ceb08113be77ab66f87b8c32ce809fb3.ppt
- Количество слайдов: 27
Bio. DCV: a grid-enabled complete validation setup for functional profiling http: //mpa. itc. it Silvano Paoli, Davide Albanese, Giuseppe Jurman, Annalisa Barla, Stefano Merler, Roberto Flor, Stefano Cozzini, James Reid, Cesare Furlanello Trieste, Feb 2006
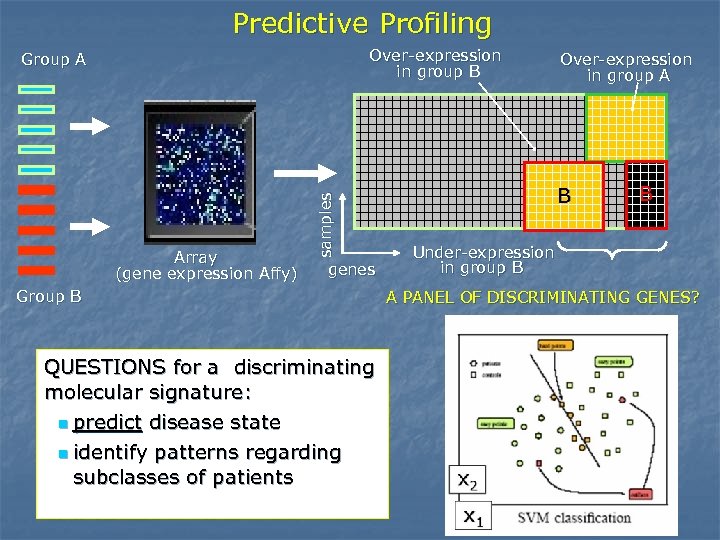
Predictive Profiling Over-expression in group B Array (gene expression Affy) samples Group A genes Group B QUESTIONS for a discriminating molecular signature: n predict disease state n identify patterns regarding subclasses of patients Over-expression in group A B B Under-expression in group B A PANEL OF DISCRIMINATING GENES?
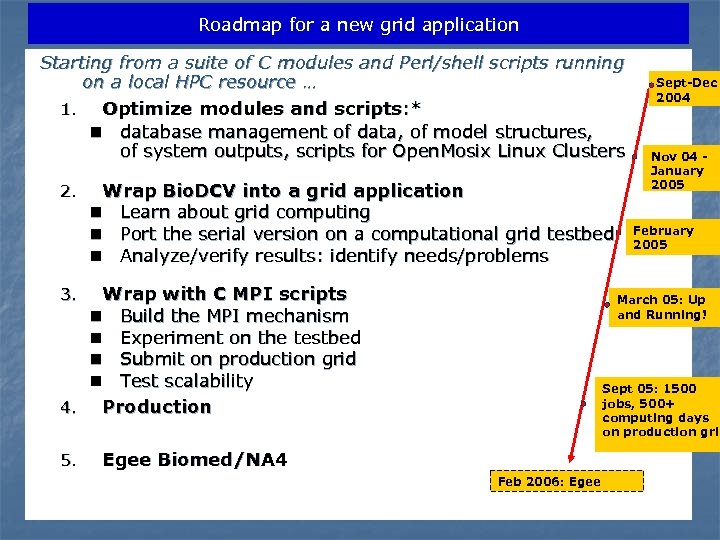
Roadmap for a new grid application Starting from a suite of C modules and Perl/shell scripts running on a local HPC resource … 1. Optimize modules and scripts: * n database management of data, of model structures, of system outputs, scripts for Open. Mosix Linux Clusters 2. Wrap Bio. DCV into a grid application n Learn about grid computing n Port the serial version on a computational grid testbed n Analyze/verify results: identify needs/problems Wrap with C MPI scripts n Build the MPI mechanism n Experiment on the testbed n Submit on production grid n Test scalability 4. Production 3. 5. Sept-Dec 2004 Nov 04 January 2005 February 2005 March 05: Up and Running! Sept 05: 1500 jobs, 500+ computing days on production grid Egee Biomed/NA 4 Feb 2006: Egee
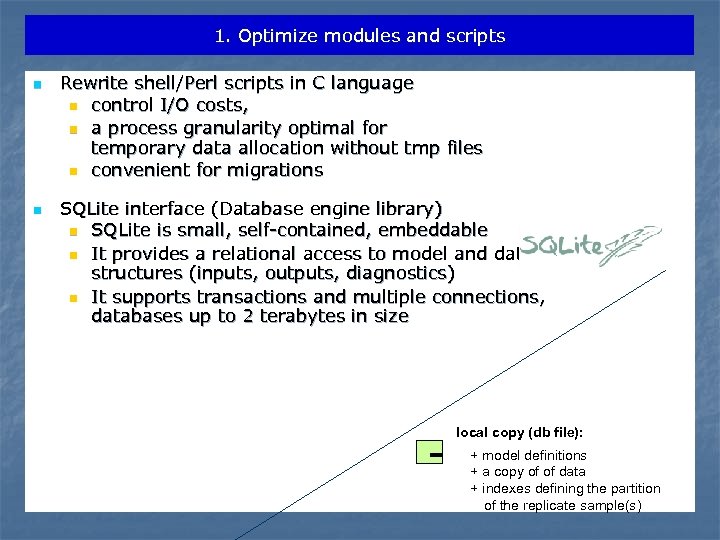
