6833e31c9e0230664606999b54f79c6e.ppt
- Количество слайдов: 32

Biocep-R Open Science in the cloud, towards a universal platform for mathematical and statistical computing Karim Chine karim. chine@m 4 x. org

Croire possible le souhaitable est aussi dangereux que de croire souhaitable le possible. Utopies sentimentales et automatismes de la technique. Nicolás Gómez Dávila Il n’y a que le solitaire qui soit capable de penser plus que des vérités tactiques. Nicolás Gómez Dávila

Extract from the Grid. Solve Description Document The emergence of Grid computing as the prototype of a next generation cyberinfrastructure for science has excited high expectations for its potential as an accelerator of discovery, but it has also raised questions about whether and how the broad population of research professionals, who must be the foundation of such productivity, can be motivated to adopt this new and more complex way of working. The rise of the new era of scientific modeling and simulation has, after all, been precipitous, and many science and engineering professionals have only recently become comfortable with the relatively simple world of the uniprocessor workstations and desktop scientific computing tools. In that world, software packages such as Matlab and Mathematica represent general-purpose scientific computing environments (SCEs) that enable users — totaling more than a million worldwide — to solve a wide variety of problems through flexible user interfaces that can model in a natural way the mathematical aspects of many different problem domains. Moreover, the ongoing, exponential increase in the computing resources supplied by the typical workstation makes these SCEs more and more powerful, and thereby tends to reduce the need for the kind of resource sharing that represents a major strength of Grid computing [1]. Certainly there are various forces now urging collaboration across disciplines and distances, and the burgeoning Grid community, which aims to facilitate such collaboration, has made significant progress in mitigating the well-known complexities of building, operating, and using distributed computing environments. But it is unrealistic to expect the transition of research professionals to the Grid to be anything but halting and slow if it means abandoning the SCEs that they rightfully view as a major source of their productivity. We therefore believe that Grid computing’s prospects for success will tend to rise and fall according to its ability to interface smoothly with the general purpose SCEs that are likely to continue to dominate the toolbox of its targeted user base.

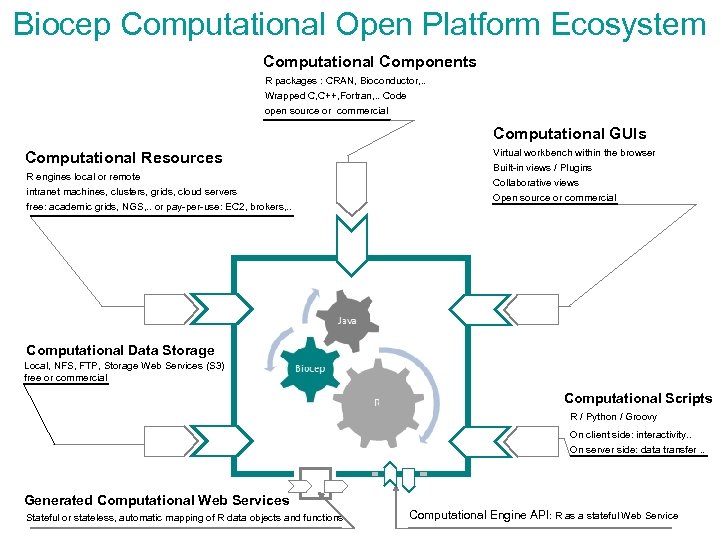
Biocep Computational Open Platform Ecosystem Computational Components R packages : CRAN, Bioconductor, . . Wrapped C, C++, Fortran, . . Code open source or commercial Computational GUIs Computational Resources R engines local or remote intranet machines, clusters, grids, cloud servers free: academic grids, NGS, . . or pay-per-use: EC 2, brokers, . . Virtual workbench within the browser Built-in views / Plugins Collaborative views Open source or commercial Computational Data Storage Local, NFS, FTP, Storage Web Services (S 3) free or commercial Computational Scripts R / Python / Groovy On client side: interactivity. . On server side: data transfer. . Generated Computational Web Services Stateful or stateless, automatic mapping of R data objects and functions Computational Engine API: R as a stateful Web Service

r. Java / JRI Java. GD Object Export / Import Layer R Server R Virtualization mapping RServices API RServices skeleton Graphic devices skels R packages skels Server Side - Personal Machine, Academic Grids, Clusters, Clouds Client Side - Internet Virtual R Workbench Internet Browser Java Applet Virtual R Workbench URL Docking Framework R Console R Graphic Device+Interactors R Workspace R Help Browser R Script Editor R Spreadsheet Groovy / Jython Script Editor

