06a39189ebebbac788ba34831177d0d4.ppt
- Количество слайдов: 48
Bio. Mart Federated Database Architecture Arek Kasprzyk EBI 9 June 2005
Bio. Mart • A join project – European Bioinformatics Institute (EBI) – Cold Spring Harbor Laboratory (CSHL) • Aim – To develop a simple and scalable data management system capable of integrating distributed data sources.

Challenges • Data sources – Large – Distributed – Different data
Requirements • User – All data accessible through a single set of interaces – Suitable for power biologists and bioinformaticians • Deployer – ‘Out of the box’ installation – Built in query optimization – Easy data federation • Architecture – Distributed – Domain agnostic – Platform independent
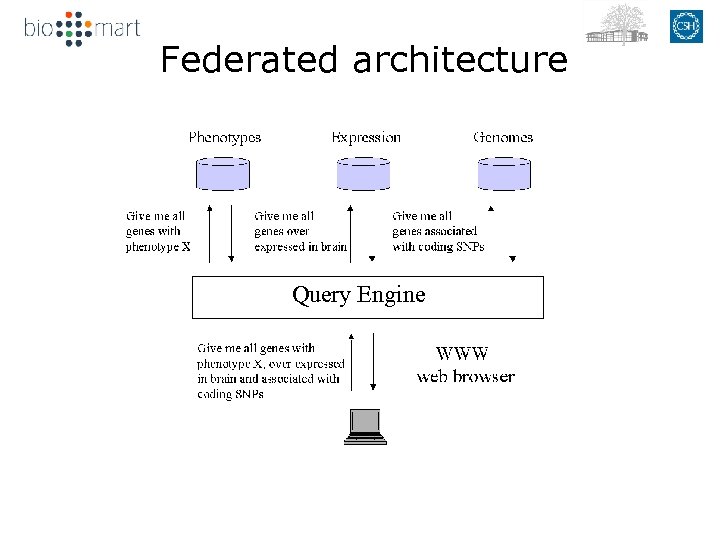
Federated architecture Query Engine
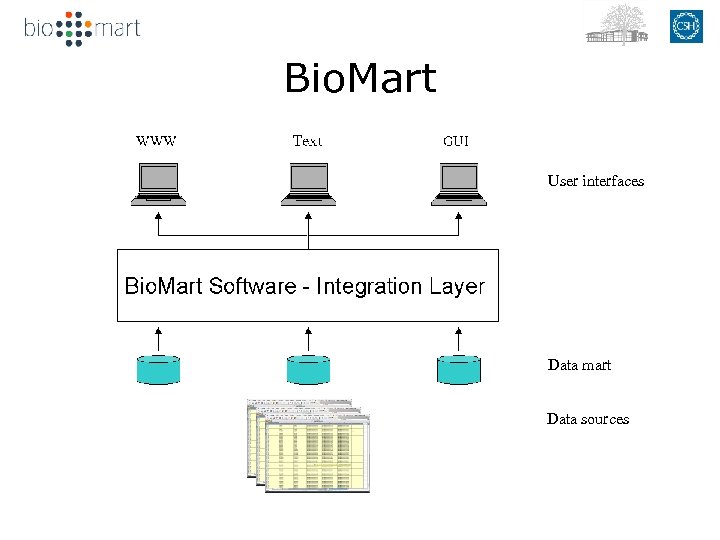
