098658571a61ed32697c517cef4a2211.ppt
- Количество слайдов: 32
BB 30055: Genes and genomes Genomes - Dr. MV Hejmadi (bssmvh)

BB 30055: Genomes - MVH Recommended texts: 1) Genetics from genes to genomes 2 e - Hartwell et al 2) Human Molecular Genetics 3 – Strachan and Read 4) Genomes 2 - TA Brown 5) Genes VII – Benjamin Lewin Special issue Journals: Nature (2001) 15 th Feb Vol 409 Science (2001) Vol 291 No 5507 Full text of both above journals available at http: //www. bath. ac. uk/library/subjects/bs/links. html#hgp
BB 30055: Genomes - MVH 3 broad areas (A)Genomes, transcriptomes, proteomes (B) Applications of the human genome project (C) Genome evolution
A) Genomes, transcriptomes, proteomes Genome projects - Human Genome Project (HGP): a history - Other genome projects: why do it - Genome organisation - insights from HGP Repeat elements Transposable elements Mitochondrial genomes Y chromosome Post-genomics -transcriptomes - proteomes
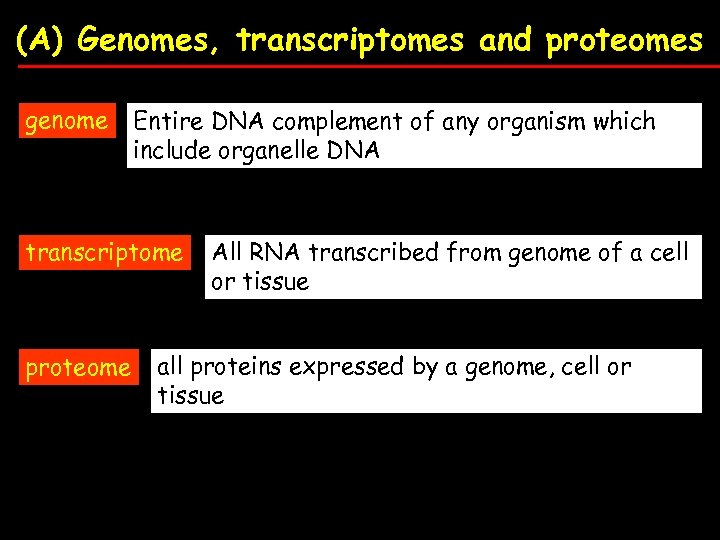
(A) Genomes, transcriptomes and proteomes genome Entire DNA complement of any organism which include organelle DNA transcriptome proteome All RNA transcribed from genome of a cell or tissue all proteins expressed by a genome, cell or tissue