Server-side, grid-enabled, collaborative spreadsheet Cells access from R Console / R scripts Server-side Data Model 1 - Data in server memory 2 - viewing –editing on client machine R functions: 1. cells. get 2. cells. put 3. cells. select Dynamically evaluted cells using R functions Paste R expressions into cells Export cells to R variables Jean Pierre Paul Macros Collaboration - User-defined actions : run user R / Groovy / Python scripts - Simultaneous viewing of the same data - Events-driven macros 1 - on cells change 2 - on R variables change by Jean, Pierre & Paul - Data Links : dock R variables in the Spreadsheet : synchronized changes - Collaborative cells editing - Broadcasted cells Selection
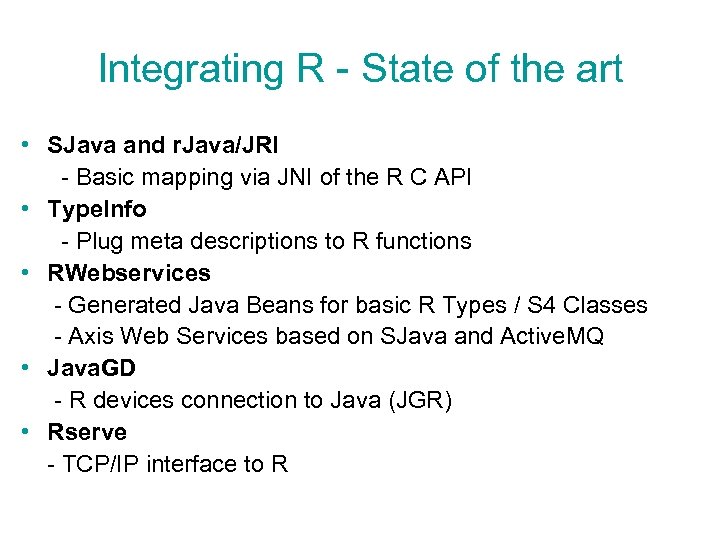
Integrating R - State of the art • SJava and r. Java/JRI - Basic mapping via JNI of the R C API • Type. Info - Plug meta descriptions to R functions • RWebservices - Generated Java Beans for basic R Types / S 4 Classes - Axis Web Services based on SJava and Active. MQ • Java. GD - R devices connection to Java (JGR) • Rserve - TCP/IP interface to R

What was missing ? • High Level Java API for Accessing R • Stateful, Resuable, Remotable R Components • Scalable, Distributed, R Based Infrastructure • Safe multiple clients framework for components usage as a poo of indistinguishable Remote Resources • User friendly Interface for the remote resources creation, tracking and debugging

What was missing ? • Generated light-weight Java proxies for R Types / S 4 Classes • On-demand mapping and deployment of R packages as RMI Components or as JAX-WS Web Services • Remotable R Graphics / Swing Components for R • Remote R components files exchange API • Semi-thick client (applet) for web based tools using R