1. Optimize modules and scripts n n Rewrite shell/Perl scripts in C language n control I/O costs, n a process granularity optimal for temporary data allocation without tmp files n convenient for migrations SQLite interface (Database engine library) n SQLite is small, self-contained, embeddable n It provides a relational access to model and data structures (inputs, outputs, diagnostics) n It supports transactions and multiple connections, databases up to 2 terabytes in size local copy (db file): + model definitions + a copy of of data + indexes defining the partition of the replicate sample(s)
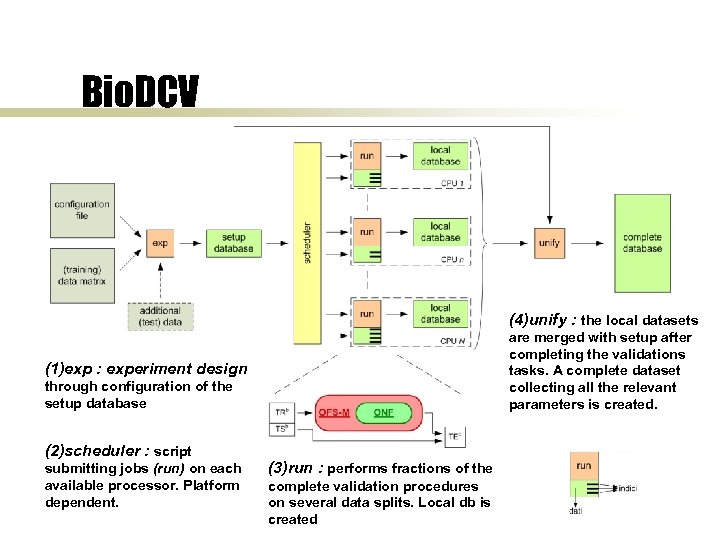
Bio. DCV (4)unify : the local datasets are merged with setup after completing the validations tasks. A complete dataset collecting all the relevant parameters is created. (1)exp : experiment design through configuration of the setup database (2)scheduler : script submitting jobs (run) on each available processor. Platform dependent. (3)run : performs fractions of the complete validation procedures on several data splits. Local db is created

2. Wrapping into a grid application n n Why porting into the grid? n Because we do not have “enough” computational resources… How to port the Bio. DCV in grid? n n PRELIMINARY n Identify a collaborator with experience in grid computing (e. g. the Egrid Project hosted at ICTP http: //www. egrid. it ) n Train human resources (SP Trieste) n Join the Egrid testbed (installing a grid site in Trento) HANDS-ON n n n Porting of the serial application on the testbed patch code as needed: code portability is mandatory to make life easier Identify requirements/problems