Bio. Mart User interfaces Data mart Data sources
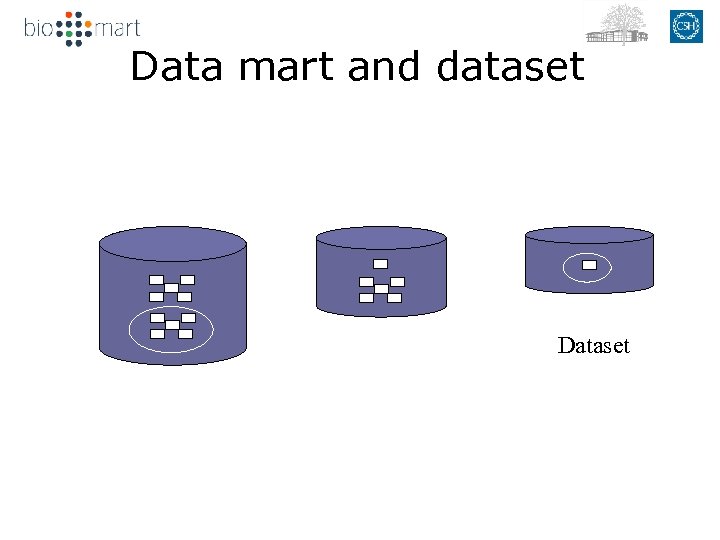
Data mart and dataset Dataset
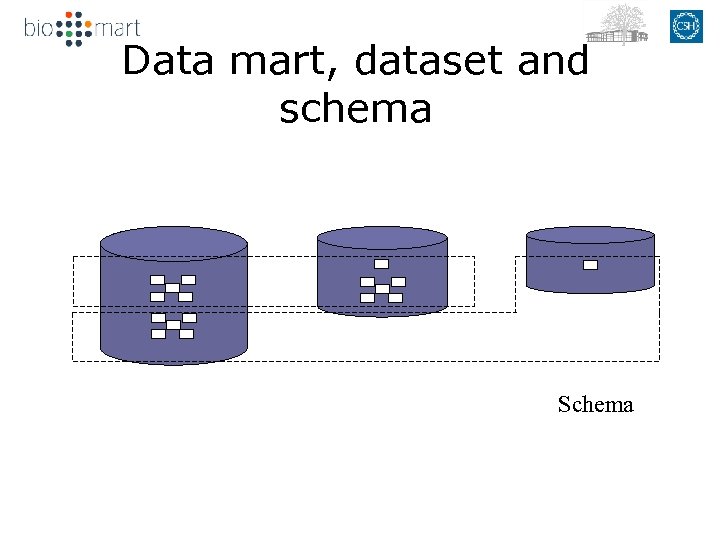
Data mart, dataset and schema Schema
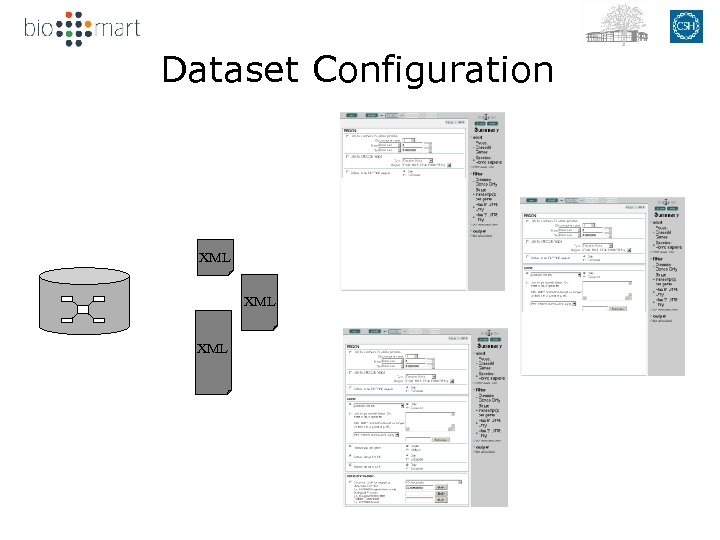
Dataset Configuration XML XML
Bio. Mart abstractions • Dataset – A subset of data organized into 1 or more tables • Attribute – A single data point – e. g. gene name • Filter – An operation on an attribute – e. g. ‘Chromosome =1’
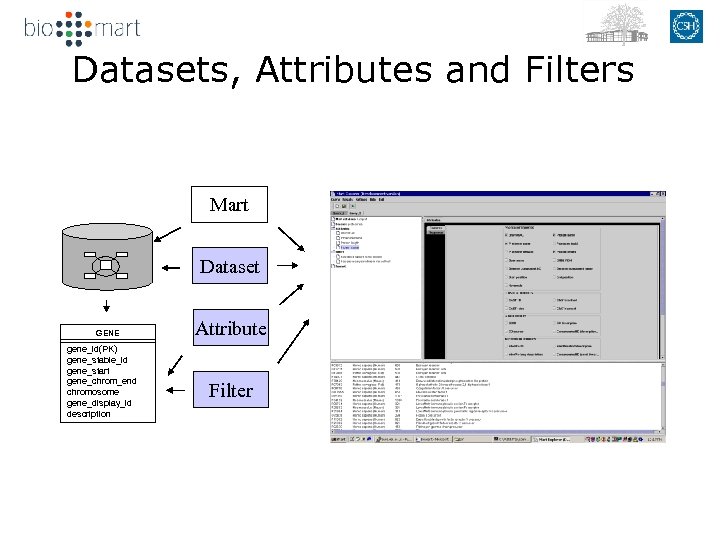
Datasets, Attributes and Filters Mart Dataset GENE gene_id(PK) gene_stable_id gene_start gene_chrom_end chromosome gene_display_id description Attribute Filter
Examples Upstream sequences for all kinases up-regulated in brain and associated with a QTL for a neurological disorder Name, chromosome position, description of all genes located on chromosome 1, expressed in lung, associated with human homologues and nonsynonymous snp changes