Why study the genome? 3 main reasons • description of sequence of every gene valuable. Includes regulatory regions which help in understanding not only the molecular activities of the cell but also ways in which they are controlled. • identify & characterise important inheritable disease genes or bacterial genes (for industrial use) • Role of intergenic sequences e. g. satellites, intronic regions etc
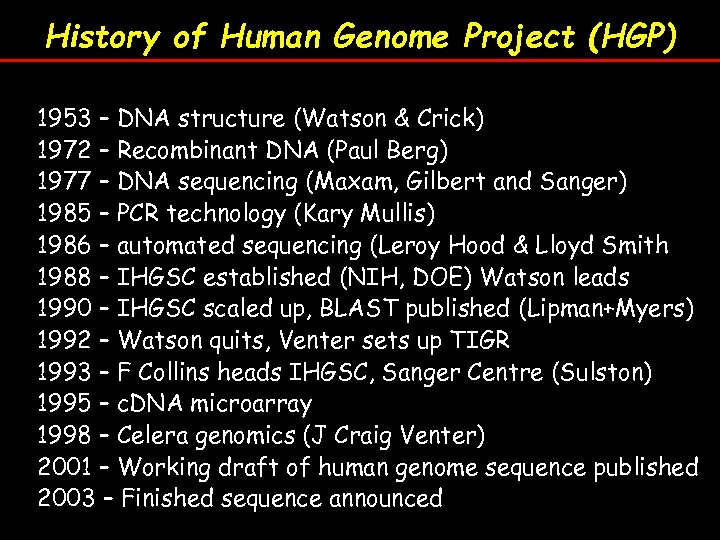
History of Human Genome Project (HGP) 1953 – DNA structure (Watson & Crick) 1972 – Recombinant DNA (Paul Berg) 1977 – DNA sequencing (Maxam, Gilbert and Sanger) 1985 – PCR technology (Kary Mullis) 1986 – automated sequencing (Leroy Hood & Lloyd Smith 1988 – IHGSC established (NIH, DOE) Watson leads 1990 – IHGSC scaled up, BLAST published (Lipman+Myers) 1992 – Watson quits, Venter sets up TIGR 1993 – F Collins heads IHGSC, Sanger Centre (Sulston) 1995 – c. DNA microarray 1998 – Celera genomics (J Craig Venter) 2001 – Working draft of human genome sequence published 2003 – Finished sequence announced

