274053e3c8edf9bccccbc468cf534f6b.ppt
- Количество слайдов: 24

Big data and Life Sciences Guy Coates Wellcome Trust Sanger Institute gmpc@sanger. ac. uk

The Sanger Institute Funded by Wellcome Trust. • • • 2 nd largest research charity in the world. ~700 employees. Based in Hinxton Genome Campus, Cambridge, UK. Large scale genomic research. • • Sequenced 1/3 of the human genome. (largest single contributor). Large scale sequencing with an impact on human and animal health. Data is freely available. • Websites, ftp, direct database access, programmatic APIs. • Some restrictions for potentially identifiable data. My team: • Scientific computing systems architects.
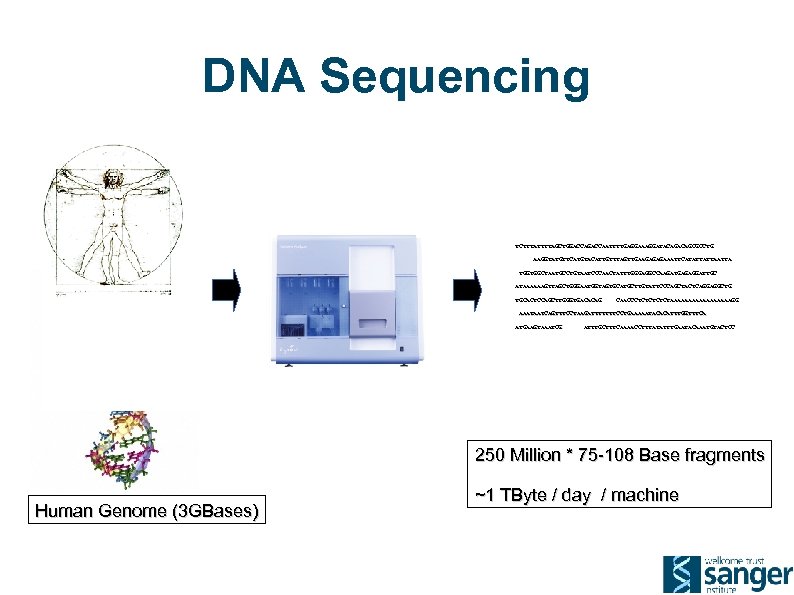
DNA Sequencing TCTTTATTTTAGCTGGACCAATTTTGAGGAAAGGATACAGCGCCTG AAGGTATGTTCATGTACATTGTTTAGTTGAAGAGAGAAATTCATATTATTA TGGTGGCTAATGCCTGTAATCCCAACTATTTGGGAGGCCAAGATGAGAGGATTGC ATAAAAAAGTTAGCTGGGAATGGTAGTGCATGCTTGTATTCCCAGCTACTCAGGAGGCTG TGCACTCCAGCTTGGGTGACACAG CAACCCTCTCTAAAAAAAAAGG AAATAATCAGTTTCCTAAGATTTTTTTCCTGAAAAATACACATTTGGTTTCA ATGAAGTAAATCG ATTTGCTTTCAAAACCTTTATATTTGAATACAAATGTACTCC 250 Million * 75 -108 Base fragments Human Genome (3 GBases) ~1 TByte / day / machine

Economic Trends: Cost of sequencing halves every 12 months. • Wrong side of Moore's Law. The Human genome project: • • • 13 years. 23 labs. $500 Million. A Human genome today: • • • 3 days. 1 machine. $8, 000. Trend will continue: • $1000 genome is probable within 2 years. • Informatics not included.
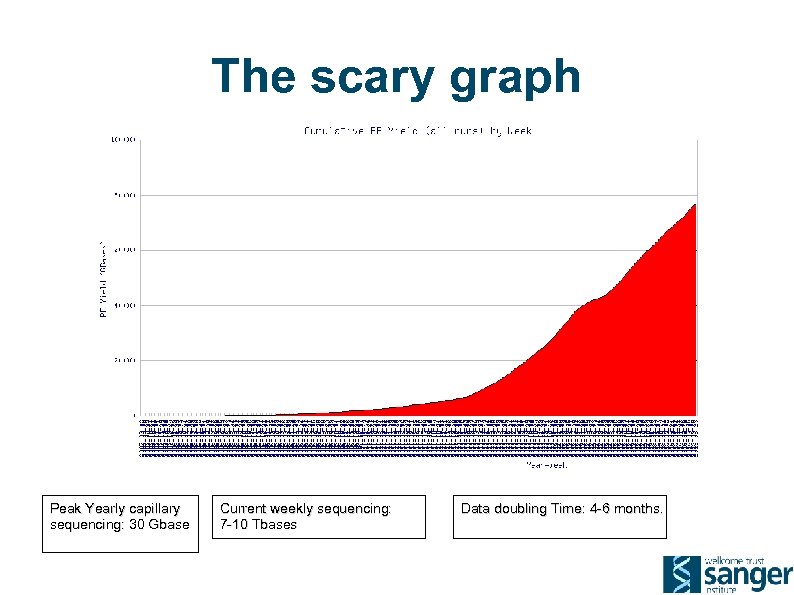
The scary graph Peak Yearly capillary sequencing: 30 Gbase Current weekly sequencing: 7 -10 Tbases Data doubling Time: 4 -6 months.