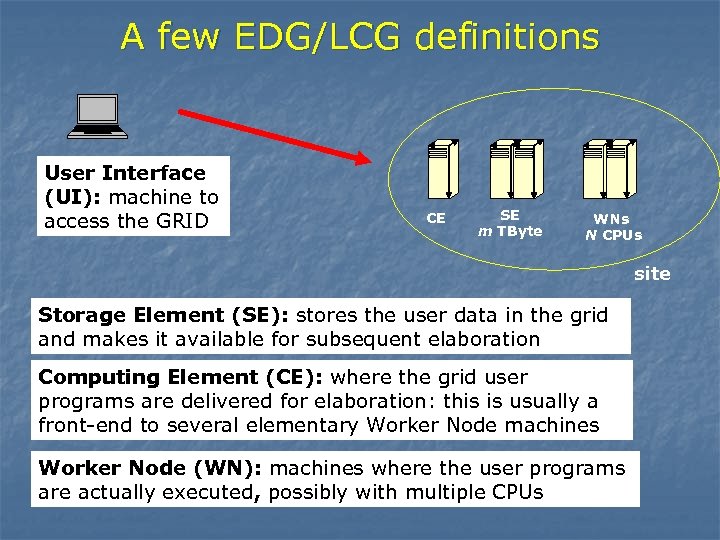
A few EDG/LCG definitions User Interface (UI): machine to access the GRID CE SE m TByte WNs N CPUs site Storage Element (SE): stores the user data in the grid and makes it available for subsequent elaboration Computing Element (CE): where the grid user programs are delivered for elaboration: this is usually a front-end to several elementary Worker Node machines Worker Node (WN): machines where the user programs are actually executed, possibly with multiple CPUs
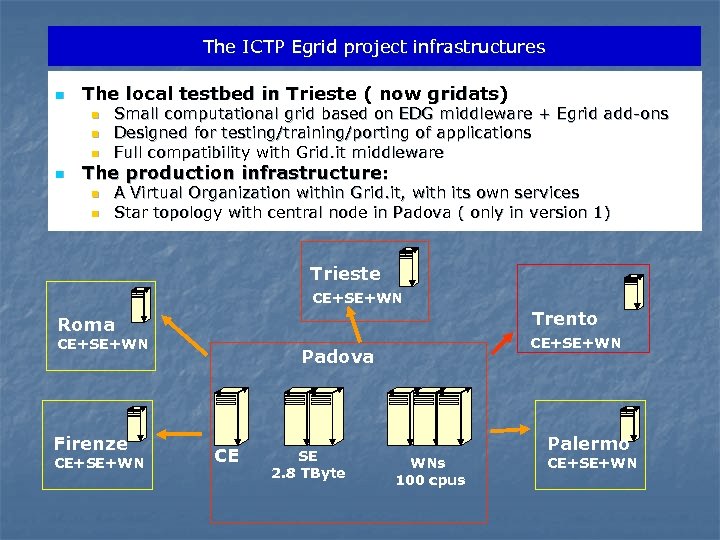
The ICTP Egrid project infrastructures n The local testbed in Trieste ( now gridats) n n Small computational grid based on EDG middleware + Egrid add-ons Designed for testing/training/porting of applications Full compatibility with Grid. it middleware The production infrastructure: n n A Virtual Organization within Grid. it, with its own services Star topology with central node in Padova ( only in version 1) Trieste CE+SE+WN Trento Roma CE+SE+WN Firenze CE+SE+WN Padova CE SE 2. 8 TByte WNs 100 cpus Palermo CE+SE+WN
Hands on n Porting the serial application n n Testing/Evaluation Problems identified: n n n Easy task due to portability (no actual work needed) No software/library dependencies Job submission overhead due to EDG mechanisms Managing multiple (~hundreds/thousands) jobs is difficult and cumbersome Answer: parallellize jobs on the GRID via MPI n n Single submission Multiple executions
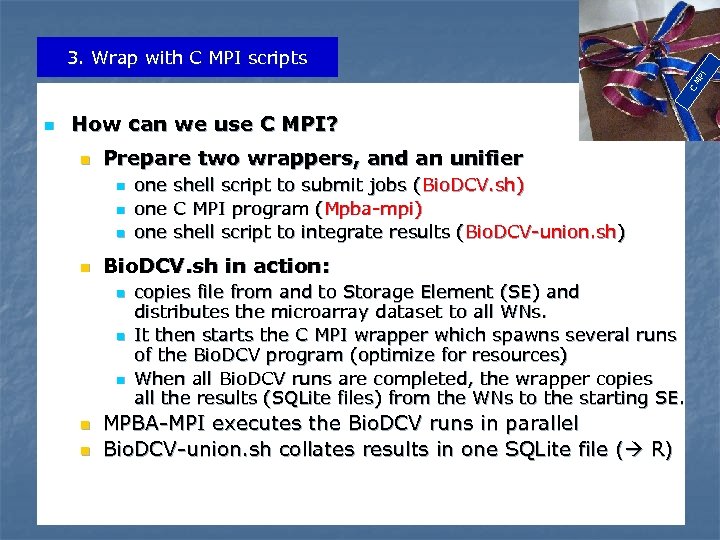