HGP Goal: Obtain the entire DNA sequence of human genome Players: (A) International Human Genome Sequence Consortium (IHGSC) - public funding, free access to all, started earlier - used mapping overlapping clones method (B) Celera Genomics – private funding, pay to view - started in 1998 - used whole genome shotgun strategy

Whose genome is it anyway? (A) International Human Genome Sequence Consortium (IHGSC) - composite from several different people generated from 10 -20 primary samples taken from numerous anonymous donors across racial and ethnic groups (B) Celera Genomics – 5 different donors (one of whom was J Craig Venter himself !!!)
Genomicists looked at two basic features of genomes: sequence and polymorphism Major challenge - to determine sequence of each chromosome in genome and identify polymorphisms – How does one sequence a 500 Mb chromosome 600 bp at a time? – How accurate should a genome sequence be? • DNA sequencing error rate is about 1% per 600 bp – How does one distinguish sequence errors from polymorphisms? • Rate of polymorphism in diploid human genome is about 1 in 500 bp – Repeat sequences may be hard to place – Unclonable DNA cannot be sequenced (30%)

Divide and conquer strategy meets most challenges • Chromosomes are broken into small overlapping pieces and cloned • Ends of clones sequenced and reassembled into original chromosome strings • Each piece is sequenced multiple times to reduce error rate – 10 -fold sequence coverage achieves a rate of error less than 1/10, 000
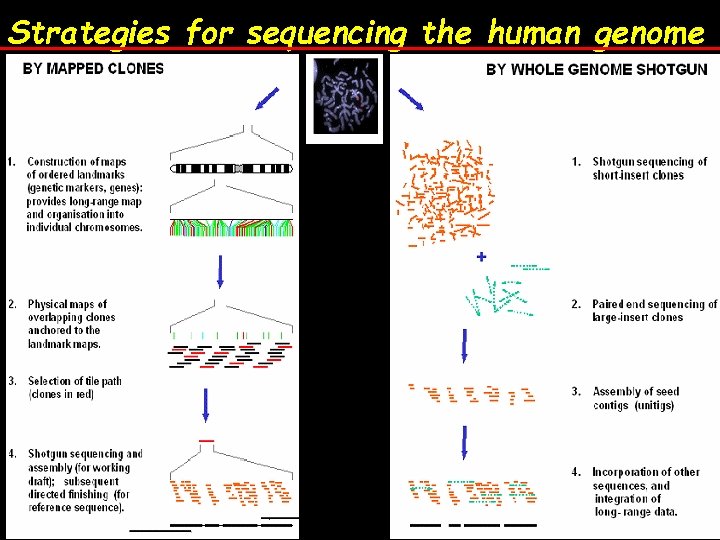
Strategies for sequencing the human genome
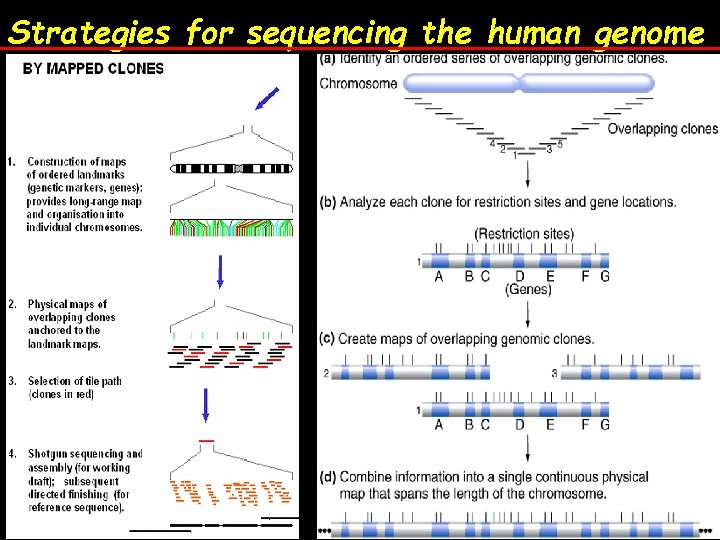
Strategies for sequencing the human genome
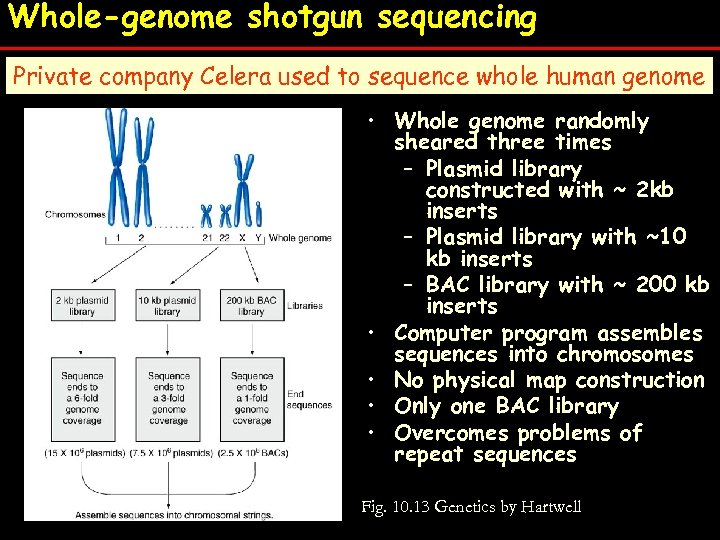
Whole-genome shotgun sequencing Private company Celera used to sequence whole human genome • • • Whole genome randomly sheared three times – Plasmid library constructed with ~ 2 kb inserts – Plasmid library with ~10 kb – Plasmid library with ~10 inserts kb inserts – BAC library with ~ 200 kb Computer program assembles sequences into chromosomes inserts No physical map construction program assembles • Computer sequences into chromosomes Only one BAC library Overcomes problems ofphysical map construction • No repeat sequences • Only one BAC library • Overcomes problems of repeat sequences Fig. 10. 13 Genetics by Hartwell Fig. 10. 13