Standard R objects mapping to Java

Generated beans for Expression. Set Generated Java Bean Proxy Class

RServices API - I public interface RServices extends Managed. Servant public String console. Submit(String expression) throws … evaluate(String expression) throws … public { get. Object(String expression) throws … get. Object. Converted(String expression) throws … get. Reference(String expression) throws … get. Object. Name(String expression) throws … RObject public void put. And. Assign(Object obj, String name) throws … public RObject put. And. Get. Reference(Object obj) throws Remote. Exception; public public RObject void call(String method. Name, Object. . . args) throws … call. And. Convert(String method. Name, Object. . . args) throws … call. And. Get. Reference(String method. Name, Object. . . args) throws … call. And. Get. Object. Name(String method. Name, Object. . . args) throws … call. And. Assign(String var. Name, String method. Name, Object. . . args)throws public RObject realize. Object. Name(RObject object. Name) throws … public Object realize. Object. Name. Converted(RObject object. Name) throws … public RObject reference. To. Object(RObject ref. Obj) throws … public boolean is. Reference(RObject obj) throws … public void assign. Reference(String name, RObject ref. Obj) throws … }
![RServices API - II public interface RServices extends Managed. Servant { public String[] list. RServices API - II public interface RServices extends Managed. Servant { public String[] list.](https://present5.com/presentation/6833e31c9e0230664606999b54f79c6e/image-14.jpg)
RServices API - II public interface RServices extends Managed. Servant { public String[] list. Packages() throws … public RPackage get. Package(String package. Name) throws … public GDDevice new. Device(int w, int h) throws … public GDDevice[] list. Devices() throws … public interface GDDevice extends Remote { public Vector<GDObject> pop. All. Graphic. Objects() throws … public void fire. Size. Changed. Event(int w, int h) throws … public void dispose() throws … … } public public String[] get. Working. Directory. File. Names() throws … File. Description get. Working. Directory. File. Description(String file. Name) throws… void create. Working. Directory. File(String file. Name) throws … void remove. Working. Directory. File(String file. Name) throws … byte[] read. Working. Directory. File. Block(String name, long off, int size)throws… void append. Block. To. Working. Directory. File(String name, byte[] block)throws… public String public byte[] get. RHelp. File. Uri(String topic, String pack) throws … get. RHelp. File(String uri) throws … public Vector<RAction> pop. RActions() throws … }

RServices API - III public interface RServices extends Managed. Servant public void start. Http. Server(int port) throws … stop. Http. Server() throws … public String python. Exec(String python. Command) throws … public RObject python. Eval(String python. Command) throws … public void python. Set(String name, Object Value) throws … public String public Object public void groovy. Exec(String groovy. Command) throws … groovy. Eval(String expression) throws … groovy. Set(String name, Object Value) throws … public void set. Call. Back(RCallback callback) throws … public public get. Status() throws … stop() throws … free. Reference(RObject ref. Obj) throws … free. All. References() throws … print(String expression) throws … source. From. Resource(String resource) throws … source. From. Buffer(String. Buffer buffer) throws … get. RNI() throws … … } String void String RNI {

Remote Resources Pooling Framework • Generic Standalone framework • Pooling of any RMI components and if combined with JNI of any library / open architecture • New Remote Object Registry based on Derby| Oracle| My. SQL • Three implementations available - rmiregistry / mono-node / single client process - rmiregistry / multinodes / single client process - database ROR / multinodes / multiple client processes • User friendly interface for the remote resources creation, tracking and debugging, nodes and pools management

Computational Engines Pools Node 1: Windows XP Pool A Pool B Pool C Node 2: Mac OS Front-end host Remote Objects Registry R-HTTP R-SOAP Node 3: 64 bits Server / Linux Parallel Computing Applications Borrow Rs Supervisor Use Rs Release Rs . NET Appli Perl Node 4 : EC 2 virtual machine 1 Scripts log. On Use R log. Off Web Application Borrow R Generate Graphics/Data Cloudbursting Release R Node 5 : EC 2 virtual machine 2 via Amazon Web Services

R Pools JVM R r. Java / JRI Supervisor Java. GD Object Export / Import Layer mapping. jar RServices API RServices skelton R packages skeltons R graphic device skelton Client Application Remote Objects Registry Borrow R Return R Pooling framework Browser( java plugins( applet ) ) Pooling framework tunneling graphic help config servlet Http Tunneling JVM Generated mapping JAX-WS servlet/artifacts Tomcat SOAP . NET, Perl. . Application

Amazon Machine Image : ami-cd 5 fb 9 a 4 JVM R Ubuntu 9. 04 – R 2. 9. 0 – java 1. 6. 0 – scilab 5. 1. 0 r. Java / JRI Remote Objects Registry Java. GD (Derby Database) Object Export / Import Layer mapping. jar RServices API RServices skelton R packages skeltons R graphic device skelton Pooling framework tunneling graphic help config servlet Generated mapping JAX-WS servlet/artifacts Tomcat Amazon Data center – US Shell’s Network SSH Tunnel : Putty, . . Virtual R Workbench / Plugins SOAP Http / Restful API Third Party Application s: Excel, Open. Office, . . Supervisor Http / Restful API . NET, Perl. . Application Browser : IE, Firefox, . .

