Applications-Formats-Vivek-Haibao.pptx
- Количество слайдов: 38

Bioinformatics Applications Vivek Krishnakumar & Haibao Tang J. Craig Venter Institute Plant Informatics Workshop July 15, 2013
Module 1: FASTA
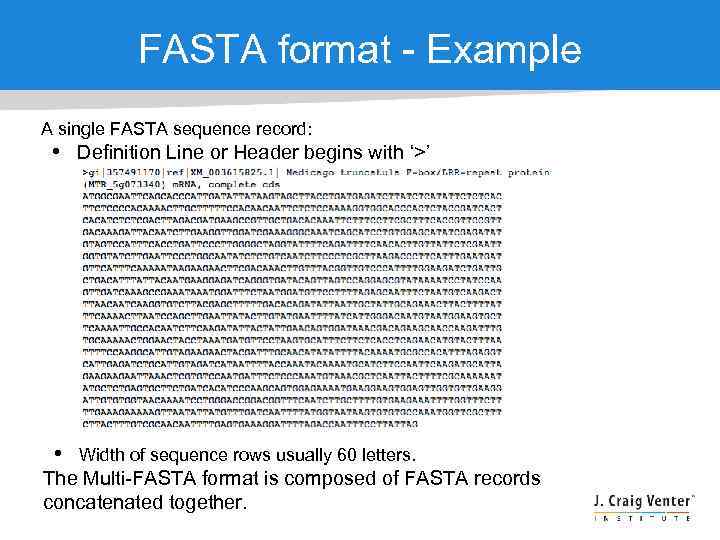
FASTA format - Example A single FASTA sequence record: • Definition Line or Header begins with ‘>’ • Width of sequence rows usually 60 letters. The Multi-FASTA format is composed of FASTA records concatenated together.
FASTA - Definition Line • • • Minimum requirement for definition line is '>' symbol followed by an identifier >Medtr 5 g 073340 Extra comments may be added after the identifier separated by a whitespace Several pre-defined conventions already exist, that are followed by the sequence databases
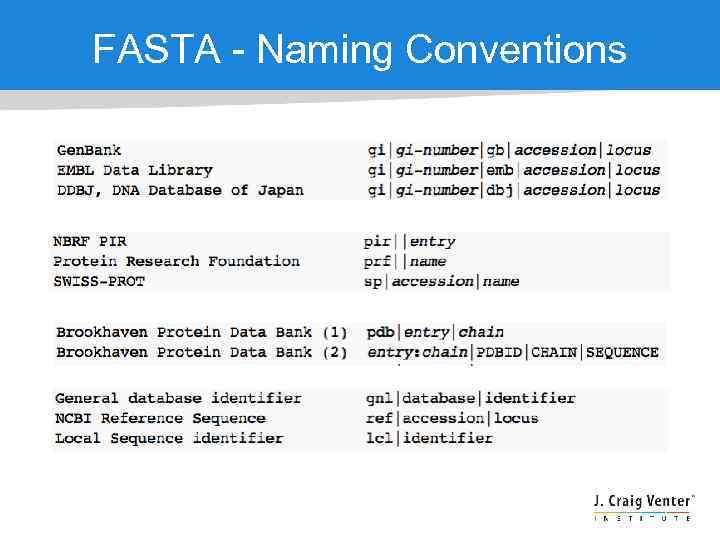
FASTA - Naming Conventions
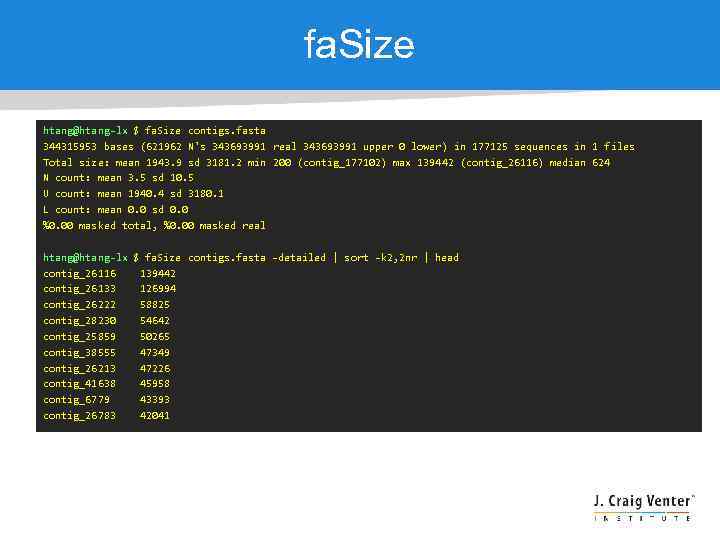
fa. Size htang@htang-lx $ fa. Size contigs. fasta 344315953 bases (621962 N's 343693991 real 343693991 upper 0 lower) in 177125 sequences in 1 files Total size: mean 1943. 9 sd 3181. 2 min 200 (contig_177102) max 139442 (contig_26116) median 624 N count: mean 3. 5 sd 10. 5 U count: mean 1940. 4 sd 3180. 1 L count: mean 0. 0 sd 0. 0 %0. 00 masked total, %0. 00 masked real htang@htang-lx $ fa. Size contigs. fasta -detailed | sort -k 2, 2 nr | head contig_26116 139442 contig_26133 126994 contig_26222 58825 contig_28230 54642 contig_25859 50265 contig_38555 47349 contig_26213 47226 contig_41638 45958 contig_6779 43393 contig_26783 42041

fa. One. Record, fa. Some. Records fa. One. Record - Extract a single record from a. FA file usage: fa. One. Record in. fa record. Name fa. Some. Records - Extract multiple fa records usage: fa. Some. Records in. fa list. File out. fa options: -exclude - output sequences not in the list file. • Does not create index like cdbindex/cdbyank (does not create extra files) • Sufficiently fast
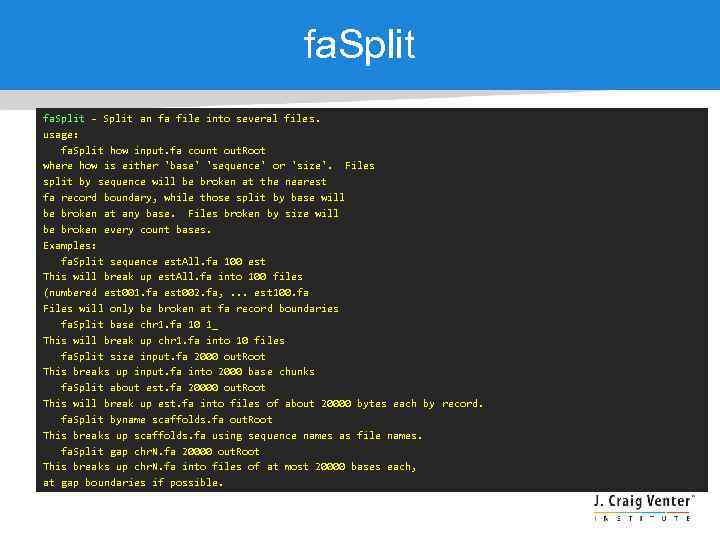