Data model FK FK PK PK FK FK
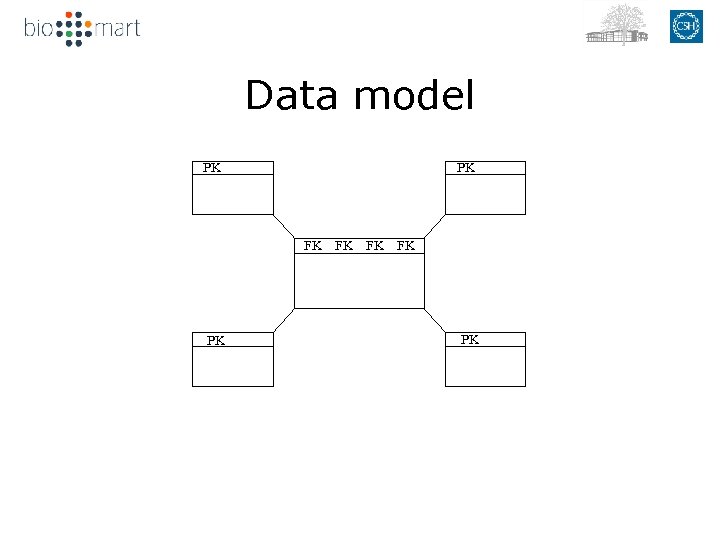
Data model PK PK FK FK FK PK PK PK FK FK
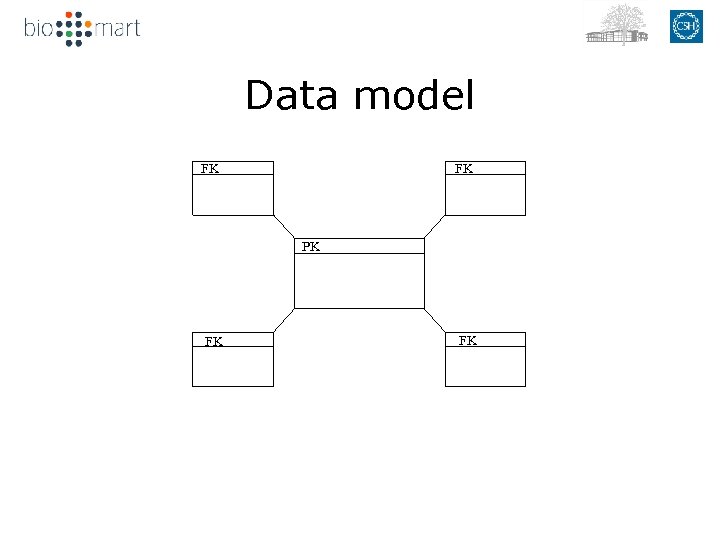
Data model FK FK PK PK FK FK
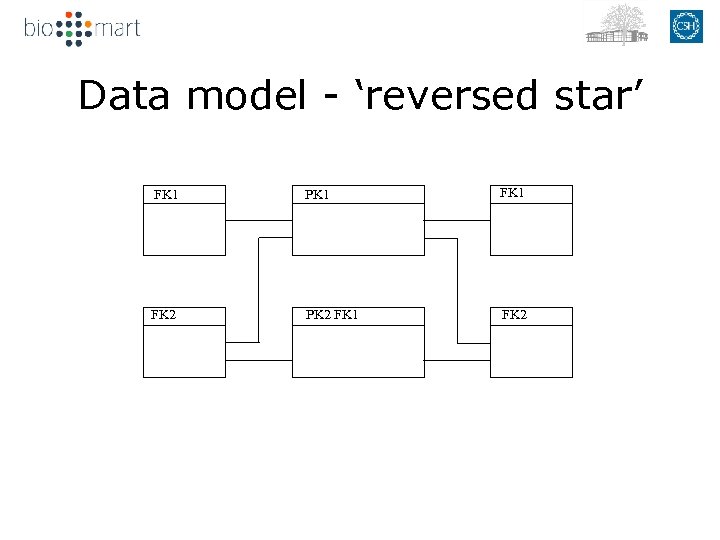
Data model - ‘reversed star’ dm FK 1 FK 2 dm FK 2 PK 1 main 1 PK 1 2 PK 2 FK 1 PK 2 PK 1 dm FK 1 FK 2
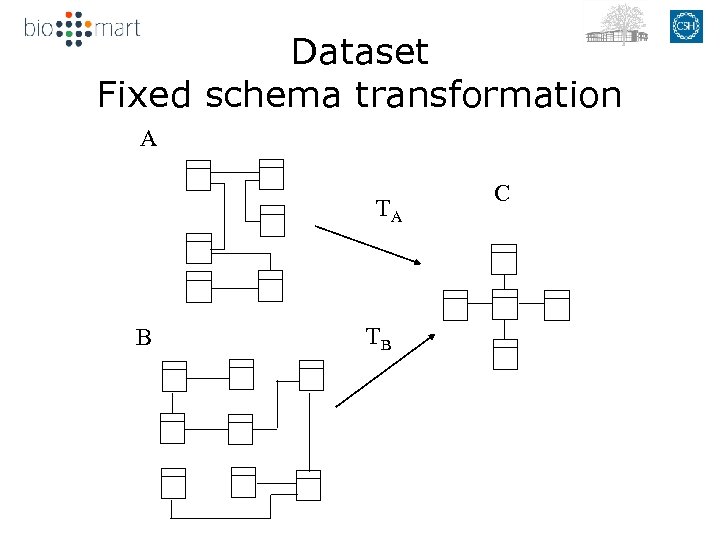
Dataset Fixed schema transformation A TA B TB C
Bio. Mart abstractions • Link – ‘common currency’ between two datasets – e. g. accession • Exportable – Potential links to export • Importable – Potential links to import
Exportables, Importables and Links Dataset 1 Links Dataset 2
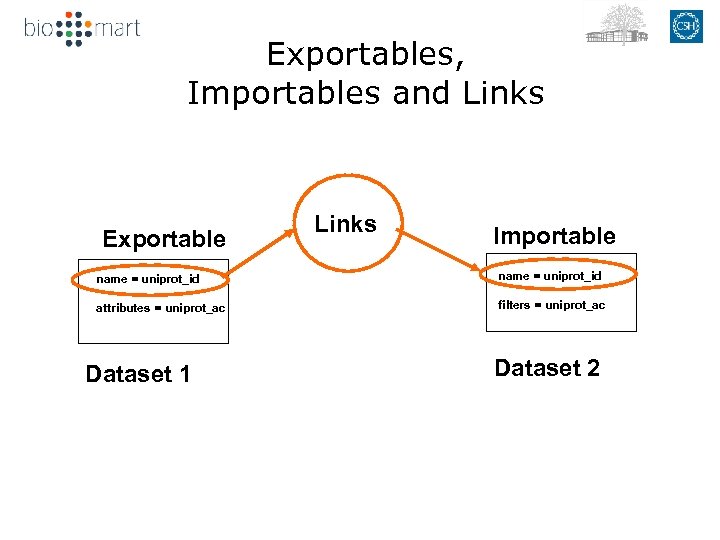