Gen III Sequencers this year?
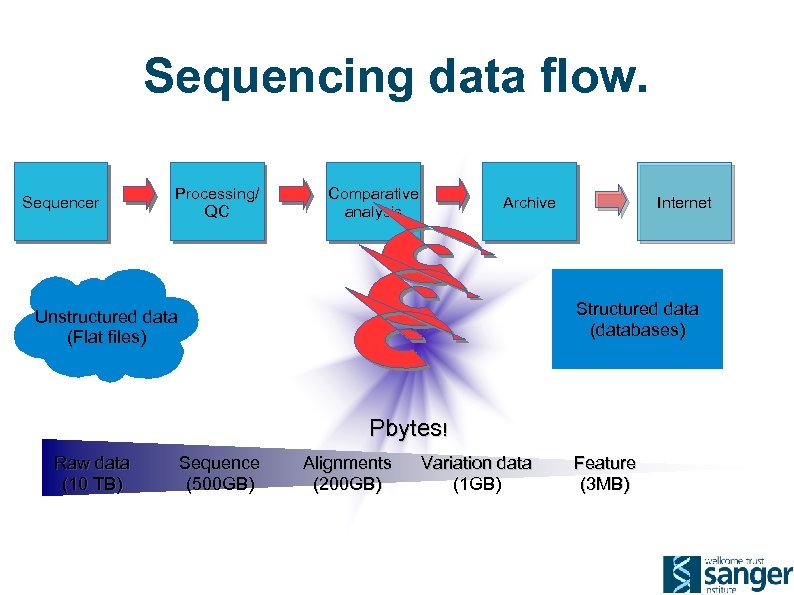
Sequencing data flow. Sequencer Processing/ QC Comparative analysis Archive Internet Structured data (databases) Unstructured data (Flat files) Pbytes! Raw data (10 TB) Sequence (500 GB) Alignments (200 GB) Variation data (1 GB) Feature (3 MB)
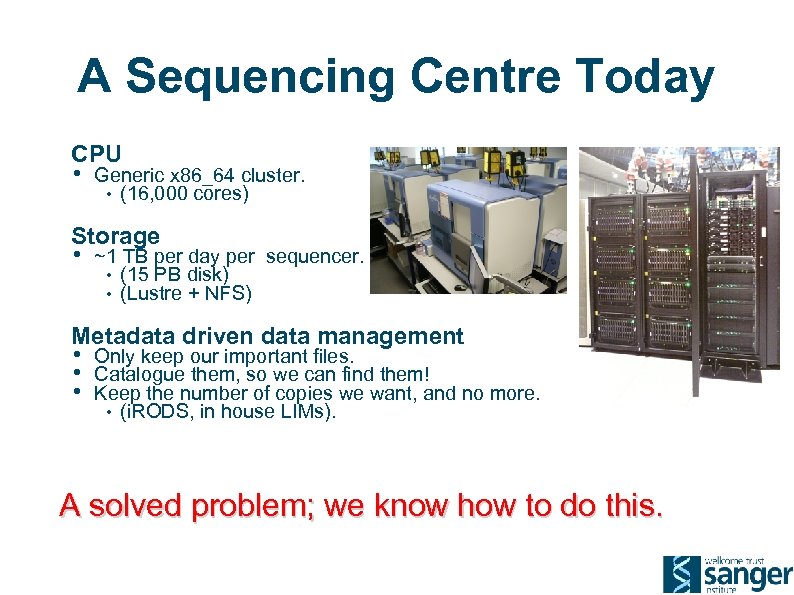
A Sequencing Centre Today CPU • Generic x 86_64 cluster. • (16, 000 cores) Storage • ~1 TB per day per sequencer. • (15 PB disk) • (Lustre + NFS) Metadata driven data management • • • Only keep our important files. Catalogue them, so we can find them! Keep the number of copies we want, and no more. • (i. RODS, in house LIMs). A solved problem; we know how to do this.