fa. Split - Split an fa file into several files. usage: fa. Split how input. fa count out. Root where how is either 'base' 'sequence' or 'size'. Files split by sequence will be broken at the nearest fa record boundary, while those split by base will be broken at any base. Files broken by size will be broken every count bases. Examples: fa. Split sequence est. All. fa 100 est This will break up est. All. fa into 100 files (numbered est 001. fa est 002. fa, . . . est 100. fa Files will only be broken at fa record boundaries fa. Split base chr 1. fa 10 1_ This will break up chr 1. fa into 10 files fa. Split size input. fa 2000 out. Root This breaks up input. fa into 2000 base chunks fa. Split about est. fa 20000 out. Root This will break up est. fa into files of about 20000 bytes each by record. fa. Split byname scaffolds. fa out. Root This breaks up scaffolds. fa using sequence names as file names. fa. Split gap chr. N. fa 20000 out. Root This breaks up chr. N. fa into files of at most 20000 bases each, at gap boundaries if possible.
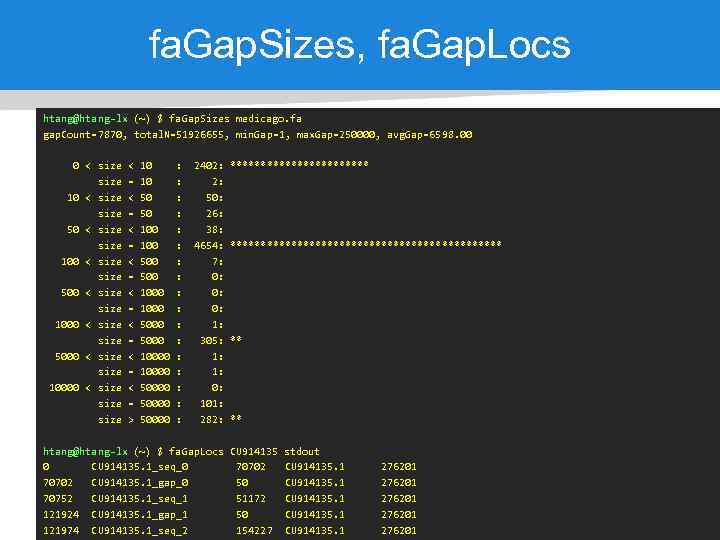
fa. Gap. Sizes, fa. Gap. Locs htang@htang-lx (~) $ fa. Gap. Sizes medicago. fa gap. Count=7870, total. N=51926655, min. Gap=1, max. Gap=250000, avg. Gap=6598. 00 0 < size 10 < size 50 < size 100 < size 500 < size 1000 < size 5000 < size 10000 < size < = < = > 10 10 50 50 100 500 1000 5000 10000 50000 : : : : : 2402: 2: 50: 26: 38: 4654: 7: 0: 0: 0: 1: 305: 1: 1: 0: 101: 282: ********************************** ** ** htang@htang-lx (~) $ fa. Gap. Locs CU 914135 stdout 0 CU 914135. 1_seq_0 70702 CU 914135. 1_gap_0 50 CU 914135. 1 70752 CU 914135. 1_seq_1 51172 CU 914135. 1 121924 CU 914135. 1_gap_1 50 CU 914135. 1 121974 CU 914135. 1_seq_2 154227 CU 914135. 1 276201 276201

Module 2: FASTQ
Format for HTS Data • • • High Throughput Sequencing (HTS) instruments produce large quantities of sequence data Requirement to store sequence quality along with raw sequence arose Thus, logical extension to FASTA was developed, called FASTQ o o Minimal representation of sequencing read Ability to store numeric quality score
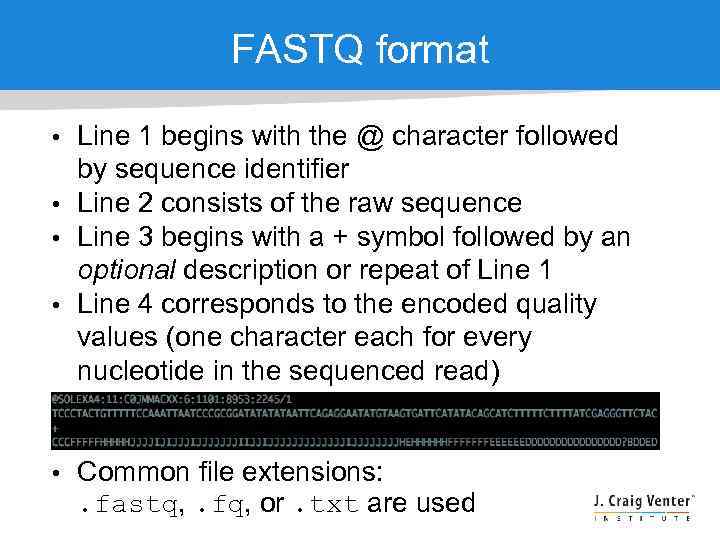