Scripting JVM r. Java / JRI File System Java. GD Object Export / Import Layer mapping. jar RServices API RServices skeleton R graphic device skel R packages skels Server Client Virtual R Workbench Create an R Server Open Swing input Dialog Client Side Groovy Script Connect to an existing R Server Use R Server import javax. swing. JOption. Pane; n=JOption. Pane. show. Input. Dialog(null, 100); n=Integer. decode(n); client. R. get. Instance(). put. And. Assign(n, "n") if (n%2==0) { Create an R Server Connect to an existing R Server <R> hist(rnorm(n)) Use R Server </R> } else { <R> Create an R Server plot(rnorm(n)) </R> } Connect to an existing R Server Embedded R Use R Server
![Parallel Computing final double[][] m=. . ; Future<Double>[] result=new Future[m. length]; Executor. Service Parallel Computing final double[][] m=. . ; Future<Double>[] result=new Future[m. length]; Executor. Service](https://present5.com/presentation/6833e31c9e0230664606999b54f79c6e/image-21.jpg)
Parallel Computing final double[][] m=. . ; Future<Double>[] result=new Future[m. length]; Executor. Service exec = Executors. new. Fixed. Thread. Pool(50); for (int i=0; i<result. length; ++i) { final double[] v=m[i]; result[i]= exec. submit( new Callable<Double>() { public Double call() throws Exception { RServices r=null; try { r=(RServices)Servant. Provider. Factory. get. Factory(). get. Servant. Provider(). borrow. Servant. Proxy(); Rnumeric mean=(RNumeric)r. call("mean", new RNumeric(v)); return mean. get. Value()[0]; } finally { Servant. Provider. Factory. get. Factory(). get. Servant. Provider(). return. Servant. Proxy(r); } } }); } while(true) { int count=0; for (int i=0; i<result. length; ++i) if (result[i]. is. Done()) ++count; if (count==result. length) break; Thread. sleep(100); } for (int i=0; i<result. length; ++i) System. out. println(result[i]. get());

Snow with Biocep From the R Console : make. Cluster(n, . . . ) stop. Cluster(cl) Starting and Stopping clusters cluster. Eval. Q(cl, expr) The expression is evaluated on the slave nodes. cluster. Apply(cl, seq, fun, . . . ) Calls the function with the first element of the list on the first node, with the element of the list on the second node, and so on. second cluster. Export(cl, list) Assigns the global values on the master of the variables named in 'list' to variables of the same names in the global environments of each node. …

Web Services Generation rws. war Script / globals. r square function(x) {return(x^2) } type. Info(square) Simultaneous. Type. Specification( Typed. Signature(x = "numeric"), return. Type = "numeric") + mapping. jar WS generator + pooling framework + R Java Bridge + JAX-WS Script / rjmap. xml <rj> <publish> <functions> <function name="square" for. Web="true"/> </functions> </publish> <scripts> <init. Script name="globals. r" embed="true"/> </scripts> </rj> Deploy R HTTP rws. war tomcat - Servlets - Generated artifacts WSDL http: //127. 0. 0. 1: 8080/ rws/r. Global. Env. Function? WSDL public static void main(String[] args) throws Exception { RGlobal. Env. Function. Web g=new RGlobal. Env. Function. Web. Service. Locator(). getr. Global. Env. Function. Web. Port(); RNumeric x=new RNumeric(); x. set. Value(new Double[]{6. 0}); System. out. println(g. square(x). get. Value()[0]); } Eclipse Web Service Client Generator Client artifacts

Workflows with Stateful Web Services Login Pwd Log. On Session. ID associated with a reserved R worker Options ES T 1 ESon 1 T 2 ESon 2 T 3 ESon 3 f ( ES ) get. Data Retrieve Data log. Off T 1, T 2, T 3 : Generated Stateful Web Services for R functions T 1, T 2 & T 3 Log. On, get. Data : R-SOAP methods ES : Expression. Set ESon 1, ESon 2, ESon 3 : Expression. Set Object Names f = T 3 o T 2 o T 1 + remove ESonx kill R Server + « Clean » R Server + Put R Server back in the Pool

R Virtualization on an LSF Cluster LSF Node 3 Shared File System 1 File System 2 LSF Node 1 LSF Submission Host LSF Node 2 create process kill process bsub –J xxx java –jar biocep-core. jar DMZ bkill –J xxx RMI Front-end Host SS H H SS R Servers Manager biocep-core Generated mapping Tunneling Sessions JAX-WS servlet/artifacts Servlet Manager Virtual R Workbench Tomcat DMZ Http tunneling SOAP Serialized Java Objects Http Tunneling Java Applications INTERNET Java, . NET, perl Applications