C MP I 3. Wrap with C MPI scripts n How can we use C MPI? n Prepare two wrappers, and an unifier n n Bio. DCV. sh in action: n n n one shell script to submit jobs (Bio. DCV. sh) one C MPI program (Mpba-mpi) one shell script to integrate results (Bio. DCV-union. sh) copies file from and to Storage Element (SE) and distributes the microarray dataset to all WNs. It then starts the C MPI wrapper which spawns several runs of the Bio. DCV program (optimize for resources) When all Bio. DCV runs are completed, the wrapper copies all the results (SQLite files) from the WNs to the starting SE. MPBA-MPI executes the Bio. DCV runs in parallel Bio. DCV-union. sh collates results in one SQLite file ( R)
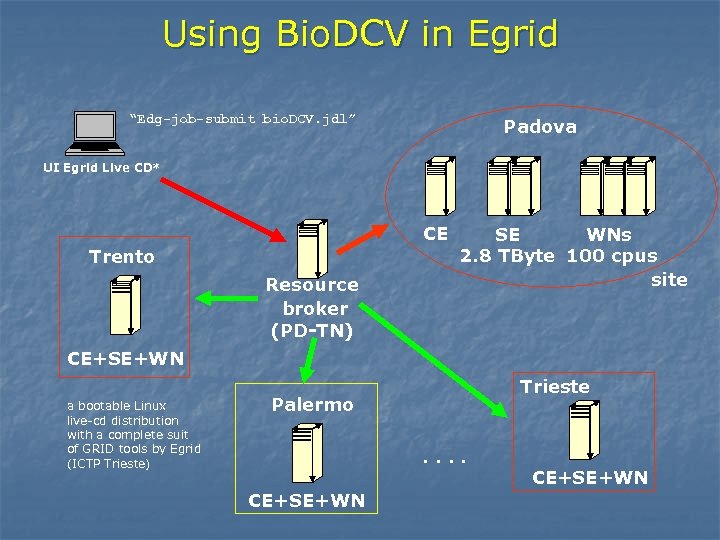
Using Bio. DCV in Egrid “Edg-job-submit bio. DCV. jdl” Padova UI Egrid Live CD* CE Trento Resource broker (PD-TN) SE WNs 2. 8 TByte 100 cpus site CE+SE+WN a bootable Linux live-cd distribution with a complete suit of GRID tools by Egrid (ICTP Trieste) Trieste Palermo. . CE+SE+WN
![A Job Description [ ] Bio. DCV. jdl Type = "Job"; Job. Type = A Job Description [ ] Bio. DCV. jdl Type = "Job"; Job. Type =](https://present5.com/presentation/ceb08113be77ab66f87b8c32ce809fb3/image-13.jpg)
A Job Description [ ] Bio. DCV. jdl Type = "Job"; Job. Type = "MPICH"; Node. Number = 64; Executable = “Bio. DCV. sh"; Arguments = “Mpba-mpi 64 lfn: /utenti/spaoli/sarcoma. db 400"; Std. Output = "test. out"; Std. Error = "test. err"; Input. Sandbox = {“Bio. DCV. sh", “Mpba-mpi", "run. sh"}; Output. Sandbox = {"test. err", "test. out", "executable. out"}; Requirements = other. Glue. CEInfo. LRMSType == "PBS" || other. Glue. CEInfo. LRMSType == "LSF";