Exportables, Importables and Links Exportable Links Importable name = uniprot_id attributes = uniprot_ac filters = uniprot_ac Dataset 1 Dataset 2
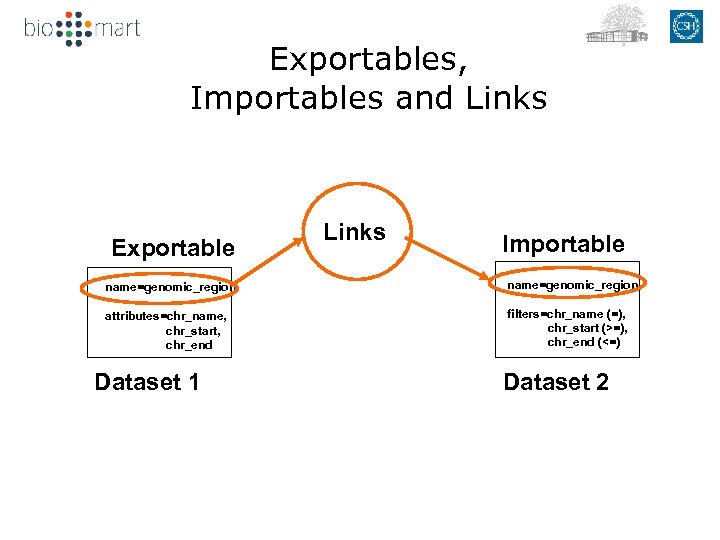
Exportables, Importables and Links Exportable Links Importable name=genomic_region attributes=chr_name, chr_start, chr_end filters=chr_name (=), chr_start (>=), chr_end (<=) Dataset 1 Dataset 2
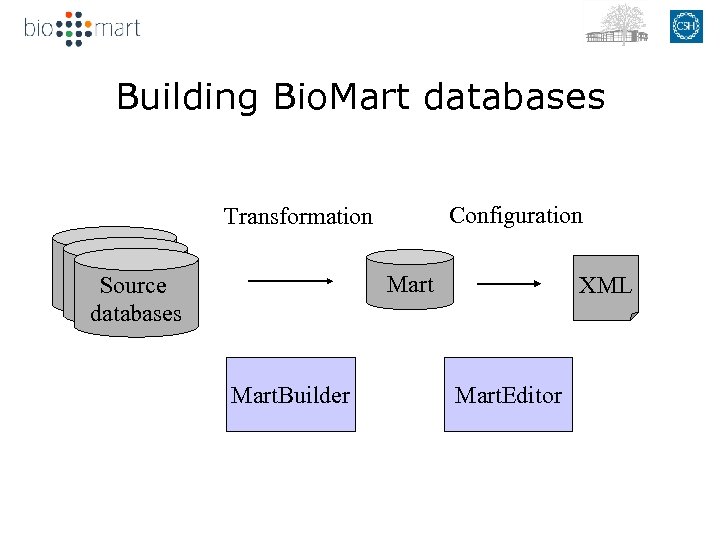
Building Bio. Mart databases Configuration Transformation Mart Source databases Mart. Builder XML Mart. Editor
Mart. Editor
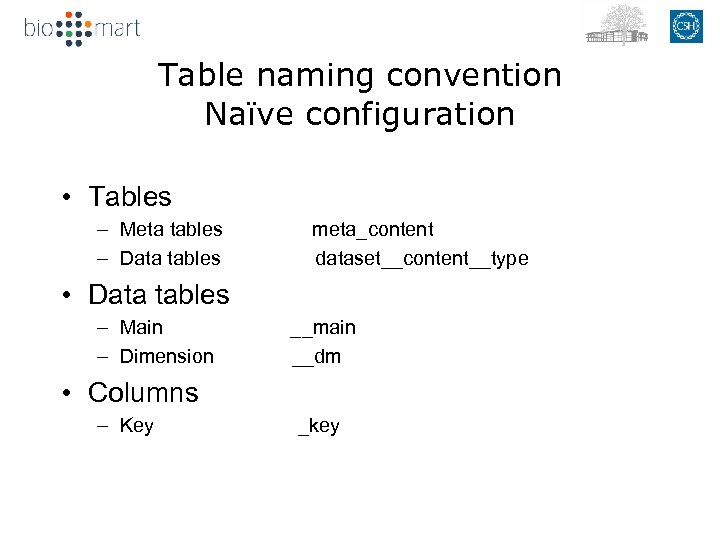
Table naming convention Naïve configuration • Tables – Meta tables – Data tables meta_content dataset__content__type • Data tables – Main – Dimension __main __dm • Columns – Key _key
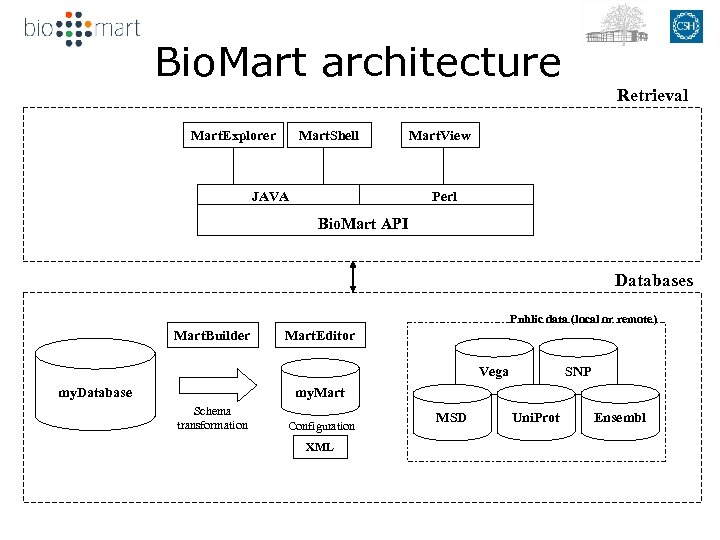