FASTQ format • Line 1 begins with the @ character followed by sequence identifier • Line 2 consists of the raw sequence • Line 3 begins with a + symbol followed by an optional description or repeat of Line 1 • Line 4 corresponds to the encoded quality values (one character each for every nucleotide in the sequenced read) • Common file extensions: . fastq, . fq, or. txt are used
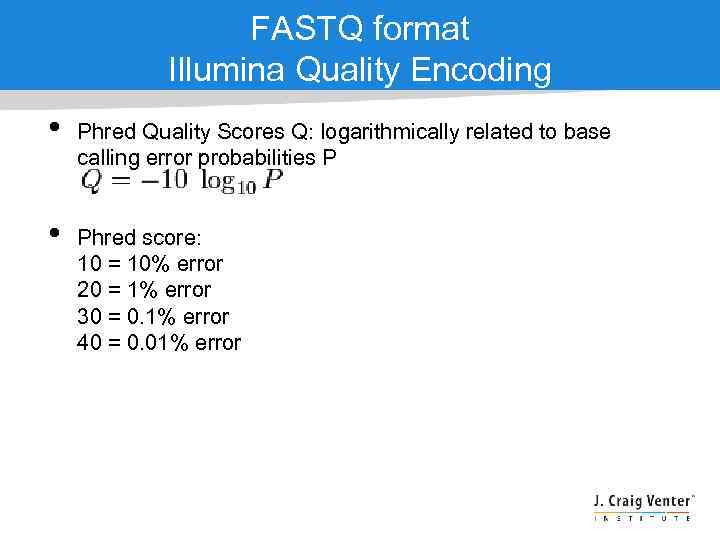
FASTQ format Illumina Quality Encoding • • Phred Quality Scores Q: logarithmically related to base calling error probabilities P Phred score: 10 = 10% error 20 = 1% error 30 = 0. 1% error 40 = 0. 01% error
FASTQ format Illumina Quality Encoding • • Illumina format encodes a quality score between 0 and 62 using ASCII 64 to 126 (as compared to Sanger which encodes 0 to 93 using ASCII 33 to 126) The phred score + some offset = ASCII code, example: Two types of offsets (phred +33, and phred +64). Most of the FASTQ files are phred +33. How do I know which offset it is? There is a quick tip

FASTQ format Illumina Phred + 33
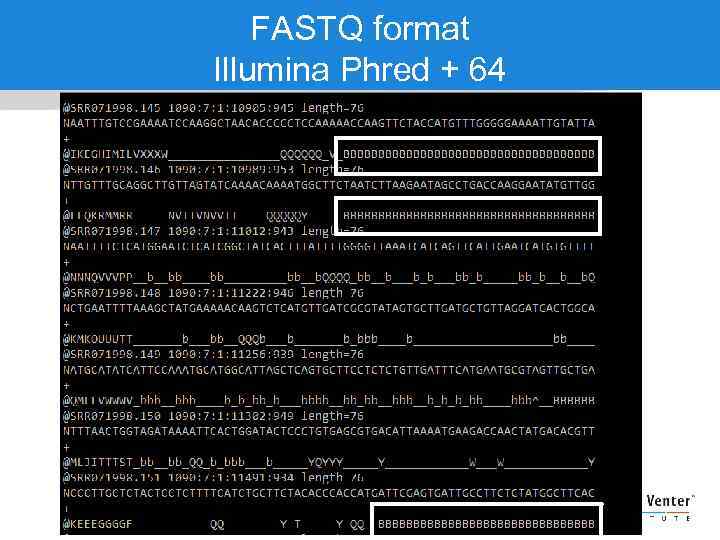
FASTQ format Illumina Phred + 64

Module 3: GFF, BED
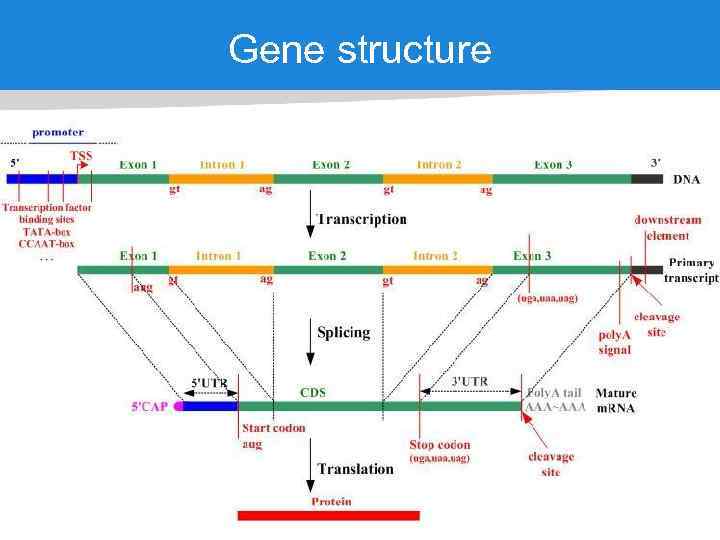
Gene structure
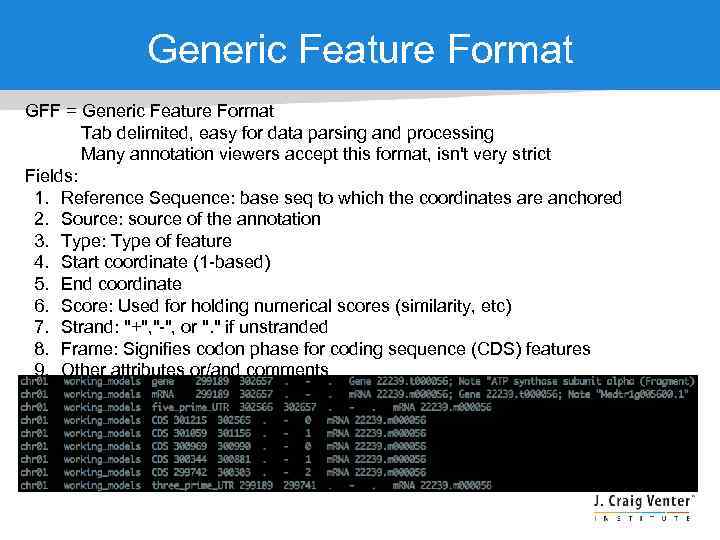
Generic Feature Format GFF = Generic Feature Format Tab delimited, easy for data parsing and processing Many annotation viewers accept this format, isn't very strict Fields: 1. Reference Sequence: base seq to which the coordinates are anchored 2. Source: source of the annotation 3. Type: Type of feature 4. Start coordinate (1 -based) 5. End coordinate 6. Score: Used for holding numerical scores (similarity, etc) 7. Strand: "+", "-", or ". " if unstranded 8. Frame: Signifies codon phase for coding sequence (CDS) features 9. Other attributes or/and comments
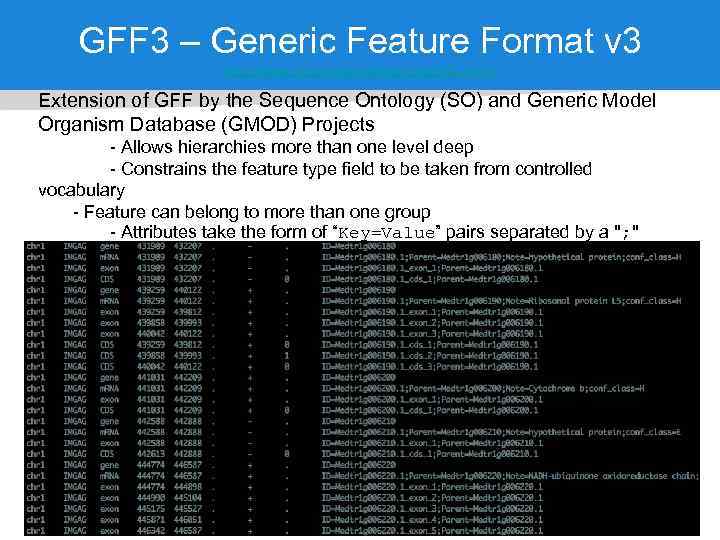
GFF 3 – Generic Feature Format v 3 http: //www. sequenceontology. org/gff 3. shtml Extension of GFF by the Sequence Ontology (SO) and Generic Model Organism Database (GMOD) Projects - Allows hierarchies more than one level deep - Constrains the feature type field to be taken from controlled vocabulary - Feature can belong to more than one group - Attributes take the form of “Key=Value” pairs separated by a "; " - Uppercase 'Keys' are reserved. Lower case 'keys' are user-defined