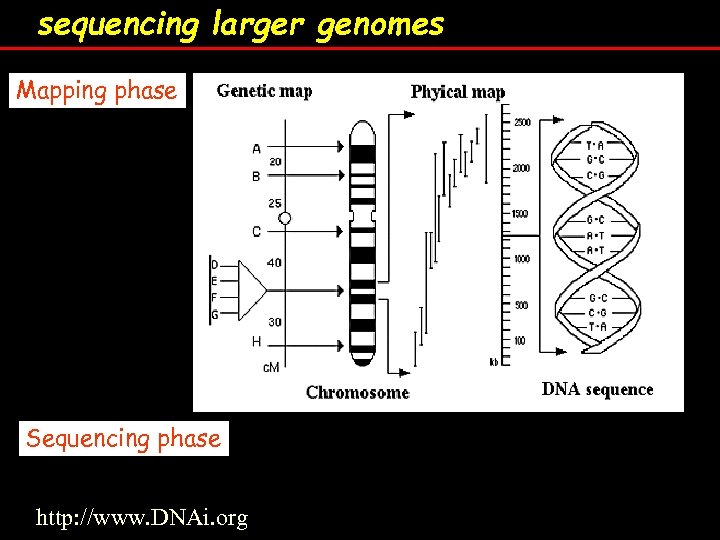
sequencing larger genomes Mapping phase Sequencing phase http: //www. DNAi. org
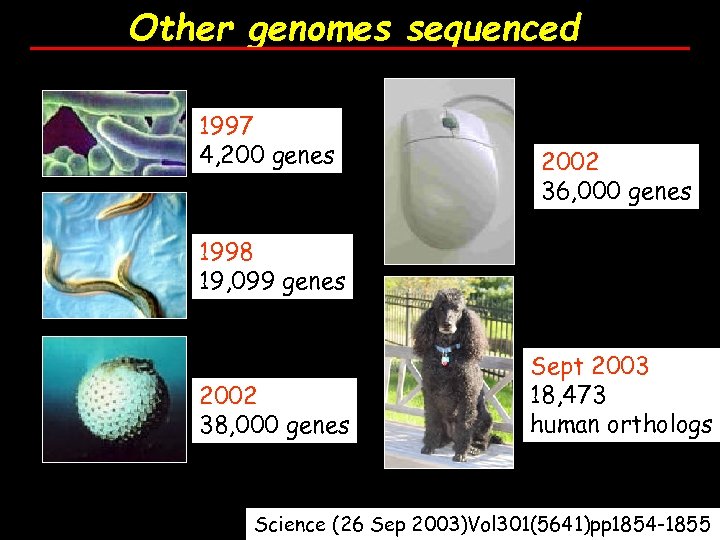
Other genomes sequenced 1997 4, 200 genes 2002 36, 000 genes 1998 19, 099 genes 2002 38, 000 genes Sept 2003 18, 473 human orthologs Science (26 Sep 2003)Vol 301(5641)pp 1854 -1855
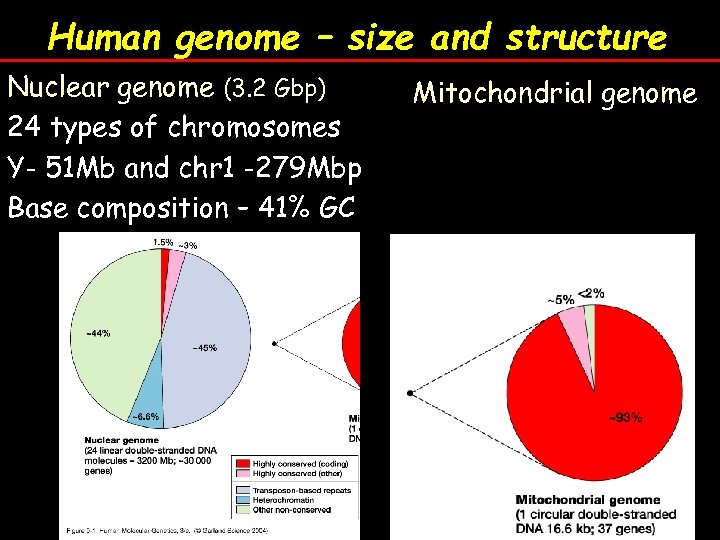
Human genome – size and structure Nuclear genome (3. 2 Gbp) 24 types of chromosomes Y- 51 Mb and chr 1 -279 Mbp Base composition – 41% GC Mitochondrial genome
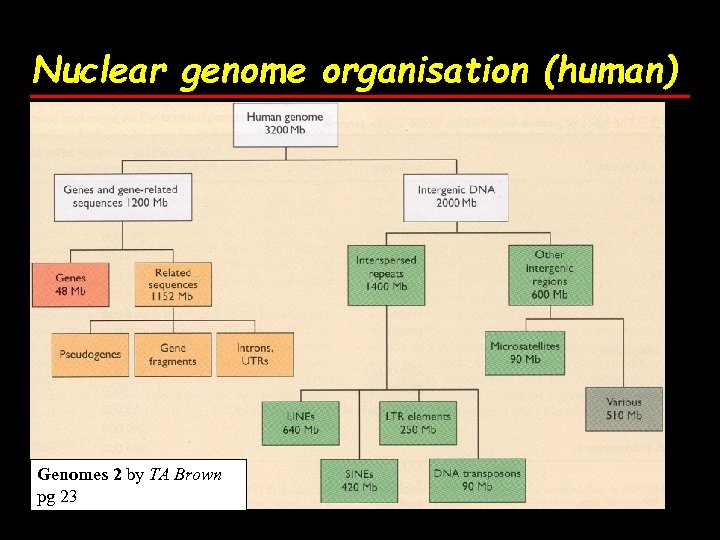
Nuclear genome organisation (human) Genomes 2 by TA Brown pg 23
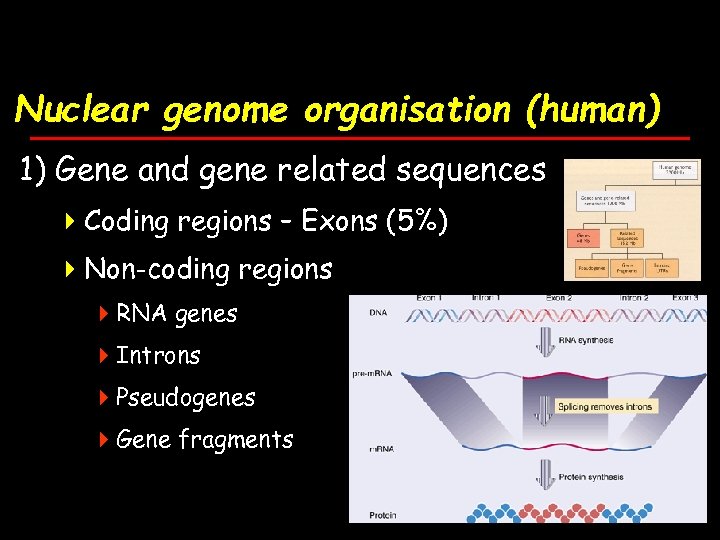
Nuclear genome organisation (human) 1) Gene and gene related sequences 4 Coding regions – Exons (5%) 4 Non-coding regions 4 RNA genes 4 Introns 4 Pseudogenes 4 Gene fragments
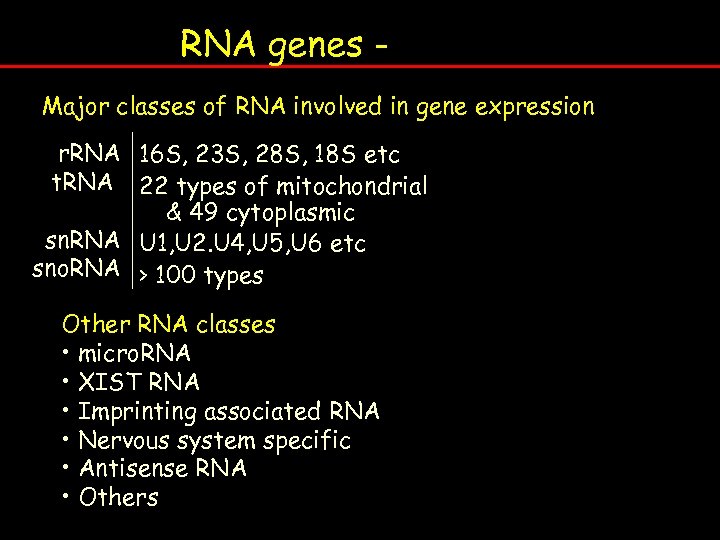
RNA genes - Nuclear genome organisation Major classes of RNA involved in gene expression (human) r. RNA 16 S, 23 S, 28 S, 18 S etc t. RNA 22 types of mitochondrial & 49 cytoplasmic sn. RNA U 1, U 2. U 4, U 5, U 6 etc sno. RNA > 100 types Other RNA classes • micro. RNA • XIST RNA • Imprinting associated RNA • Nervous system specific • Antisense RNA • Others