/usr/local/Cluster-Apps/biocep/. . PBS Node 3 NFS 1 National Grid Service Oxford’s Cluster NFS 2 List, Get, Put PBS Node 1 PBS Node 2 PBS Submission Host Pool Manager Daemon ngs. oerc. ox. ac. uk bind Naming Registry create, kill RMI, Port XX 000: XX 300 5 ports / Engine SSH, Port 22 List + security token xen-ngs 001. oerc. ox. ac. uk RMI Over SSL Xen virtual machine p ooku , Loo kup L List, Emailer Daemon R Servers SMTP Server Manager Recipient biocep-core Generated mapping Tunneling Sessions JAX-WS servlet/artifacts Servlet Manager (3) Login via SSL Mutual Auth (4) HTTPS tunneling – Invoke Obj. SOAP Https Tunneling Java Applications Java, . NET, perl Applications (1) Authenticate with e-science certificate / Submit to NGS : $BIOCEP_HOME/submit. Server dupont@oxford. ac. uk dupont_publickey (2) Get email (Java Web Start URL) : Virtual R Workbench URL + R server name INTERNET Tomcat

Netbeans 6 – Visual GUI builder GUI Plugins my. Plugin. jar Compile + my. View 1 + my. View 2 + descriptor. xml Import Plugin Virtual R Workbench Upload plugin Plugins Repository Browse Repository * my. Plugin *my. Dashboard * Klimt * i. Plots * Mondrian *E. Profiler Download Plugin

FTP INTERNET Collaborative R File System workspace Server Amazon S 3 ACADEMIC GRIDS, NGS, EC 2, INTRANET LSF, INTRANET HOST. . DMZ Tunneling Sessions Servlet Manager Tomcat DMZ Same R session for U 1, U 2 & U 3 Broadcasted Main R Graphic Device INTERNET Broadcasted console + chat User 1 Collaborative Script editor Collaborative Spreadsheet Same virtual workspace for U 1, U 2 & U 3 User 2 User 3

Ease Of Use - I Reasonable Pre-requirementsanstalled : to run the workbench and connectservers on remote hosts Java 5 and R>=2. 5 accessibles from the command line : to run R servers, generate mappings & Web Services, run the miniature virtualisation and the R-SOAP Web Apps. . All-in-one Highly Productive Workbench Docking framework, spreadsheets, syntax highlighting enabled editors, objects viewer, help browser, storage views, zooming system on R graphics, settings persistence. . Easy Computational Resource Acquisition Provide nothing to run R servers on local machine Provide HOST / PORT / LOGIN / PWD to run R Servers on remote hosts (SSH) Provide URL & (LOGIN/PWD or X. 509 Certificate) to Connect to Grid Rs or Cluster Rs Easy Scripting Simple API for running/connecting to R servers Embeddable R code (<R> </R>) within scripts Automatic conversion from/to R Objects for common data types(standard, arrays, collections)

Ease Of Use -II Easy Plugins Integration Import local file / Browse Plugins repository and choose a plugin « Push button » Web Services Generation/Web Services Deployment Add Type. Info to your function / add your function name to an XML / run biocep-tools Deploy: java –port=80 –cp biocep-core. jar Http. Server rvirtual. war My. Web. Services. war Self-contained jar & war files distribution : biocep. jar biocep-core. jar biocep-tools. jar rvirtual. war rws. war Configurationless Parallel Computing from R console : make. Cluster(n, . . ), stop. Cluster(cl), cluster. Eval. Q(cl, expr), cluster. Apply(cl, seq, fun, . . ). . .

