5fd6e326ea588269bfd9c8d9afc4f786.ppt
- Количество слайдов: 14

Bio. Pat. ML Pattern sharing for the Genomic Sciences Stefan Maetschke, Michael Towsey and James M. Hogan MQUTe. R Microsoft QUT e. Research Centre Queensland University of Technology, Australia 2008 Microsoft e. Science Workshop 7 -9 December Indianapolis

The Bio. Pat. ML project includes: u A comprehensive pattern description language u Web services for pattern storage and searching u Integration with the semantic web
Unifying the Description of Patterns in Biological Sequences Bio. Pat. ML supports: 1. DNA, RNA, AA sequences 2. Principled aggregation of different pattern types e. g. motifs, gaps, loops 3. Hierarchical patterns 4. Pattern libraries 5. Integrated scoring of pattern matches 6. Some existing pattern databases e. g. Prosite Bio. Pat. ML exploits the advantages of XML and RDF.
![Simple Patterns <Motif alphabet=“DNA” motif=“TA[AT]AAW” /> C A G A T A A T Simple Patterns <Motif alphabet=“DNA” motif=“TA[AT]AAW” /> C A G A T A A T](https://present5.com/presentation/5fd6e326ea588269bfd9c8d9afc4f786/image-4.jpg)
Simple Patterns <Motif alphabet=“DNA” motif=“TA[AT]AAW” /> C A G A T A A T T C C A G A T A <Motif alphabet=“DNA” motif=“TA[AT]AAW” name=“Pribnow-box” threshold=“ 0. 5” />
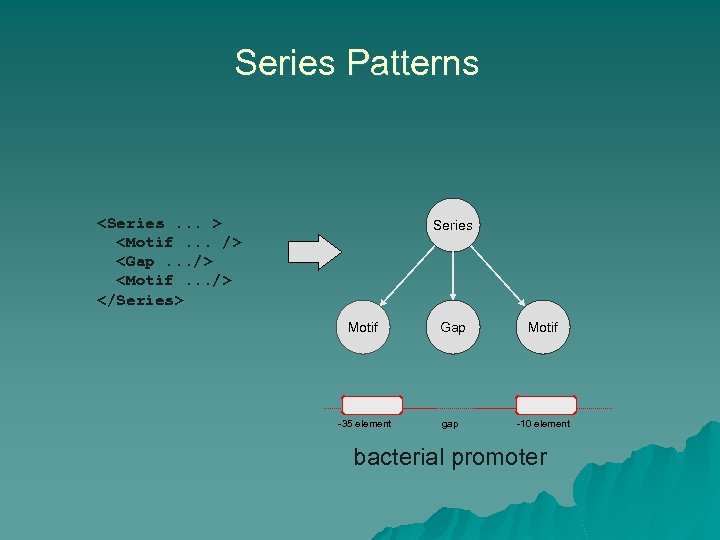
Series Patterns <Series. . . > <Motif. . . /> <Gap. . . /> <Motif. . . /> </Series> Series Motif Gap TTGACA -35 element Motif TATAAT gap -10 element bacterial promoter

