2] SEQUENCE FROM N. A. (ADH-S ALLELE). MEDLINE; 81247357. [NCBI, Ex. PASy, Israel, Japan] Benyajati C. , Place A. R. , Powers D. A. , Sofer W. ; "Alcohol dehydrogenase gene of Drosophila melanogaster: relationship of intervening sequences to functional domains in the protein. "; Proc. Natl. Acad. Sci. U. S. A. 78: 2717 -2721(1981). [3] SEQUENCE FROM N. A. (ADH-S ALLELE). MEDLINE; 91200630. [NCBI, Ex. PASy, Israel, Japan] Kreitman M. , Hudson R. R. ; "Inferring the evolutionary histories of the Adh and Adh-dup loci in Drosophila melanogaster from patterns of polymorphism and divergence. "; Genetics 127: 565 -582(1991). [4] SEQUENCE FROM N. A. (ADH-S ALLELE). STRAIN=CANTON-S; Brogna S. , Ashburner M. ; Submitted (DEC-1996) to the EMBL/ Gen. Bank/DDBJ databases. [5] SEQUENCE OF 140 -255 FROM N. A. (ADH-F ALLELE). MEDLINE; 81124290. [NCBI, Ex. PASy, Israel, Japan] Benyajati C. , Wang N. , Reddy A. , Weinberg E. , Sofer W. ; "Alcohol dehydrogenase in Drosophila: isolation and characterization of messenger RNA and c. DNA clone. "; Nucleic Acids Res. 8: 5649 -5667(1980). [6] SEQUENCE (ADH-F ALLELE). MEDLINE; 84256516. [NCBI, Ex. PASy, Israel, Japan] Thatcher D. R. ; "The complete amino acid sequence of three alcohol dehydrogenase alleloenzymes (Adh. N-11, Adh. S and Adh. UF) from the fruitfly Drosophila melanogaster. "; Biochem. J. 187: 875 -886(1980). [7] ERRATUM. Thatcher D. R. ; Biochem. J. 191: 895 -895(1980). [8] SEQUENCE OF 207 -224 (ADH-FCH. D. ALLELE). MEDLINE; 81247428. [NCBI, Ex. PASy, Israel, Japan] Chambers G. K. , Laver W. G. , Campbell S. , Gibson J. B. ; "Structural analysis of an electrophoretically cryptic alcohol dehydrogenase variant from an Australian population of Drosophila melanogaster. "; Proc. Natl. Acad. Sci. U. S. A. 78: 3103 -3107(1981). [9] SEQUENCE FROM N. A. (ADH-F ALLELE). RC STRAIN=KA 12, AND RI 32; RX MEDLINE; 92077396. [NCBI, Ex. PASy, Israel, Japan] RA Laurie C. C. , Bridgham J. T. , Choudhary M. ; RT "Associations between DNA sequence variation and variation in RT expression of the Adh gene in natural populations of Drosophila RT melanogaster. "; RL Genetics 129: 489 -499(1991). RN [10] RP MUTAGENESIS. RX MEDLINE; 90212596. [NCBI, Ex. PASy, Israel, Japan] RA Chen Z. , Lu L. , Shirley M. , Lee W. R. , Chang S. H. ; RT "Site-directed mutagenesis of glycine-14 and two 'critical' cysteinyl RT residues in Drosophila alcohol dehydrogenase. "; RL Biochemistry 29: 1112 -1118(1990). RN [11] RP MUTAGENESIS OF TYR-152 AND LYS-156. RX MEDLINE; 93213802. [NCBI, Ex. PASy, Israel, Japan] RA Chen Z. , Jiang J. C. , Lin Z. -G. , Lee W. R. , Baker M. E. , Chang S. H. ; RT "Site-specific mutagenesis of Drosophila alcohol dehydrogenase: RT evidence for involvement of tyrosine-152 and lysine-156 in RT catalysis. "; RL Biochemistry 32: 3342 -3346(1993). RN [12] RP MUTAGENESIS OF GLY-129; GLY-132; TYR-152; LYS-156; AND GLY 183. RX MEDLINE; 93202283. [NCBI, Ex. PASy, Israel, Japan] RA Cols N. , Marfany G. , Atrian S. , Gonzalez-Duarte R. ; RT "Effect of site-directed mutagenesis on conserved positions of RT Drosophila alcohol dehydrogenase. "; RL FEBS Lett. 319: 90 -94(1993). RN [13] RP MUTAGENESIS OF TYR-152. RX MEDLINE; 92371633. [NCBI, Ex. PASy, Israel, Japan] RA Albalat R. , Gonzalez-Duarte R. , Atrian S. ; RT "Protein engineering of Drosophila alcohol dehydrogenase. The RT hydroxyl group of Tyr 152 is involved in the active site of the RT enzyme. "; RL FEBS Lett. 308: 235 -239(1992). CC -!- CATALYTIC ACTIVITY: ALCOHOL + NAD(+) = ALDEHYDE OR KETONE + NADH. CC -!- ENZYME REGULATION: INHIBITED BY 2, 2, 2 -TRIFLUOROETHANOL AND CC PYRAZOLE. CC -!- SUBUNIT: HOMODIMER. CC -!- POLYMORPHISM: VIRTUALLY ALL NATURAL POPULATIONS OF THIS SPECIES CC ARE POLYMORPHIC FOR 2 ELECTROPHORETICALLY DISTINGUISHABLE ALLELES, CC ADH-S AND ADH-F. THE SEQUENCE OF THE ADH-S ALLELE IS SHOWN. CC -!- SIMILARITY: BELONGS TO THE SHORT-CHAIN DEHYDROGENASES/REDUCTASES CC (SDR) FAMILY. CC -------------------------------------CC This SWISS-PROT entry is copyright. It is produced through a collaboration CC between the Swiss Institute of Bioinformatics and the EMBL outstation CC the European Bioinformatics Institute. There are no restrictions on its CC use by non-profit institutions as long as its content is in no way CC modified and this statement is not removed. Usage by and for commercial CC entities requires a license agreement (See http: //www. isb-sib. ch/announce/ CC or send an email to license@isb-sib. ch). CC -------------------------------------DR EMBL; M 36580; AAA 28331. 1; -. [EMBL / Gen. Bank / DDBJ] [Co. Ding. Sequence] DR EMBL; Z 00030; CAA 77330. 1; -. [EMBL / Gen. Bank / DDBJ] [Co. Ding. Sequence] DR EMBL; J 01066; AAB 59183. 1; -. [EMBL / Gen. Bank / DDBJ] [Co. Ding. Sequence] DR EMBL; M 17827; AAA 28341. 1; -. [EMBL / Gen. Bank / DDBJ] [Co. Ding. Sequence] DR EMBL; M 17828; AAA 28342. 1; -. [EMBL / Gen. Bank / DDBJ] [Co. Ding. Sequence] DR EMBL; M 19547; AAA 70210. 1; -. [EMBL / Gen. Bank / DDBJ] [Co. Ding. Sequence] DR EMBL; M 17830; AAA 28343. 1; -. [EMBL / Gen. Bank / DDBJ] [Co. Ding. Sequence] DR EMBL; M 17831; AAA 28344. 1; -. [EMBL / Gen. Bank / DDBJ] [Co. Ding. Sequence] DR EMBL; M 17832; AAA 28345. 1; -. [EMBL / Gen. Bank / DDBJ] [Co. Ding. Sequence] DR EMBL; M 17833; AAA 28346. 1; -. [EMBL / Gen. Bank / DDBJ] [Co. Ding. Sequence] DR EMBL; M 17834; AAA 28347. 1; -. [EMBL / Gen. Bank / DDBJ] [Co. Ding. Sequence] DR EMBL; M 17835; AAA 28348. 1; -. [EMBL / Gen. Bank / DDBJ] [Co. Ding. Sequence] DR EMBL; M 17836; AAA 28349. 1; -. [EMBL / Gen. Bank / DDBJ] [Co. Ding. Sequence] DR EMBL; M 17837; AAA 70212. 1; -. [EMBL / Gen. Bank / DDBJ] [Co. Ding. Sequence] DR EMBL; X 60791; CAA 43204. 1; -. [EMBL / Gen. Bank / DDBJ] [Co. Ding. Sequence] DR EMBL; X 60792; CAA 43205. 1; -. [EMBL / Gen. Bank / DDBJ] [Co. Ding. Sequence] DR EMBL; U 20765; AAA 88817. 1; -. [EMBL / Gen. Bank / DDBJ] [Co. Ding. Sequence] DR EMBL; X 78384; CAA 55151. 1; -. [EMBL / Gen. Bank / DDBJ] [Co. Ding. Sequence] DR EMBL; X 98338; CAA 66981. 1; -. [EMBL / Gen. Bank / DDBJ] [Co. Ding. Sequence] DR PIR; A 00343; DEFFA. DR PIR; S 17083. DR PIR; S 17085. DR FLYBASE; FBgn 0000055; Adh. DR INTERPRO; IPR 002198; -. DR INTERPRO; IPR 002424; -. DR INTERPRO; IPR 002425; -. DR INTERPRO; IPR 003030; -. DR PFAM; PF 00106; adh_short; 1. DR PFAM; PF 00663; adh_short_C; 1. DR PRINTS; PR 00080; SDRFAMILY. DR PRINTS; PR 01167; INSADHFAMILY. DR PRINTS; PR 01168; ALCDHDRGNASE. DR PROSITE; PS 00061; ADH_SHORT; 1. DR PRODOM [Domain structure / List of seq. sharing at least 1 domain] DR BLOCKS; P 00334. DR DOMO; P 00334. DR PROTOMAP; P 00334. DR PRESAGE; P 00334. DR DIP; P 00334. DR SWISS-2 DPAGE; GET REGION ON 2 D PAGE. KW Oxidoreductase; NAD; Acetylation. FT INIT_MET 0 0 FT MOD_RES 1 1 ACETYLATION. FT NP_BIND 11 34 NAD (BY SIMILARITY). FT ACT_SITE 152 FT VARIANT 192 K -> T (IN ADH-F ALLELE). FT VARIANT 214 P -> S (IN ADH-FCH. D ALLELE). FT MUTAGEN 14 14 G->V: COMPLETE LOSS OF ACTIVITY. FT MUTAGEN 14 14 G->A: 31% DECREASE IN ACTIVITY. FT MUTAGEN 129 G->C: COMPLETE LOSS OF ACTIVITY. FT MUTAGEN 132 G->I: COMPLETE LOSS OF ACTIVITY. FT MUTAGEN 135 C->A: NO DECREASE IN ACTIVITY. FT MUTAGEN 152 Y->F, H, E, Q: COMPLETE LOSS OF ACTIVITY. FT MUTAGEN 152 Y->C: RETAINS ONLY 0. 25% ACTIVITY. FT MUTAGEN 156 K->I: COMPLETE LOSS OF ACTIVITY. FT MUTAGEN 156 K->R: RETAINS ONLY 2. 2% ACTIVITY. FT MUTAGEN 183 G->L: COMPLETE LOSS OF ACTIVITY. FT MUTAGEN 218 C->A: NO DECREASE IN ACTIVITY. SQ SEQUENCE 255 AA; 27630 MW; CA 7 DF 929 E 0007296 CRC 64; SFTLTNKNVI FVAGLGGIGL DTSKELLKRD LKNLVILDRI ENPAAIAELK AINPKVTVTF YPYDVTVPIA ETTKLLKTIF AQLKTVDVLI NGAGILDDHQ IERTIAVNYT GLVNTTTAIL DFWDKRKGGP GGIICNIGSV TGFNAIYQVP VYSGTKAAVV NFTSSLAKLA PITGVTAYTV NPGITRTTLV HKFNSWLDVE PQVAEKLLAH PTQPSLACAE NFVKAIELNQ NGAIWKLDLG TLEAIQWTKH WDSGI // 20