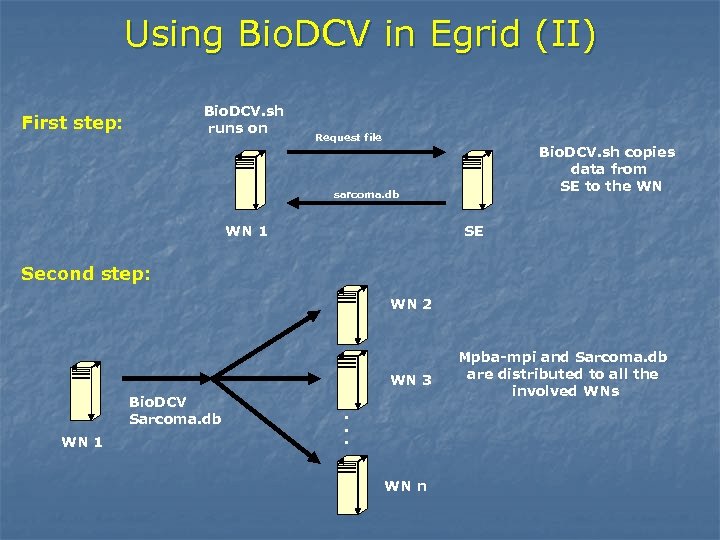
Using Bio. DCV in Egrid (II) Bio. DCV. sh runs on First step: Request file Bio. DCV. sh copies data from SE to the WN sarcoma. db WN 1 SE Second step: WN 2 WN 3 WN 1 . . . Bio. DCV Sarcoma. db WN n Mpba-mpi and Sarcoma. db are distributed to all the involved WNs
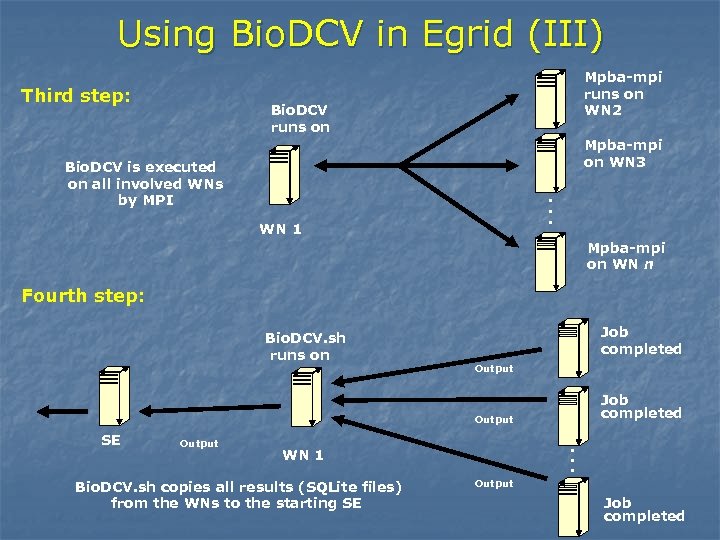
Using Bio. DCV in Egrid (III) Third step: Mpba-mpi runs on WN 2 Bio. DCV runs on Mpba-mpi on WN 3 . . . Bio. DCV is executed on all involved WNs by MPI WN 1 Mpba-mpi on WN n Fourth step: Bio. DCV. sh runs on Job completed Output SE WN 1 Bio. DCV. sh copies all results (SQLite files) from the WNs to the starting SE . . . Output Job completed
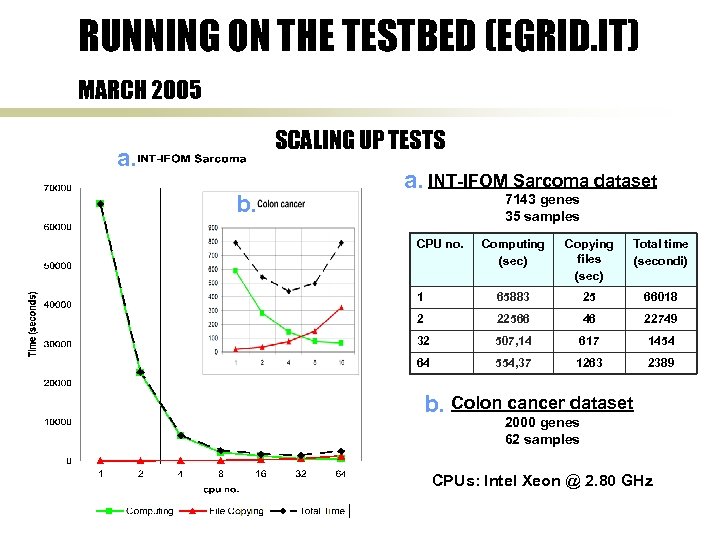
RUNNING ON THE TESTBED (EGRID. IT) MARCH 2005 SCALING UP TESTS a. b. a. INT-IFOM Sarcoma dataset 7143 genes 35 samples CPU no. Computing (sec) Copying files (sec) Total time (secondi) 1 65883 25 66018 2 22566 46 22749 32 507, 14 617 1454 64 554, 37 1263 2389 b. Colon cancer dataset 2000 genes 62 samples CPUs: Intel Xeon @ 2. 80 GHz
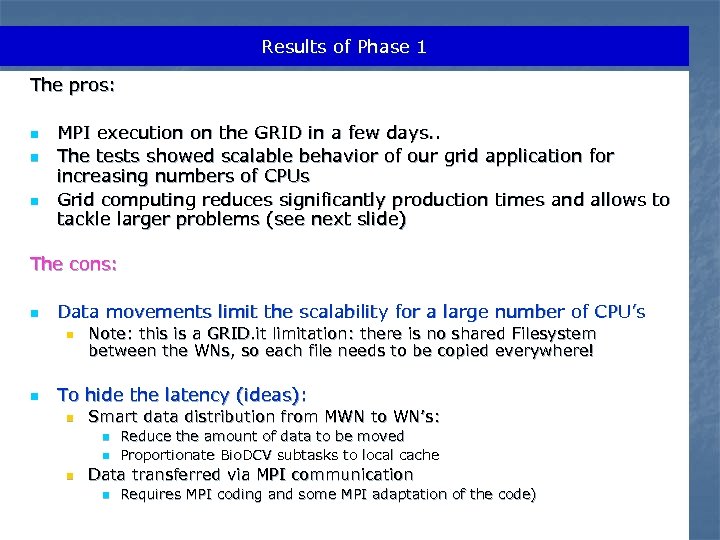
Results of Phase 1 The pros: n n n MPI execution on the GRID in a few days. . The tests showed scalable behavior of our grid application for increasing numbers of CPUs Grid computing reduces significantly production times and allows to tackle larger problems (see next slide) The cons: n Data movements limit the scalability for a large number of CPU’s n n Note: this is a GRID. it limitation: there is no