Bio. Mart architecture Retrieval Mart. Explorer Mart. Shell JAVA Mart. View Perl Bio. Mart API Databases Public data (local or remote) Mart. Builder Mart. Editor Vega SNP my. Mart my. Database Schema transformation Configuration XML MSD Uni. Prot Ensembl
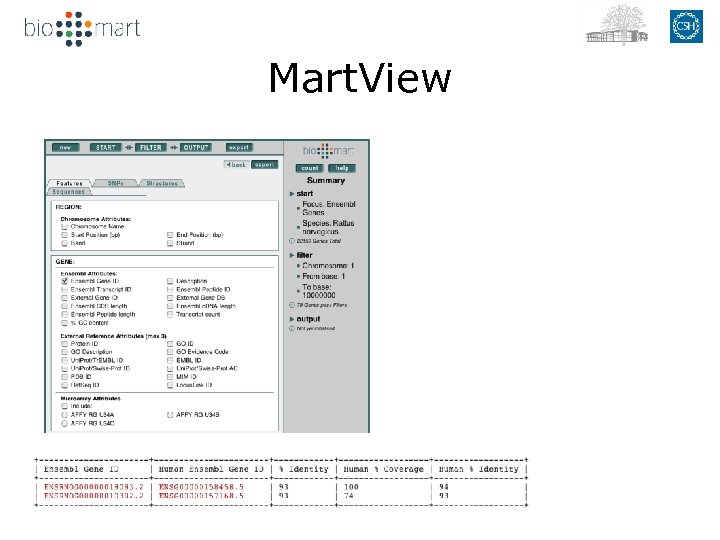
Mart. View
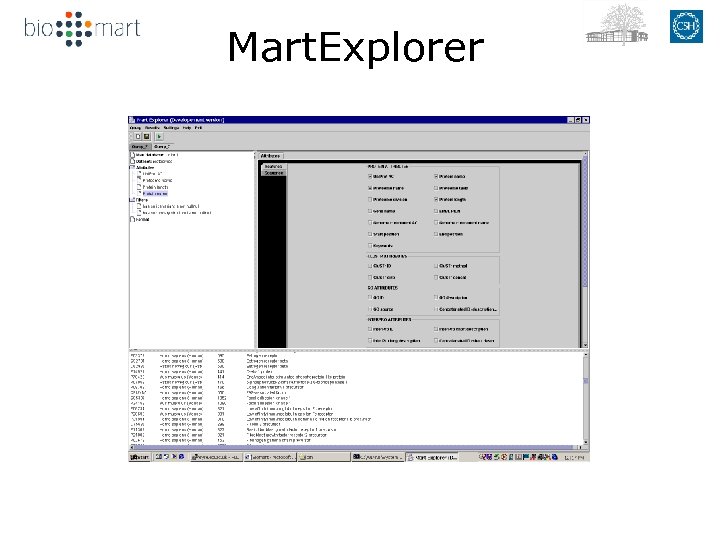
Mart. Explorer
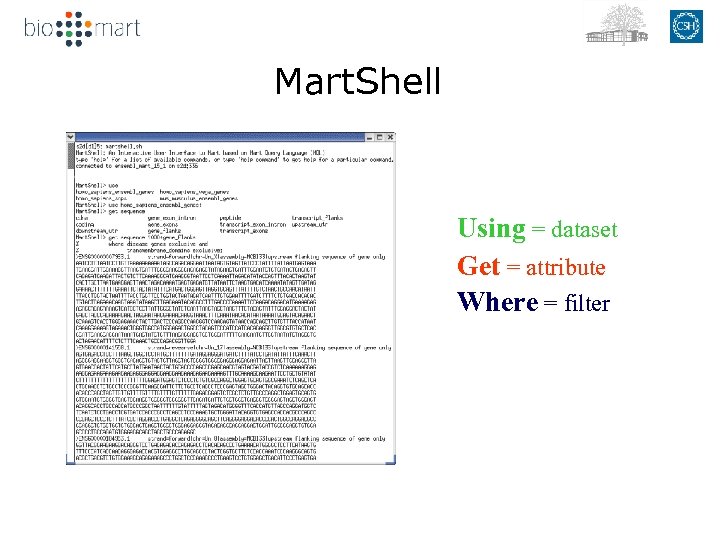
Mart. Shell Using = dataset Get = attribute Where = filter
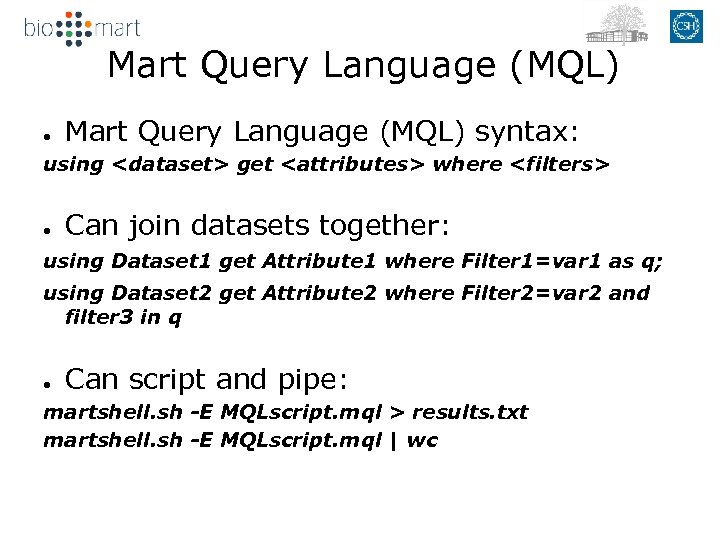
Mart Query Language (MQL) ● Mart Query Language (MQL) syntax: using <dataset> get <attributes> where <filters> ● Can join datasets together: using Dataset 1 get Attribute 1 where Filter 1=var 1 as q; using Dataset 2 get Attribute 2 where Filter 2=var 2 and filter 3 in q ● Can script and pipe: martshell. sh -E MQLscript. mql > results. txt martshell. sh -E MQLscript. mql | wc