Computer Science in Bioinformatics • FASTA • BLAST 21
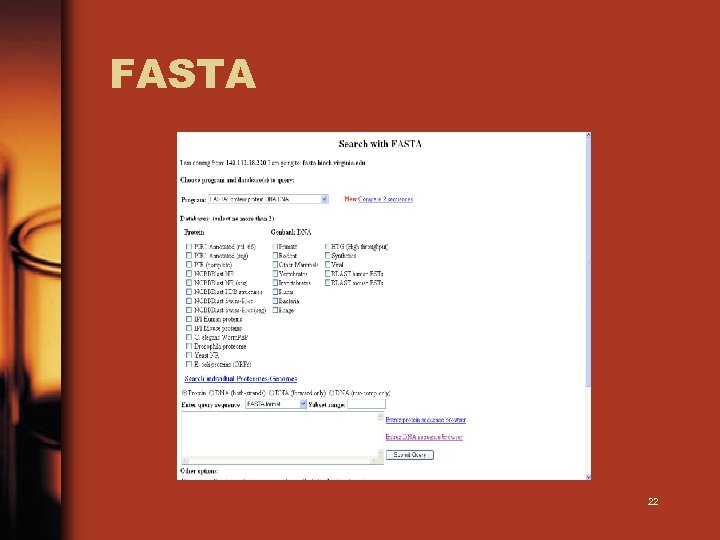
FASTA 22