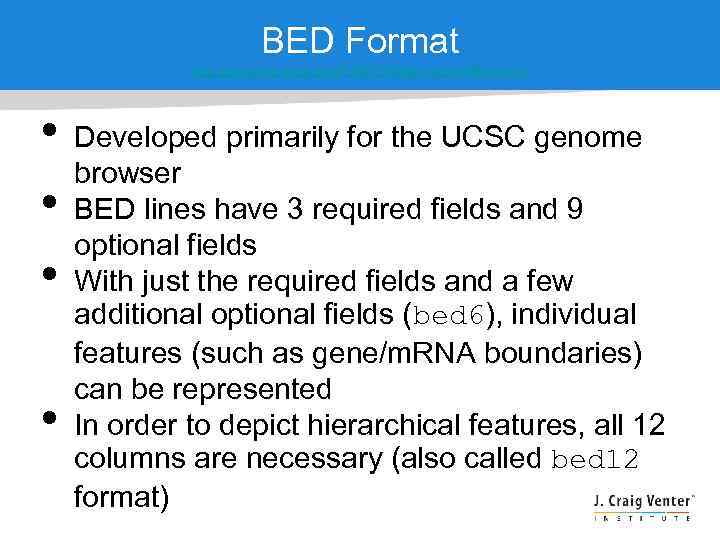
BED Format http: //genome. ucsc. edu/FAQformat. html#format 1 • • Developed primarily for the UCSC genome browser BED lines have 3 required fields and 9 optional fields With just the required fields and a few additional optional fields (bed 6), individual features (such as gene/m. RNA boundaries) can be represented In order to depict hierarchical features, all 12 columns are necessary (also called bed 12 format)

BED Format - Description Three required BED fields are: 1. chrom - The name of the chromosome/reference (e. g. chr 3, scaffold_1) 2. chrom. Start - The starting position of the feature (0 -based) 3. chrom. End - The ending position of the feature The 9 additional optional BED fields are: 1. name - Defines the name of the BED line 2. score - A score between 0 and 1000 3. strand - Defines the strand - either '+' or '-'. 4. thick. Start - The starting position at which the feature is drawn thickly (for example, the start codon in gene displays). 5. thick. End - The ending position at which the feature is drawn thickly (for example, the stop codon in gene displays) 6. item. Rgb - An RGB value of the form R, G, B (e. g. 255, 0, 0) 7. block. Count - The number of blocks (exons) in the BED line 8. block. Sizes - A comma-separated list of the block sizes, corresponding to block. Count 9. block. Starts - A comma-separated list of block starts, relative to chrom. Start, corresponding to block. Count
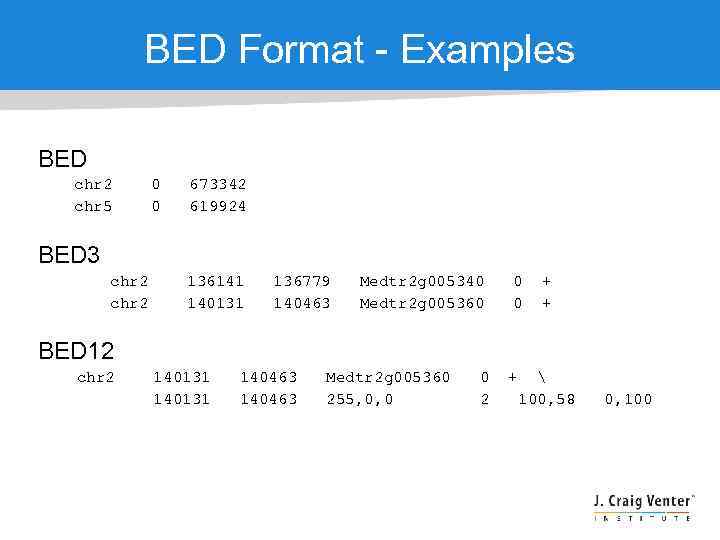
BED Format - Examples BED chr 2 chr 5 0 0 673342 619924 BED 3 chr 2 136141 140131 136779 140463 Medtr 2 g 005340 Medtr 2 g 005360 0 0 + + BED 12 chr 2 140131 140463 Medtr 2 g 005360 255, 0, 0 0 2 + 100, 58 0, 100

BEDTOOLS • Author: Aaron Quinlan, UVA • BED format (first three columns `chr`, `start`, `end` are required), 0 -based #chromosome Mt_chr 01 Mt_chr 01 start 150050 154858 155200 162535 164428 167402 169440 171722 end 154771 155160 162479 164388 166156 169400 171646 172427 name Medtr 1 g 005000 Medtr 1 g 005010 Medtr 1 g 005020 Medtr 1 g 005030 Medtr 1 g 005040 Medtr 1 g 005050 Medtr 1 g 005060 Medtr 1 g 005070 score 1000 1000 strand. . . + + - • BEDTOOLS operates on genomic coordinates and uses “Bin indexing” to speed up range queries