Acknowledgements ACS: Madi Nassiri Amazon: Simone Brunozzi, Deepak Singh AT&T Research Labs: Simon Urbanek ATUGE: Imen Essafi, Béchir Tourki, Ilyes Gouja, Hatem. Hachicha, Amine Elleuch Auckland Centre for e. Research: Nick Jones Banca d'Italia: Giuseppe Bruno Bio-IT World: Kevin Davies BNP Parisbas: Ousseynou Nakoulima Cambridge Healthtech Institute: Cindy Crowninshield City University of New York: Mario Morales, Makram Talih Columbia University: Omar Besbes Dassault Systèmes: Omri Ben Ayoun, Patrick Johnson Dataspora: Michael E. Driscoll EDF: Alejandro Ribes EBI: Alvis Brazma, Wolfgang Huber, Kimmo Kallio, Misha Kapushesky, Michael Kleen, Alberto Labarga, Philippe Rocca-Serra, Ugis Sarkans, Kirsten Williams, Eamonn Maguire EPFL: Darlene Goldstein ESPRIT: Farouk Kammoun, Tahar. Benlakhdar e-Taalim: Nadhir Douma ETH Zürich: Yohan Chalabi, Diethelm Würtz, Martin Mächler European Commission: Konstantinos Glinos, Enric Mitjana, Monika Kacik, Ioannis Sagias FHCRC: Martin Morgan, Nianhua Li, Seth Falcon Google: Olivier Bosquet FVG LLC: Lisa Wood Harvard University: Tim Clark, Sudeshna Das, Douglas Burke, Paolo Ciccarese IBM: Jean-Louis Bernaudin, Pascal Sempe, Loic Simon, Lea A Deleris, Alex Fleischer, Alain Chabrier Imperial College London: Asif Akram, Vasa Curcin, John Darlington, Brian Fuchs Indiana University: Michael Grobe INRIA: David Monteau, Christian Saguez, Claude Gomez, Sylvestre Ledru JISC: John Wood, David Flanders Johnson & Johnson - Janssen Pharmaceutica: Patrick Marichal KXEN: Eric Marcade Lancaster University: Robert Crouchley, Daniel Grose Leibniz Universität Hannover: Kornelius Rohmeier LIAMA: Baogang Hue, Kang Cai Limagrain: Zivan Karaman Mekentosj: Alexander Griekspoor, Matt Wood Microsoft: Eric Le Marois, Tony Hey Mubadala: Ghazi Ben Amor Nature Publishing Group: Ian Mulvany, Steve Scott NCe. SS: Peter Halfpenny, Rob Procter, Marzieh Asgari-Targhi, Alex Voss, Yu. Wei Lin, Mercedes Argüello Casteleiro, Wei Jie, Meik Poschen, Katy Middlebrough, Pascal Ekin, June Finch, Farzana Latif, Elisa Pieri, Frank O'Donnell New York Java User Group: Frank D Greco Oe. RC: Dimitrina Spencer, Matteo Turilli, David Wallom, Steven Young OMII-UK: Neil Chue Hong, Steve Brewer Open. Analytics: Tobias Verbeke Oracle: Dominique van Deth, Andrew Bond OSS Watch: Ross Gardler Platform Computing: Christopher Smith Royal Society: James Wilsdon San Diego Supercomputer Center: Nancy R. Wilkins. Diehr Sanger Institute: Lars Jorgensen, Phil Butcher Shell: Wayne. W. Jones, Nigel Smith Société Générale: Anis Maktouf Stanford University: John Chambers, Balasubramanian Narasimhan, Gunter Walther SYSTEM@TIC: Karim Azoum Technische Universität Dortmund: Uwe Ligges, Bernd Bischl Technoforge: Pierre-Antoine Durgeat Tekiano: Samy Ben Naceur Télécom-Paris. Tech: Isabelle Demeure, Georges Hebrail, Nesrine Gabsi The Generations Network: Jim Porzak Total: Yannick Perigois Tunisian Ministry of Communication Technologies: Naceur Ammar, Lamia Chaffai-Sghaier, Mohamed Saïd Ouerghi, Syrine Tlili Tunisian Ecole Polytechnique: Riadh Robbana UC Berkeley: Noureddine El Karoui, Terry Speed UC Davis: Rudy Beran, Debashis Paul, Duncan Temple Lang UCL: Daniel Jeffares UCLA: Ivo Dinov, Jeroen Ooms UC San Diego: Anthony Gamst UCSF: Tena Sakai Université Catholique de Louvain: Christian Ritter University of Cambridge: Ian Roberts, Robert Mac. Innis Peter Murray-Rust, Jim Downing University of Manchester: Carole Goble, Len Gill, Simon Peters, Richard D Pearson, Iain Buchan, John Ainsworth University of Plymouth: Paul Hewson University of Split: Ivica Puljak UTK: Ajay Ohri World Bank Group-IFC: Oualid Ammar Yahoo: Laurent Mirguet, Rob Weltman Independant: Charles Dallas, Romain François

www. biocep. net
6833e31c9e0230664606999b54f79c6e.ppt