shared Filesystem between the WNs, so each file needs to be copied everywhere! To hide the latency (ideas): n Smart data distribution from MWN to WN’s: n n n Reduce the amount of data to be moved Proportionate Bio. DCV subtasks to local cache Data transferred via MPI communication n Requires MPI coding and some MPI adaptation of the code)
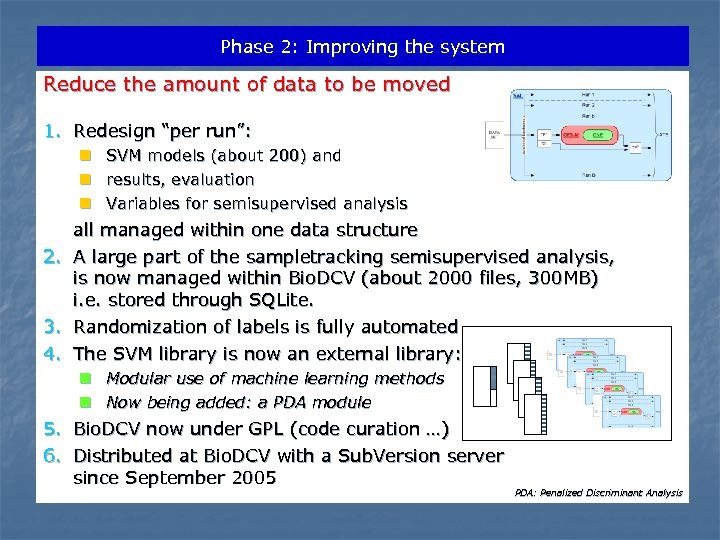
Phase 2: Improving the system Reduce the amount of data to be moved 1. Redesign “per run”: n SVM models (about 200) and n results, evaluation n Variables for semisupervised analysis 2. 3. 4. all managed within one data structure A large part of the sampletracking semisupervised analysis, is now managed within Bio. DCV (about 2000 files, 300 MB) i. e. stored through SQLite. Randomization of labels is fully automated The SVM library is now an external library: n Modular use of machine learning methods n Now being added: a PDA module 5. Bio. DCV now under GPL (code curation …) 6. Distributed at Bio. DCV with a Sub. Version server since September 2005 PDA: Penalized Discriminant Analysis
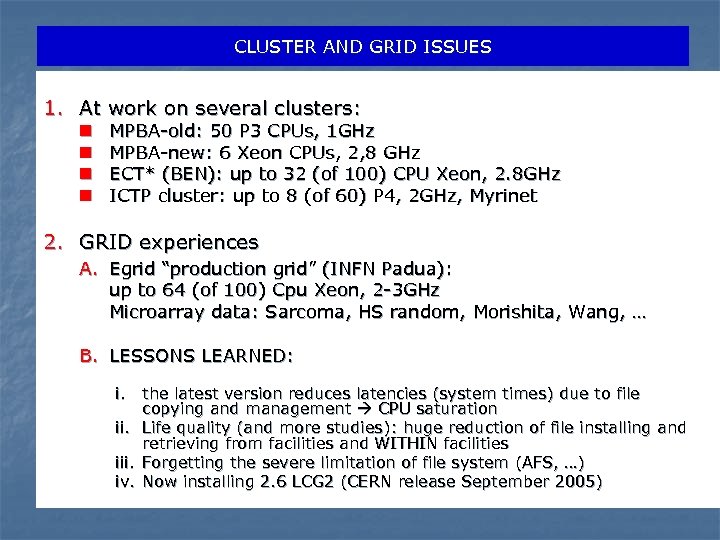
CLUSTER AND GRID ISSUES 1. At work on several clusters: n n MPBA-old: 50 P 3 CPUs, 1 GHz MPBA-new: 6 Xeon CPUs, 2, 8 GHz ECT* (BEN): up to 32 (of 100) CPU Xeon, 2. 8 GHz ICTP cluster: up to 8 (of 60) P 4, 2 GHz, Myrinet 2. GRID experiences A. Egrid “production grid” (INFN Padua): up to 64 (of 100) Cpu Xeon, 2 -3 GHz Microarray data: Sarcoma, HS random, Morishita, Wang, … B. LESSONS LEARNED: i. the latest version reduces latencies (system times) due to