This is not big data

This is not big data either. . .
Proper Big Data We want to compute across all the data. • • Sequencing data (of course). Patient records, treatment and outcomes. Why? • • Cancer: tie in genetics, patient outcomes and treatments. Pharma: high failure rate due to genetic factors in drug response. Infectious disease epidemiology. Rare genetic diseases. Many genetic effects are small • Million member cohorts to get good signal: noise.
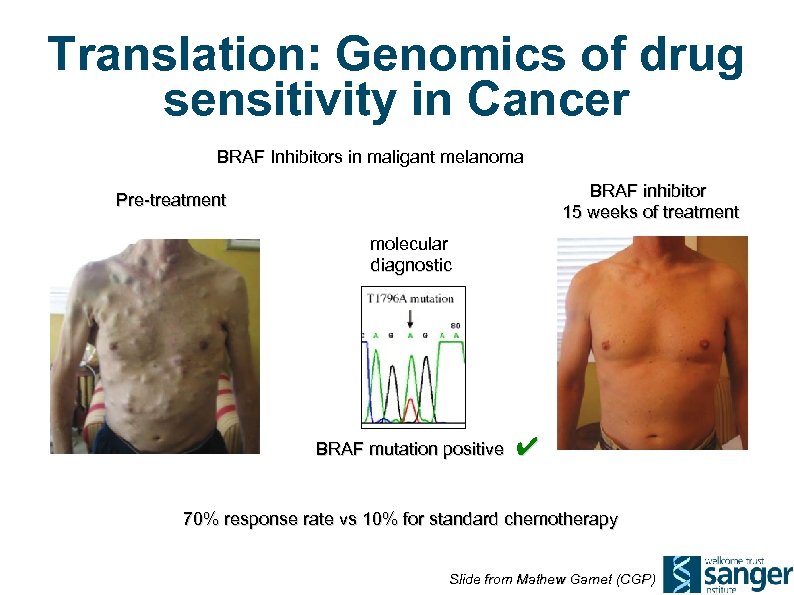
Translation: Genomics of drug sensitivity in Cancer BRAF Inhibitors in maligant melanoma BRAF inhibitor 15 weeks of treatment Pre-treatment molecular diagnostic BRAF mutation positive ✔ 70% response rate vs 10% for standard chemotherapy Slide from Mathew Garnet (CGP)
Current Data Archives EBI ERA / NCBI SRA store results of all sequencing experiments. • Public data availability: A good thing (tm) • 1. 6 Pbases Problems • • • Archives are “dark”. You can put data in, but you can't do anything with it. In order to analyse the data, you need to download it all. • 100 s of Tbytes Situation replicated at local Institute level too. • eg How does CRI get hold of their data currently held at Sanger?

The Vision Global Alliance for sharing genomic and clinical data • 70 research institutes & hospitals (including Sanger, Broad, EBI, BGI, Cancer Research UK) Million cancer genome warehouse • (UC Berkeley)
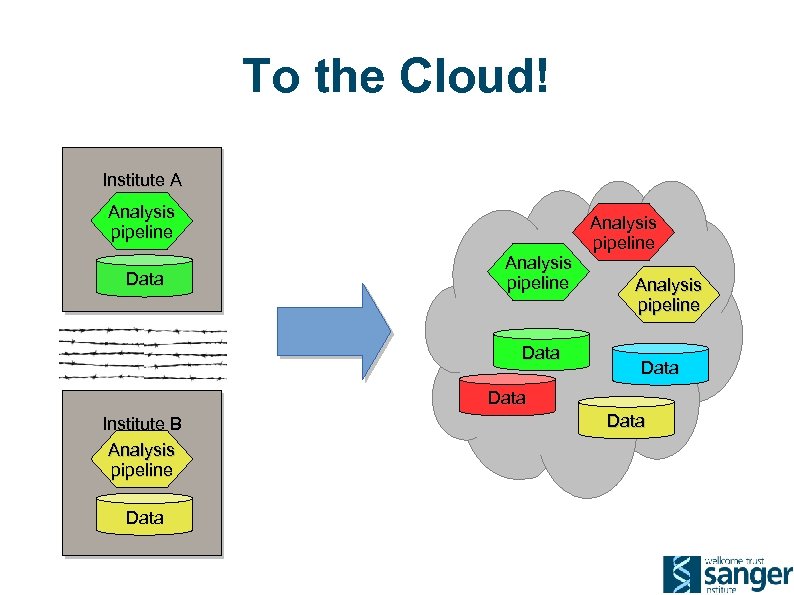
To the Cloud! Institute A Analysis pipeline Data Institute B Analysis pipeline Data