Third party software • Bioconductor (bioma. Rt) – Bio. Mart schema • Taverna – Bio. Mart java library • DAS Pro. Server – Bio. Mart perl library
bioma. Rt
Taverna

Pro. Server • No programming • DAS request and responses defined by Exportables and Importables and configured by Mart. Editor • DAS 1
Bio. Mart deployers • Large scale data federation (EBI) • Optimising access to a large database (Ensembl, Worm. Base) • Connecting priopriatery datasets to public data (Pasteur, Unilever, Serono, Sanofi-Aventis, Dev. Gen etc …)
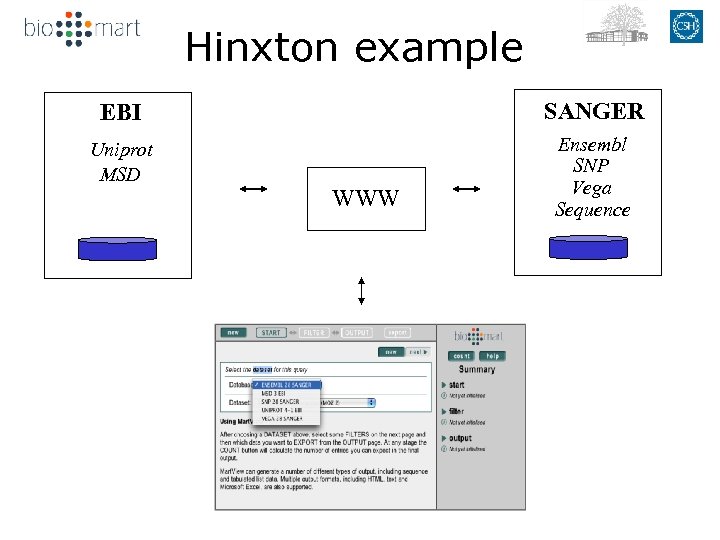
Hinxton example EBI SANGER Uniprot MSD Ensembl SNP Vega Sequence WWW
Bio. Mart deployers • Large scale data federation (Hinxton) • Optimising access to a large database (Ensembl, Worm. Base, Array. Express) • Connecting priopriatery datasets to public data (Pasteur, Unilever, Serono, Sanofi-Aventis, Dev. Gen etc …)