BEDTOOLS functionalities • Find out what’s overlapping between sets of features. (intersect. Bed) • Find closest genomic features. (closest. Bed, window. Bed) • Merge overlapping features. (merge. Bed) • Computing coverage for alignments based on genome features. (coverage. Bam, bam. To. Bed) • Calculating the depth and breadth of sequence coverage across defined "windows" in a genome. (coverage. Bed) • Sequence output. (fasta. From. Bed, mask. Fasta. From. Bed)
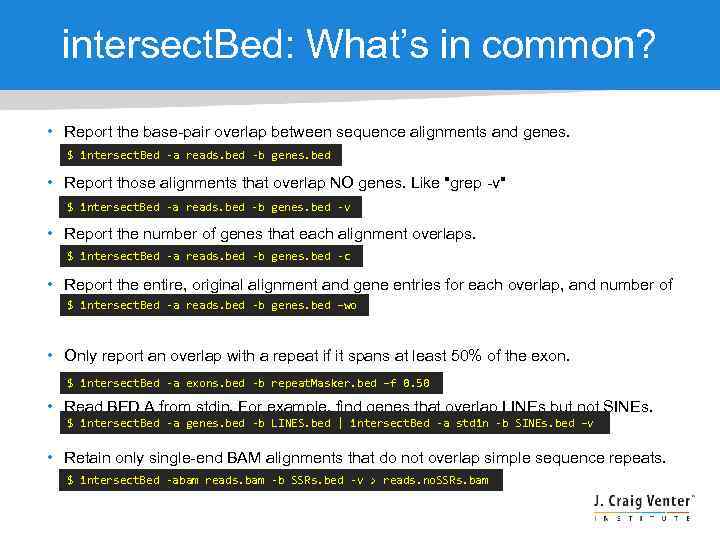
intersect. Bed: What’s in common? • Report the base-pair overlap between sequence alignments and genes. $ intersect. Bed -a reads. bed -b genes. bed • Report those alignments that overlap NO genes. Like "grep -v" $ intersect. Bed -a reads. bed -b genes. bed -v • Report the number of genes that each alignment overlaps. $ intersect. Bed -a reads. bed -b genes. bed -c • Report the entire, original alignment and gene entries for each overlap, and number of $ intersect. Bed -a reads. bed -b genes. bed –wo overlapping bases. • Only report an overlap with a repeat if it spans at least 50% of the exon. $ intersect. Bed -a exons. bed -b repeat. Masker. bed –f 0. 50 • Read BED A from stdin. For example, find genes that overlap LINEs but not SINEs. $ intersect. Bed -a genes. bed -b LINES. bed | intersect. Bed -a stdin -b SINEs. bed –v • Retain only single-end BAM alignments that do not overlap simple sequence repeats. $ intersect. Bed -abam reads. bam -b SSRs. bed -v > reads. no. SSRs. bam

window. Bed, closest. Bed: What’s in there? • window. Bed • Report all genes that are within 10000 bp upstream or downstream of CNVs. $ window. Bed -a CNVs. bed -b genes. bed -w 10000 • Report all genes that are within 10000 bp upstream or 5000 bp downstream of CNVs. $ window. Bed -a CNVs. bed -b genes. bed –l 10000 –r 5000 • Report all SNPs that are within 5000 bp upstream or 1000 bp downstream of genes. Define upstream and downstream based on strand. $ window. Bed -a genes. bed –b