Libraries of Patterns (Bio. Pat. ML resource: uri=biopatml/promoter. bpl) <Definition name=“sigma 70” > <Definitions> < Definition name=“-35 element” /> <Motif motif=“TTGACA” alphabet=“DNA” /> </Definition> < Definition name=“-10 element” /> <Motif motif=“TATAAT” alphabet=“DNA” /> </Definition> <Definition name=“Promoter” > </Definitions> <Void /> <Import uri=“biopatml/promoter. bpl” </Definition> </Definitions> <Series. . . > <Use definition=“sigma 70. -35 element” /> <Gap min=“ 13” max=’ 21” /> <Use definition=“sigma 70. -10 element” /> </Series> </Definition>
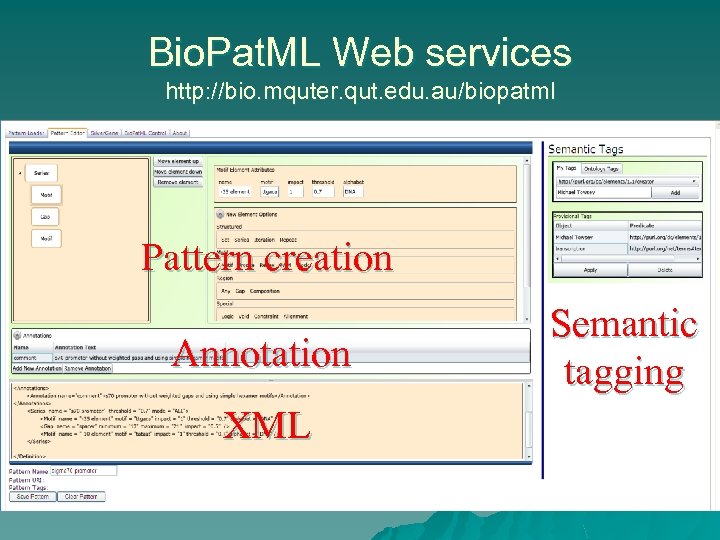
Bio. Pat. ML Web services http: //bio. mquter. qut. edu. au/biopatml Pattern creation Annotation XML Semantic tagging
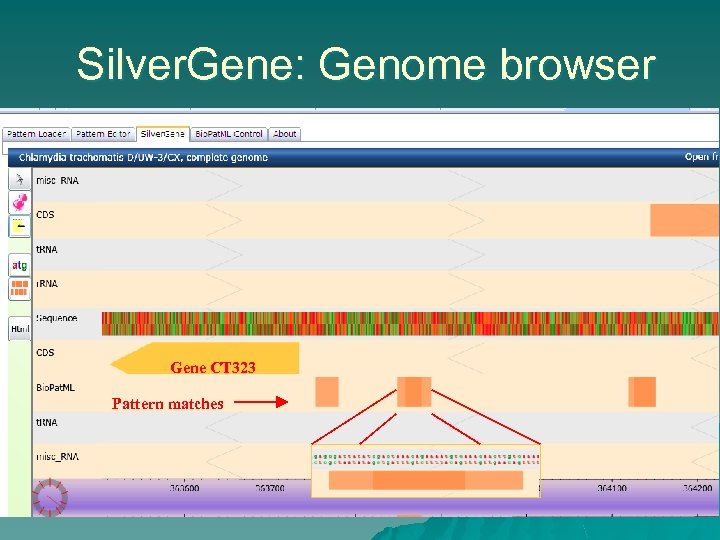
Silver. Gene: Genome browser Gene CT 323 Pattern matches
Bio. Pat. ML in the Semantic Web u Bio. Pat. ML is part of the Bio 2 RDF project u Bio 2 RDF is an initiative of Quebec Genomics Centre and Université Laval u Described as "a new integrated way to surf genomic knowledge"
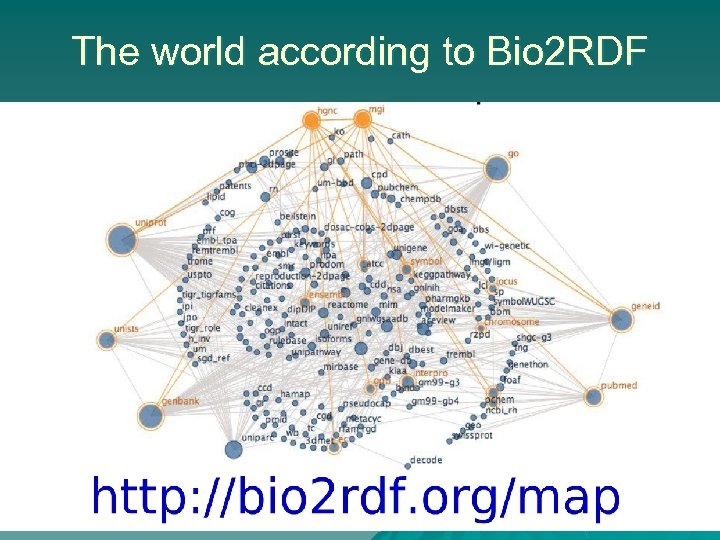
The world according to Bio 2 RDF
Bio. Pat. ML in the Semantic Web u Bio. Pat. ML in Bio 2 RDF – created a name space and terms – http: //bio 2 rdf. org/ns/biopatml u Created an RDF database of Bio. Pat. ML patterns – encapsulate Bio. Pat. ML patterns as RDF literals – RDF tagging and search
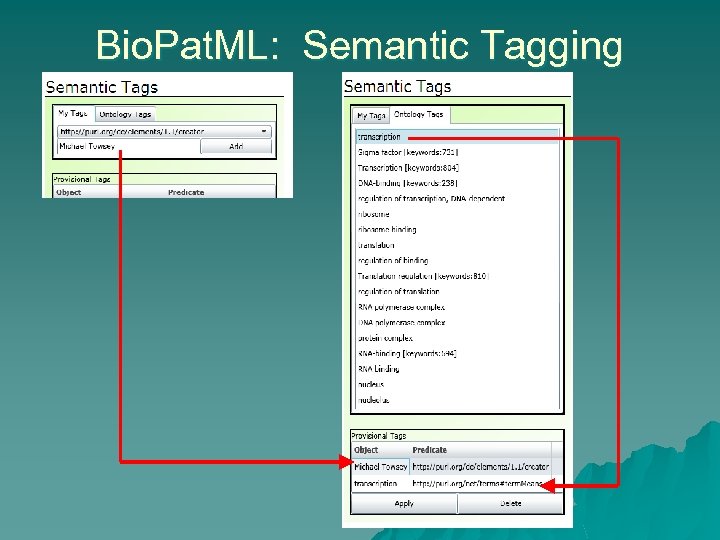
Bio. Pat. ML: Semantic Tagging
Bio. Pat. ML Resources http: //bio. mquter. qut. edu. au/biopatml http: //www. mquter. qut. edu. au/bio http: //bio 2 rdf. org/ns/biopatml http: //bio 2 rdf. org (web demo) (Bio. Pat. ML manual) (namespace & terms) (Bio 2 RDF home page)
Bioinformatics team at MQUTER Michael Towsey Jiro Sumitomo Lawrence Buckingham Chris Bowles Xin-Yi Chua Peter Ansell Jim Hogan Scott Mann
5fd6e326ea588269bfd9c8d9afc4f786.ppt