Worm. Base
Ensembl
Array. Express
Bio. Mart deployers • Large scale data federation (Hinxton) • Optimising access to a large database (Ensembl, Worm. Base) • Federating user data with public data (Pasteur, INRA, Bayer, Unilever, Serono, Sanofi-Aventis, Dev. Gen, Solexa etc …)
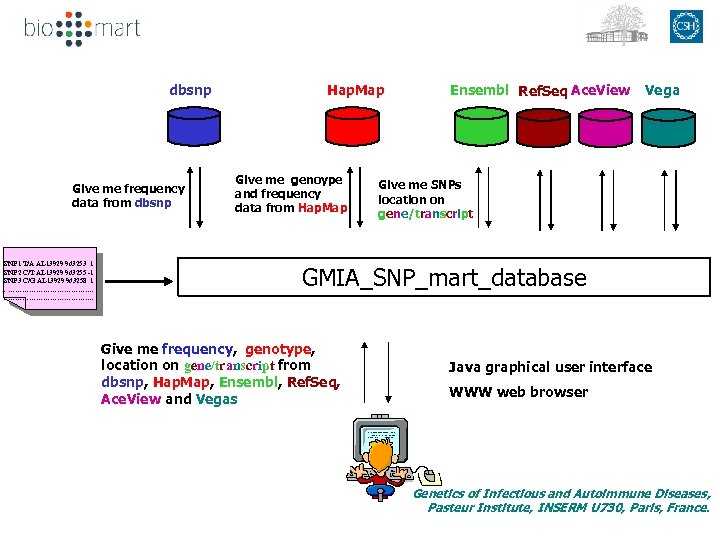
dbsnp Give me genoype and frequency data from Hap. Map Give me frequency data from dbsnp SNP 1 T/A AL 13929 963253 1 SNP 2 C/T AL 13929 963255 -1 SNP 3 C/G AL 13929 963258 1. ………………………………. Hap. Map Ensembl Ref. Seq Ace. View Vega Give me SNPs location on gene/transcript GMIA_SNP_mart_database Give me frequency, genotype, location on gene/transcript from dbsnp, Hap. Map, Ensembl, Ref. Seq, Ace. View and Vegas Java graphical user interface WWW web browser Genetics of Infectious and Autoimmune Diseases, Pasteur Institute, INSERM U 730, Paris, France.
… what next ?
Bio. Mart model • Already applied – – – – Ensembl Vega SNP Uniprot MSD Array. Express Worm. Base Variety of ‘in house’ projects • In development – Hap. Map
Summary • Bio. Mart interface – Batch queries – ‘Data mining’ – Large annotation • Bio. Mart software – Set up your own database – Make your database scalable and responsive – Federate with other data

Where are we? • 0. 2 released in february • 0. 3 to be released in june – Platforms • Mysql • Oracle • Postgres

Acknowledgments • Bio. Mart – Damian Smedley (EBI) – Darin London (EBI) – Will Spooner (CSHL) • Contributors – – – Arne Stabenau (Ensembl) Andreas Kahari (Ensembl) Craig Melsopp (Ensembl) Katerina Tzouvara (Uniprot) Paul Donlon (Unilever)
06a39189ebebbac788ba34831177d0d4.ppt