FASTA Algorithm: WATSNANDCRICK BASEBALLANDCRICKET 23
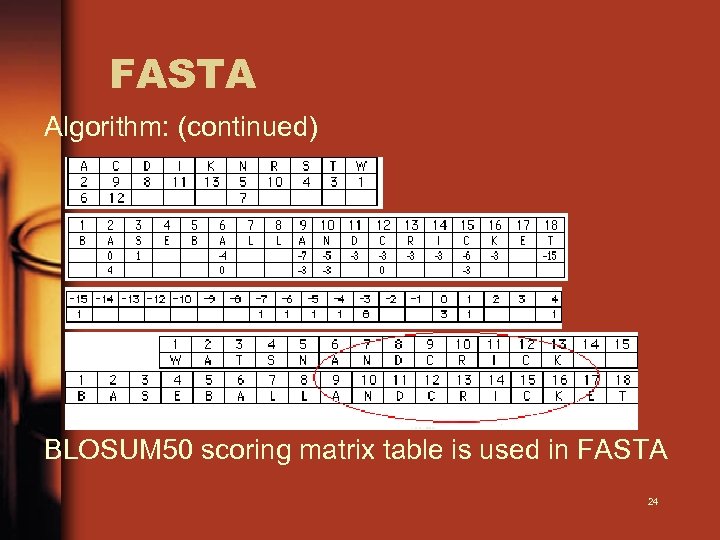
FASTA Algorithm: (continued) BLOSUM 50 scoring matrix table is used in FASTA 24