file copying and management CPU saturation ii. Life quality (and more studies): huge reduction of file installing and retrieving from facilities and WITHIN facilities iii. Forgetting the severe limitation of file system (AFS, …) iv. Now installing 2. 6 LCG 2 (CERN release September 2005)
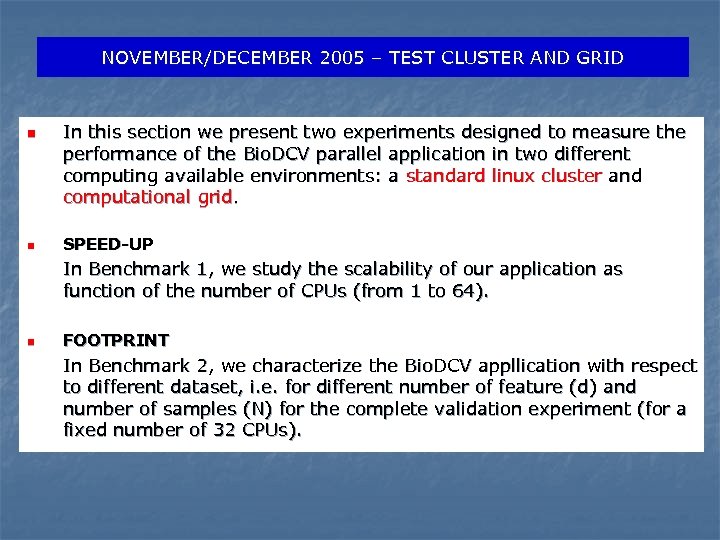
NOVEMBER/DECEMBER 2005 – TEST CLUSTER AND GRID n n In this section we present two experiments designed to measure the performance of the Bio. DCV parallel application in two different computing available environments: a standard linux cluster and computational grid. SPEED-UP In Benchmark 1, we study the scalability of our application as function of the number of CPUs (from 1 to 64). n FOOTPRINT In Benchmark 2, we characterize the Bio. DCV appllication with respect to different dataset, i. e. for different number of feature (d) and number of samples (N) for the complete validation experiment (for a fixed number of 32 CPUs).
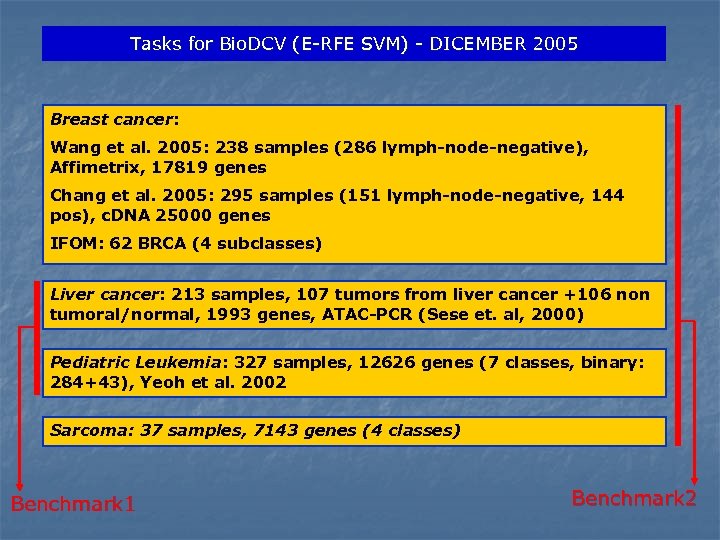
Tasks for Bio. DCV (E-RFE SVM) - DICEMBER 2005 Breast cancer: Wang et al. 2005: 238 samples (286 lymph-node-negative), Affimetrix, 17819 genes Chang et al. 2005: 295 samples (151 lymph-node-negative, 144 pos), c. DNA 25000 genes IFOM: 62 BRCA (4 subclasses) Liver cancer: 213 samples, 107 tumors from liver cancer +106 non tumoral/normal, 1993 genes, ATAC-PCR (Sese et. al, 2000) Pediatric Leukemia: 327 samples, 12626 genes (7 classes, binary: 284+43), Yeoh et al. 2002 Sarcoma: 37 samples, 7143 genes (4 classes) Benchmark 1 Benchmark 2
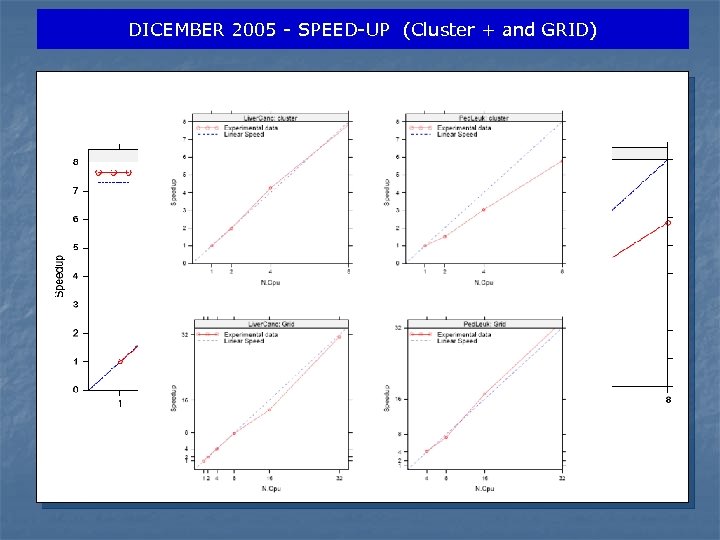