How do we get there?
Code & Algorithms Bioinformatics code: • • Integer not FP heavy. Single threaded. Large memory footprints. Interpreted languages. Not a good fit for future computing architectures. Expensive to run on public clouds. • Memory footprint leads to unused cores. Out of scope for a data talk, but still an important point.
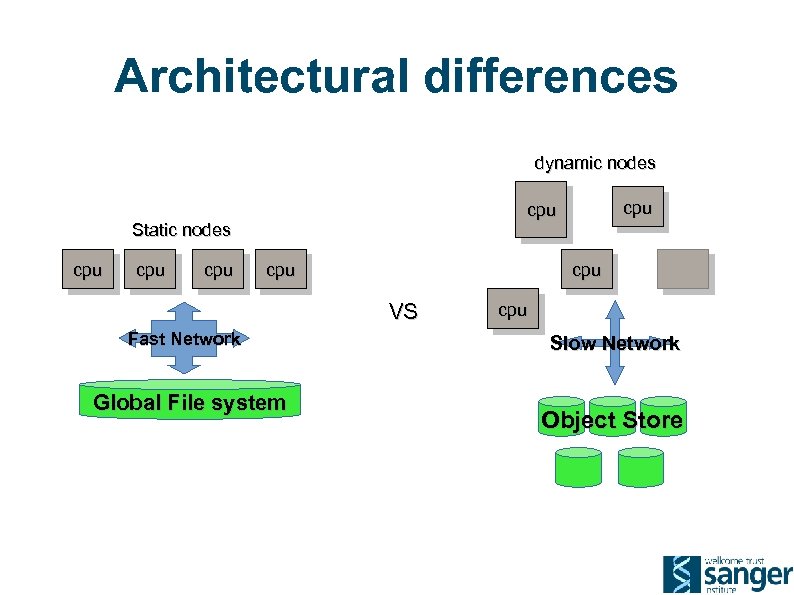
Architectural differences dynamic nodes Static nodes cpu cpu VS Fast Network Global File system cpu Slow Network Object Store

Whose Cloud? A VM is just a VM, right? • • Clouds are supposed to be programmable. Nobody wants to re-write a pipeline when they move clouds. Storage: • • Posix: • (lustre/GPFS/EMC)? Object: • Low level: AWS S 3, Openstack SWIFT, Ceph/rados • High level: Data management layer (eg i. RODS)? Cloud Interoperability? • Do we need is more standards? ! Pragmatic approach: • First person to make one that actually works, wins.
Moving data Data still has to get from our instruments to the Cloud. Good news: • Lots of products out there for wide area data movement. Bad news: • UDT / UDR We are currently using all of them(!) Network bandwidth still a problem. • • Research institutes have fast data networks. What about your GP's surgery? genetorrent rsync / ssh

Identity Access Unlikely that data archives are going to allow anonymous access. • Who are you? Federated identify providers. • • • Is everyone signed up to the same federation? Does it include the right mix of cross-national cooperation? Does your favourite bit of software support federated IDs? Janet Moonshot
The LAW Legal • • Theory: anonymised data can be stored and accessed without jumping through hoops. Practice: Risk of re-identification. Becomes easier the more data you have. • Medical records are hard to anonymise and still be useful. Ethical • Medical consent process adds more restrictions above data-protection law. • Limits data use & access even if anonymised. Controlled data access? • • No ad-hoc analysis. Access via restricted API only (“trusted intermediary model”). Policy development ongoing. • Cross juristiction for added fun.

Summary We know where we want to get to. • No shortage of Vision There are lots of interesting tools and technologies out there. • • • Getting them to work coherently together will be a challenge. Prototyping efforts are underway. Need to leverage expertese and experience in other fields. Not simply technical issues: • • Significant policy issues need to be worked out. We have to bring the public along.

Acknowledgements ISG: • • • James Beal Helen Brimmer Pete Clapham Global Alliance whitepaper: http: //www. sanger. ac. uk/about/press/assets/130605 -white-paper. pdf Million Cancer Genome Warehouse whitepaper http: //www. eecs. berkeley. edu/Pubs/Tech. Rpts/2012/EECS-2012 -211. html
274053e3c8edf9bccccbc468cf534f6b.ppt