BLAST 25

BLAST Algorithm: For protein A S T N C F : A matched with itself gives a score of 2 S matched with itself gives a score of 2 T matched with itself yields a score of 3 N matched with itself yields a score of 2 ASTN can only be 9 or less, smaller than the minimum requirement for the score, 17. 26

BLAST Algorithm: (continued) For protein A S T N C F : S matched with itself gives a score of 2 T matched with itself yields a score of 3 N matched with itself yields a score of 2 C matched with itself yields a score of 12 The total score of the word formed by these four amino acids is 19, and it exceeds 17. Substitutions of amino acids have a scoring of 1. Therefore, there are some query sequences that generate many other query sequences. 27

BLAST Algorithm: (continued) Every time an exact match occurs, BLAST attempts to extend the seed by looking for matches in either direction. However, the algorithm is capable of missing sequences, because it doesn't take gaps into account. For example, consider the sequences: ACDEEFGH ACDEFGH A C D E can be located but it will not be extended to F G H, because E is not a good match. Similarly, E F G H can be identified but A C D will not be traced. BLOSUM 62 scoring index is used in BLAST 28

The Future of Bioinformatics • Data collection and organization--New concepts for databases. • Prediction of biological functions--Computational tools are needed to analyze the collected data in the most efficient manner. • Data Labyrinth--Computational tools that integrate the scattered information is needed. 29

The Future of Bioinformatics 30
b42a24f86c319284ea3464da36d13017.ppt