snps. bed –l 5000 –r 1000 -sw • closest. Bed • Note: By default, if there is a tie for closest, all ties will be reported. closest. Bed allows overlapping features to be the closest. • Find the closest ALU to each gene. $ closest. Bed -a genes. bed -b ALUs. bed • Find the closest ALU to each gene, choosing the first ALU in the file if there is a tie. $ closest. Bed -a genes. bed -b ALUs. bed –t first
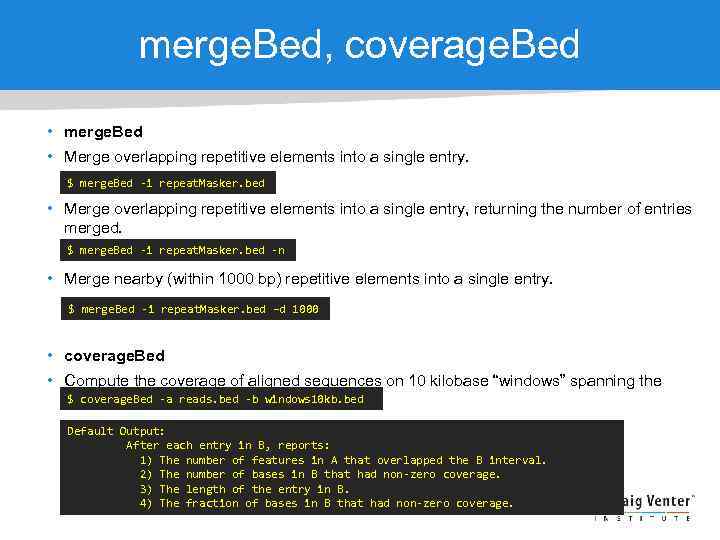
merge. Bed, coverage. Bed • merge. Bed • Merge overlapping repetitive elements into a single entry. $ merge. Bed -i repeat. Masker. bed • Merge overlapping repetitive elements into a single entry, returning the number of entries merged. $ merge. Bed -i repeat. Masker. bed -n • Merge nearby (within 1000 bp) repetitive elements into a single entry. $ merge. Bed -i repeat. Masker. bed –d 1000 • coverage. Bed • Compute the coverage of aligned sequences on 10 kilobase “windows” spanning the $ coverage. Bed -a reads. bed -b windows 10 kb. bed genome. Default Output: After each entry in B, reports: 1) The number of features in A that overlapped the B interval. 2) The number of bases in B that had non-zero coverage. 3) The length of the entry in B. 4) The fraction of bases in B that had non-zero coverage.
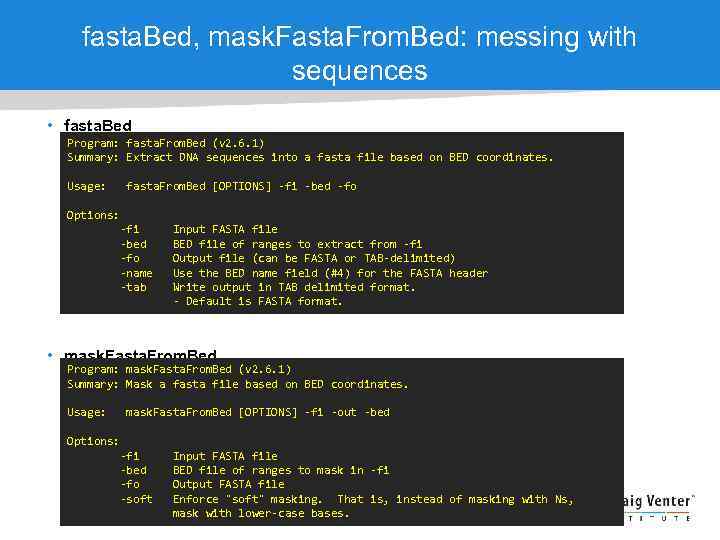
fasta. Bed, mask. Fasta. From. Bed: messing with sequences • fasta. Bed Program: fasta. From. Bed (v 2. 6. 1) Summary: Extract DNA sequences into a fasta file based on BED coordinates. Usage: fasta. From. Bed [OPTIONS] -fi -bed -fo Options: -fi -bed -fo -name -tab Input FASTA file BED file of ranges to extract from -fi Output file (can be FASTA or TAB-delimited) Use the BED name field (#4) for the FASTA header Write output in TAB delimited format. - Default is FASTA format. • mask. Fasta. From. Bed Program: mask. Fasta. From. Bed (v 2. 6. 1) Summary: Mask a fasta file based on BED coordinates. Usage: mask. Fasta. From. Bed [OPTIONS] -fi -out -bed Options: -fi -bed -fo -soft Input FASTA file BED file of ranges to mask in -fi Output FASTA file Enforce "soft" masking. That is, instead of masking with Ns, mask with lower-case bases.