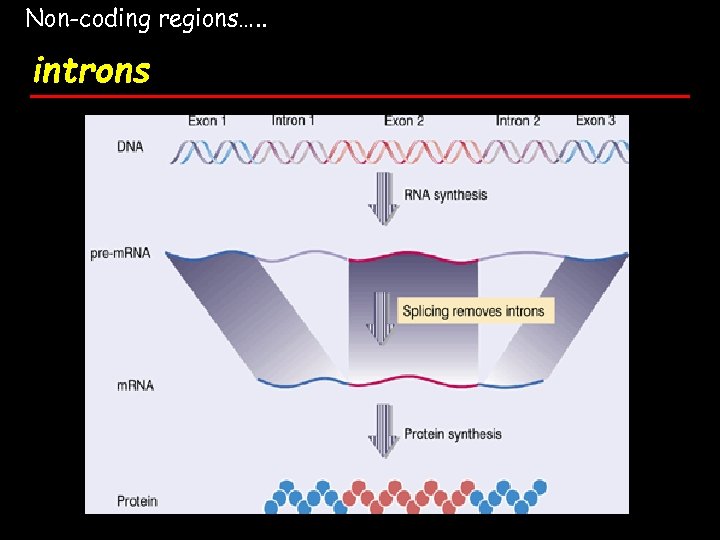
Non-coding regions…. . introns
Non-coding regions…. . Pseudogenes ( ) A non functional copy of most or all of a gene Inactivated by mutations that may cause either 8 inhibition of signal for initiation or transcription 8 prevent splicing at exon-intron boundary 8 premature termination of translation Human Mol Gen 3 by Strachan & Read pgs 262 -264
Non-coding regions…. . Pseudogenes ( ) Different classes include 4 Non-processed: 4 contain non functional copies of genomic DNA sequence incl exons and introns 4 arise from gene duplication events E. g. rabbit pseudogene b 2
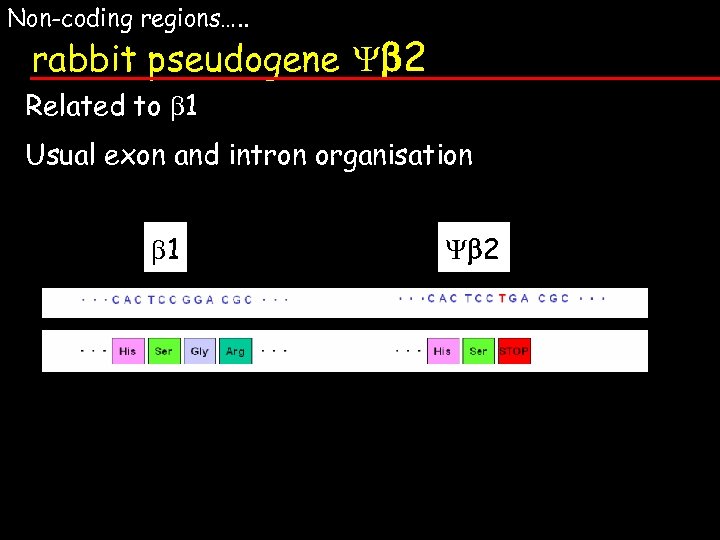
Non-coding regions…. . rabbit pseudogene b 2 Related to b 1 Usual exon and intron organisation b 1 b 2
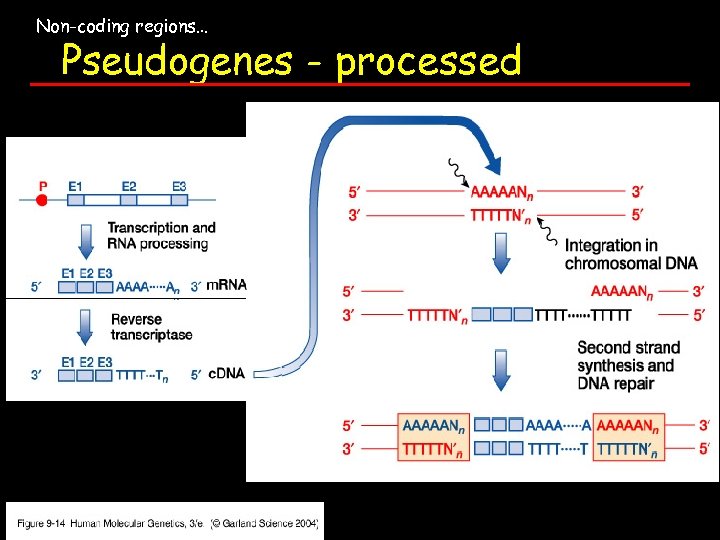
Non-coding regions… Pseudogenes - processed