DICEMBER 2005 - SPEED-UP (Cluster + and GRID)
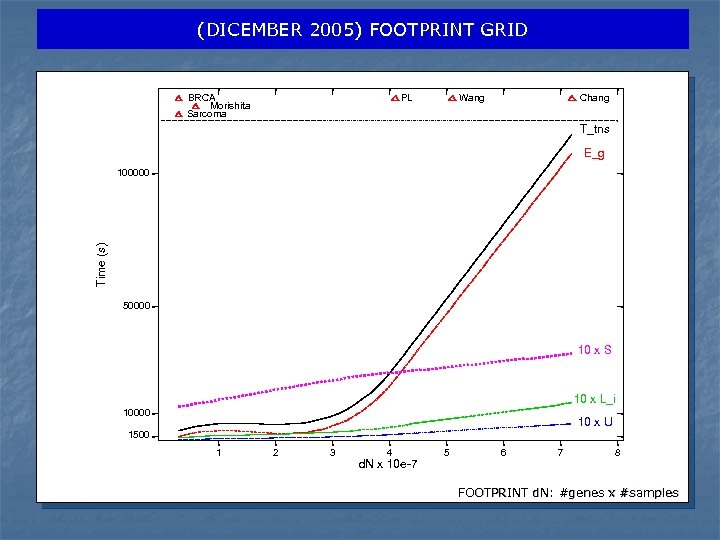
(DICEMBER 2005) FOOTPRINT GRID BRCA Morishita Sarcoma PL Wang Chang T_tns E_g Time (s) 100000 50000 10 x S 10 x L_i 10000 10 x U 1500 1 2 3 4 d. N x 10 e-7 5 6 7 8 FOOTPRINT d. N: #genes x #samples

NOVEMBER/DECEMBER 2005 –DISCUSSION n Two experiments for 139 CPU day in Egrid infrastructure n In benchmark 1, we obtain a speed-up curve very close to linear n In Benchmark 2, effective execution time increases linearly with the dataset footprint, i. e. the product of number of genes and number of samples n Performance penalty payed for data transfer from WNs and queuing policy (best effort) respect on a local linux cluster n We will investigate if substituting an MPI approach with a model that submit N different single CPU jobs n Bio. DCV system on LCG/EGEE computational grid can be used in pratical large scale experiments n Next is porting our system under EGEE’s Biomed VO.
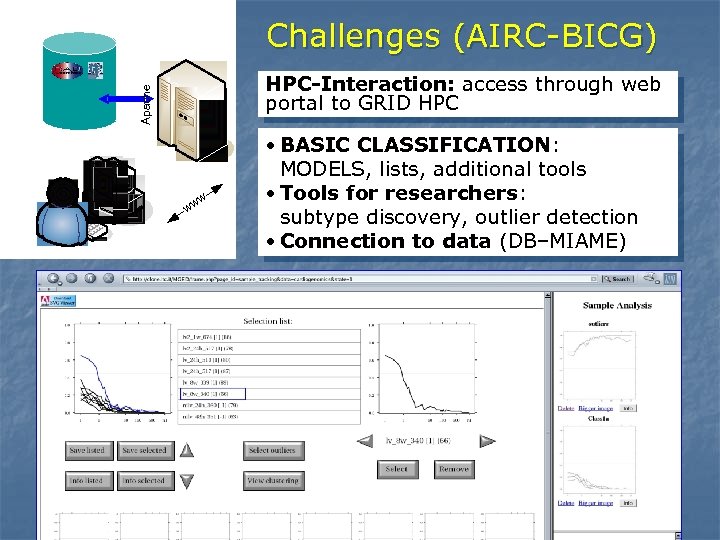
Challenges (AIRC-BICG) Apache HPC-Interaction: access through web portal to GRID HPC w ww • BASIC CLASSIFICATION: MODELS, lists, additional tools • Tools for researchers: subtype discovery, outlier detection • Connection to data (DB–MIAME)
Acknowledgments ITC-irst, Trento Alessandro Soraruf ICTP E-GRID Project, Trieste Angelo Leto Cristian Zoicas Riccardo Murri Ezio Corso Alessio Terpin IFOM-FIRC and INT, Milano Manuela Gariboldi Marco A. Pierotti Grants: BICG (AIRC) Democritos Data: IFOM-FIRCC Cardiogenomics PGA
ceb08113be77ab66f87b8c32ce809fb3.ppt