8ab5bb59eab87d245159f78e9a9cd37c.ppt
- Количество слайдов: 31

Background: survey of multiple microbial genome browsers
Scale of a genome browser • One model genome, or a few representatives of one species • Flybase, Wormbase • Several genomes of a certain type • Geno. List (microbial genomes) • Psuedomonas (7 species, 18 strains) • Hundreds or thousands – Separated into organism families • Geno. Scope (the platform shared by Ne. Me. Sys) • UCSC
genolist home
genolist search
genolist results
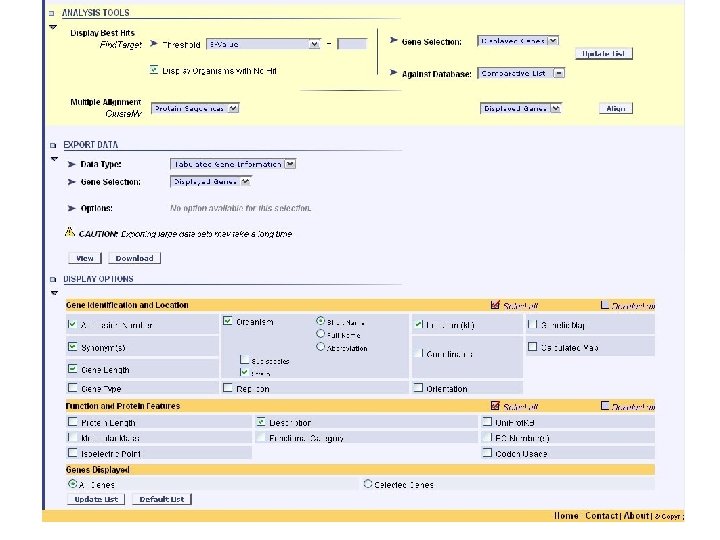
genolist analysis tools
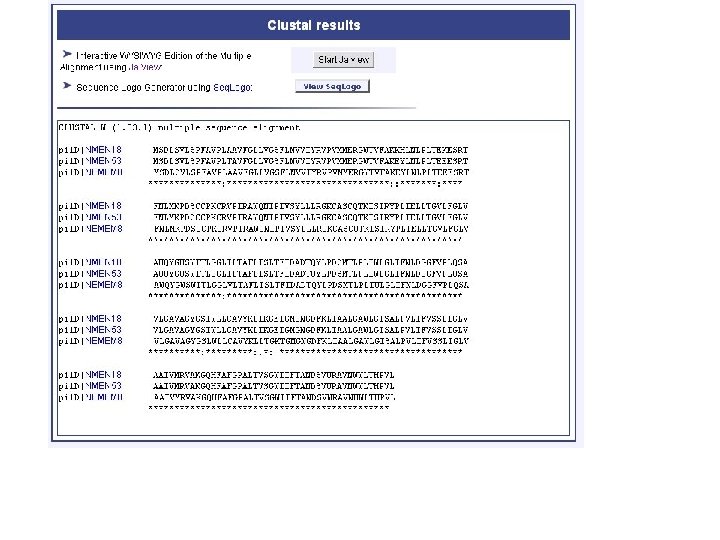
genolist alignment

genolist seqlogo
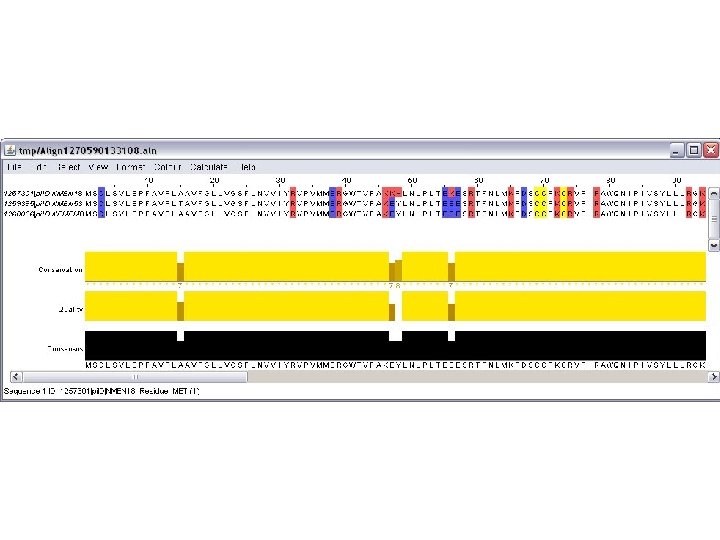
genolist jalview
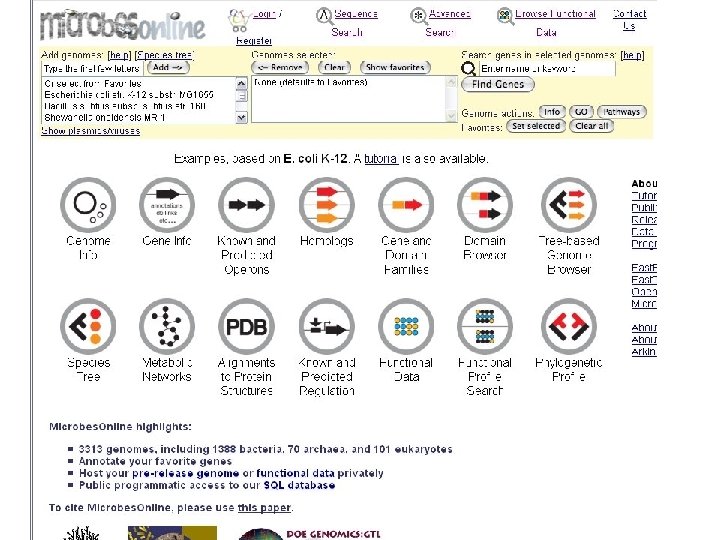
microbes home
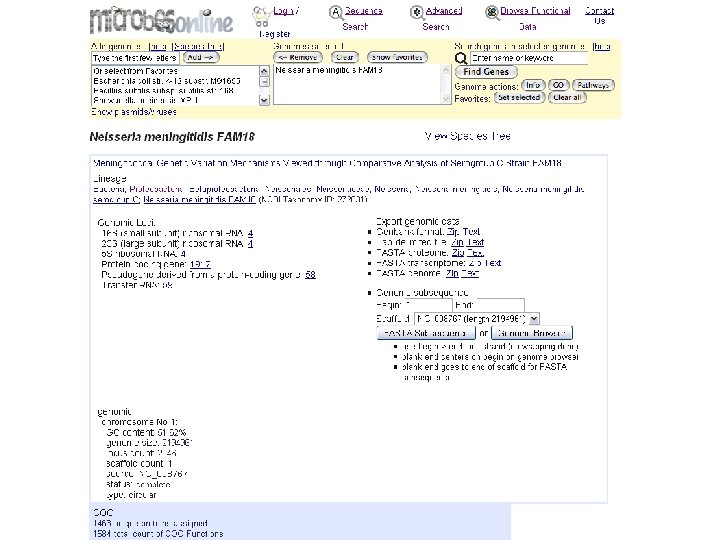