Module 4: SAM, BAM
Sequence Alignment Format • • With the advent of HTS technologies, several requirements arose: o need for a generic alignment format to store read alignments o support for short and long reads generated by different sequencing platforms o compact file size o random access ability o ability to store various types of alignments (clipped, spliced, multi-mapped, padded) As a result, SAM (Sequence Alignment/Map) format evolved
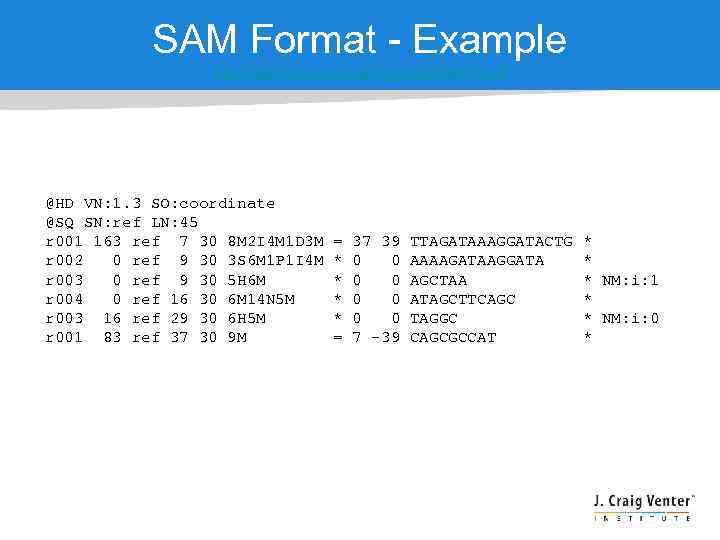
SAM Format - Example http: //samtools. sourceforge. net/SAM 1. pdf @HD VN: 1. 3 SO: coordinate @SQ SN: ref LN: 45 r 001 163 ref 7 30 8 M 2 I 4 M 1 D 3 M r 002 0 ref 9 30 3 S 6 M 1 P 1 I 4 M r 003 0 ref 9 30 5 H 6 M r 004 0 ref 16 30 6 M 14 N 5 M r 003 16 ref 29 30 6 H 5 M r 001 83 ref 37 30 9 M = * * = 37 39 0 0 0 0 7 -39 TTAGATAAAGGATACTG AAAAGATAAGGATA AGCTAA ATAGCTTCAGC TAGGC CAGCGCCAT * * * NM: i: 1 * * NM: i: 0 *
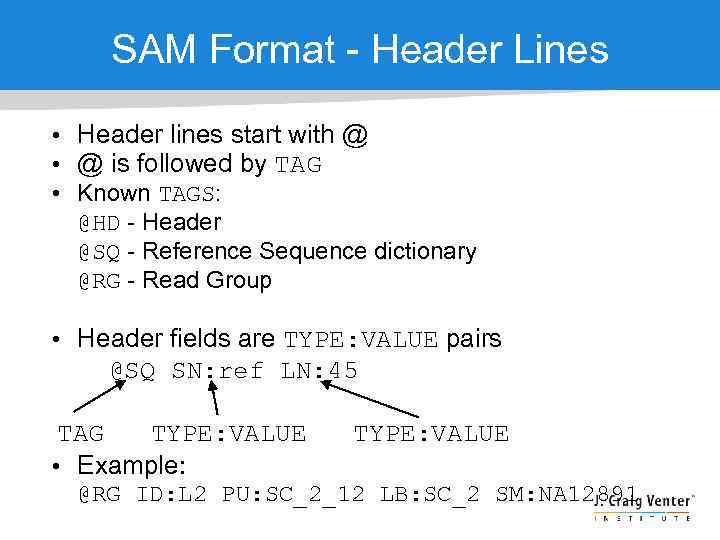
SAM Format - Header Lines • Header lines start with @ • @ is followed by TAG • Known TAGS: @HD - Header @SQ - Reference Sequence dictionary @RG - Read Group • Header fields are TYPE: VALUE pairs @SQ SN: ref LN: 45 TAG TYPE: VALUE • Example: TYPE: VALUE @RG ID: L 2 PU: SC_2_12 LB: SC_2 SM: NA 12891
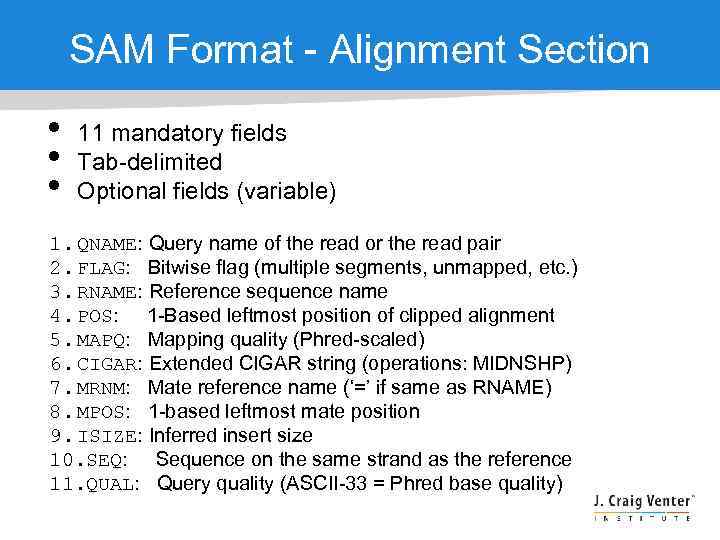
SAM Format - Alignment Section • • • 11 mandatory fields Tab-delimited Optional fields (variable) 1. QNAME: Query name of the read or the read pair 2. FLAG: Bitwise flag (multiple segments, unmapped, etc. ) 3. RNAME: Reference sequence name 4. POS: 1 -Based leftmost position of clipped alignment 5. MAPQ: Mapping quality (Phred-scaled) 6. CIGAR: Extended CIGAR string (operations: MIDNSHP) 7. MRNM: Mate reference name (‘=’ if same as RNAME) 8. MPOS: 1 -based leftmost mate position 9. ISIZE: Inferred insert size 10. SEQ: Sequence on the same strand as the reference 11. QUAL: Query quality (ASCII-33 = Phred base quality)