Non-coding regions… Pseudogenes - processed non functional copies of exonic sequences of an active gene. Thought to arise by genomic insertion of a c. DNA as a result of retroposition 4 Expressed processed: processed pseudogene integrated adjacent to a promoter site Contribute to overall repetitive elements
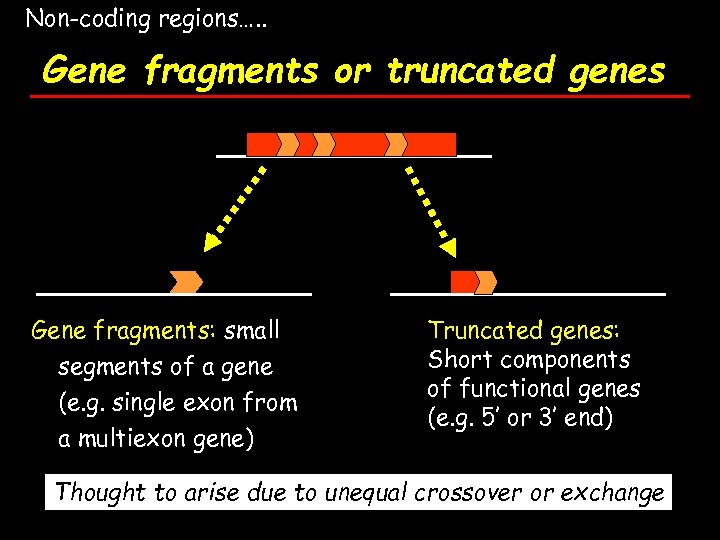
Non-coding regions…. . Gene fragments or truncated genes Gene fragments: small segments of a gene (e. g. single exon from a multiexon gene) Truncated genes: Short components of functional genes (e. g. 5’ or 3’ end) Thought to arise due to unequal crossover or exchange
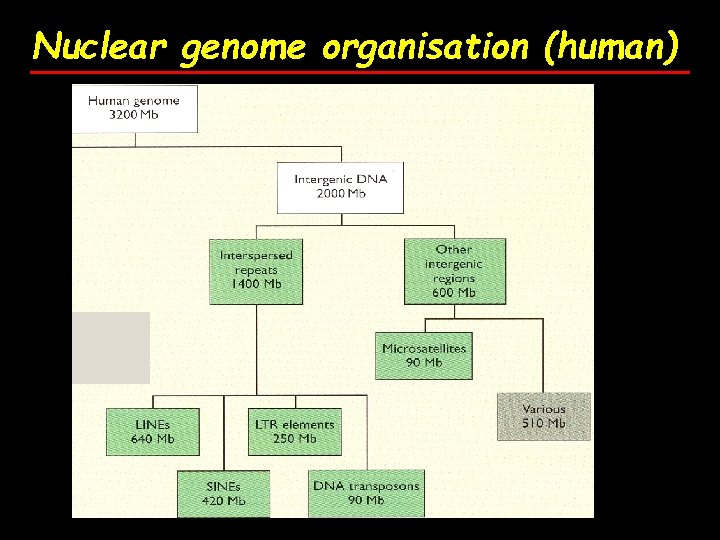
Nuclear genome organisation (human)
Nuclear genome organisation (human) 2) Extragenic (intergenic) DNA (~62% of genome) A) Unique or low copy number sequences B) Repetitive sequences (~ 53%)

A) Unique or low copy number sequences Non –coding, non repetitive and single copy sequences of no known function or significance
B) Repetitive sequences Significance Evolutionary ‘signposts’ 4 Passive markers for mutation assays 4 Actively reorganise gene organisation by creating, shuffling or modifying existing genes Chromosome structure and dynamics Provide tools for medical, forensic, genetic analysis
098658571a61ed32697c517cef4a2211.ppt