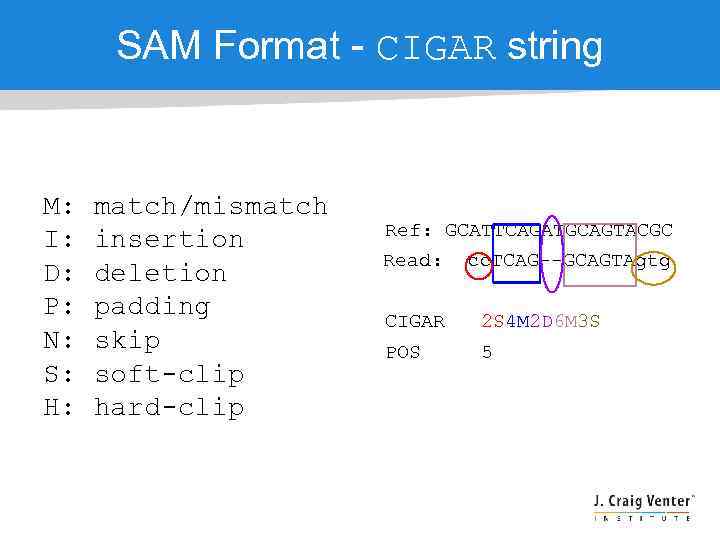
SAM Format - CIGAR string M: I: D: P: N: S: H: match/mismatch insertion deletion padding skip soft-clip hard-clip Ref: GCATTCAGATGCAGTACGC Read: cc. TCAG--GCAGTAgtg CIGAR 2 S 4 M 2 D 6 M 3 S POS 5
SAM Format Compressed Binary Version • • • Known as BAM Binary, indexed representation of SAM Uses BGZF (Blocked GZip Format) compression Storage space requirements ~27% of original SAM/BAM can be manipulated using SAMTOOLS package
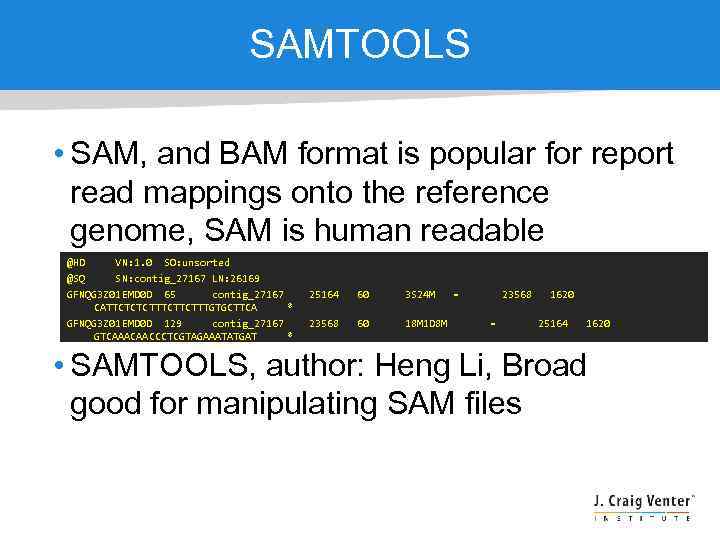
SAMTOOLS • SAM, and BAM format is popular for report read mappings onto the reference genome, SAM is human readable @HD VN: 1. 0 SO: unsorted @SQ SN: contig_27167 LN: 26169 GFNQG 3 Z 01 EMD 0 D 65 contig_27167 CATTCTCTCTTCTTTGTGCTTCA * GFNQG 3 Z 01 EMD 0 D 129 contig_27167 GTCAAACAACCCTCGTAGAAATATGAT * 25164 60 3 S 24 M 23568 60 18 M 1 D 8 M = 23568 = 1620 25164 1620 • SAMTOOLS, author: Heng Li, Broad good for manipulating SAM files
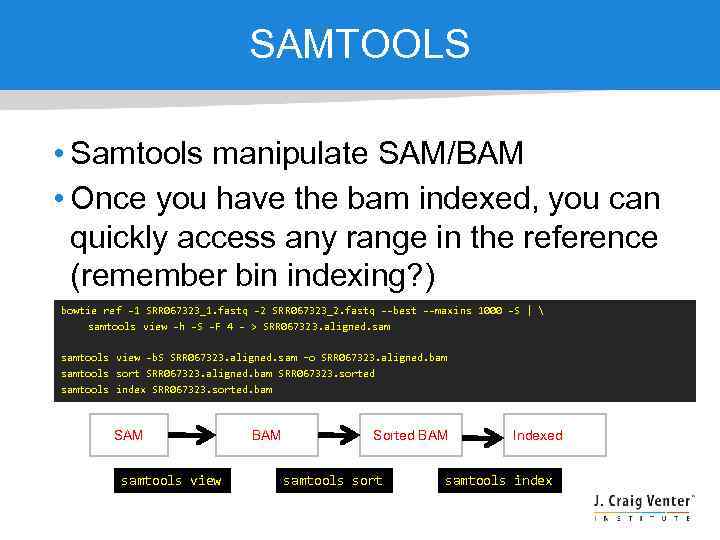
SAMTOOLS • Samtools manipulate SAM/BAM • Once you have the bam indexed, you can quickly access any range in the reference (remember bin indexing? ) bowtie ref -1 SRR 067323_1. fastq -2 SRR 067323_2. fastq --best --maxins 1000 -S | samtools view -h -S -F 4 - > SRR 067323. aligned. samtools view -b. S SRR 067323. aligned. sam –o SRR 067323. aligned. bam samtools sort SRR 067323. aligned. bam SRR 067323. sorted samtools index SRR 067323. sorted. bam SAM samtools view BAM Sorted BAM samtools sort Indexed BAM samtools index
Applications-Formats-Vivek-Haibao.pptx