microbes genome info top
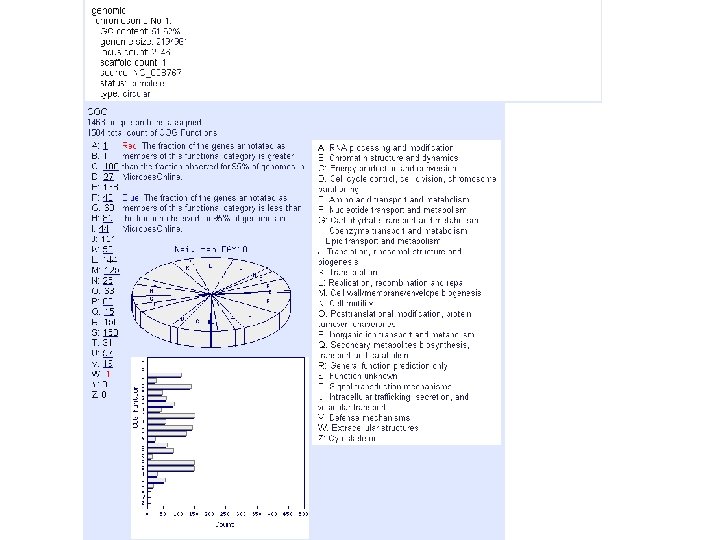
microbes genome info foot
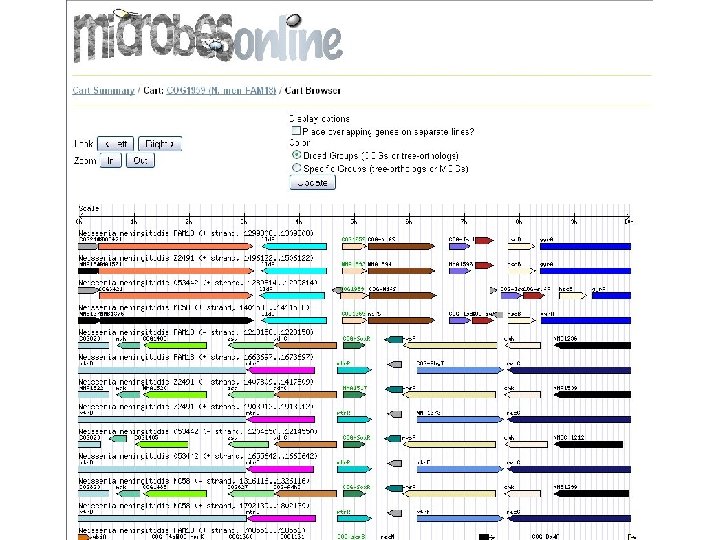
microbes browser
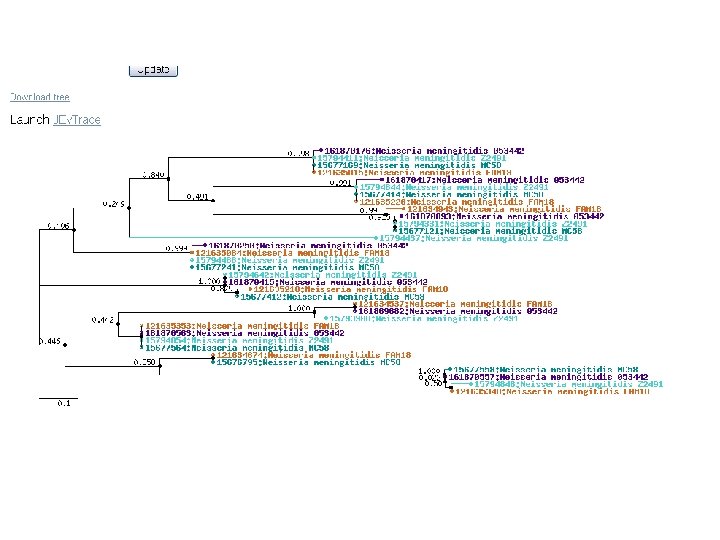
microbes phylo tree
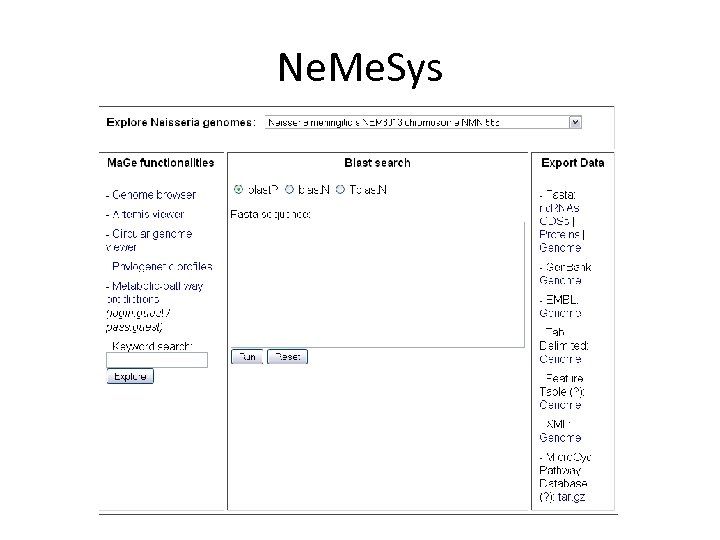
Ne. Me. Sys
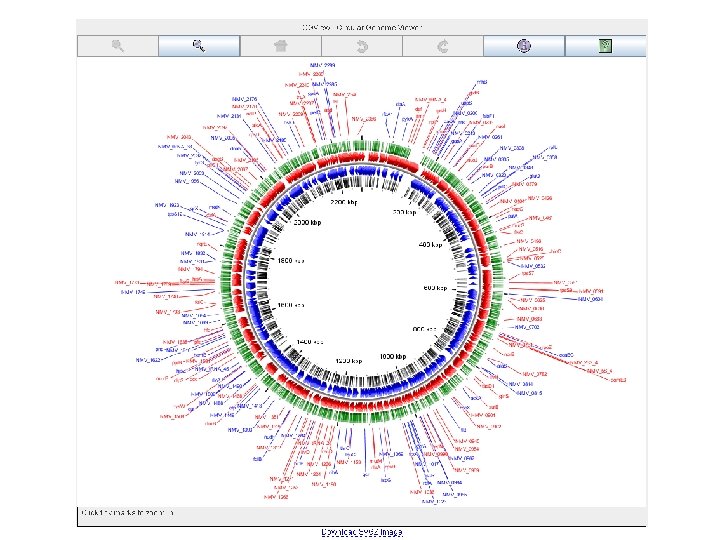
nemesys circular genome viewer
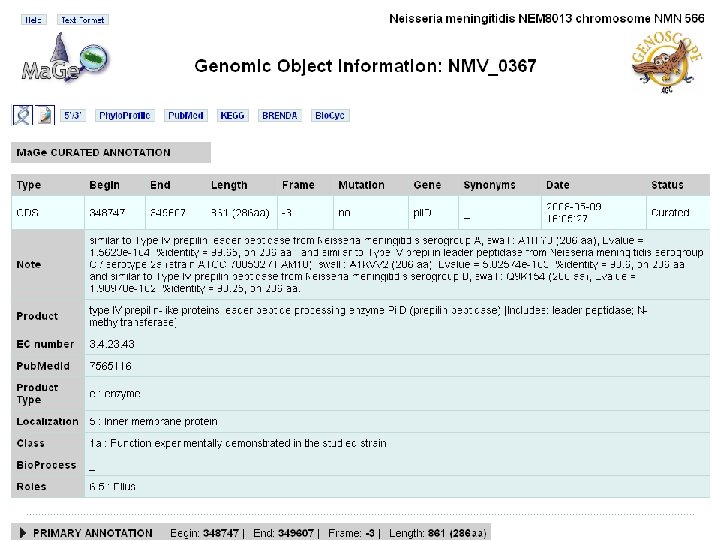
nemesys details
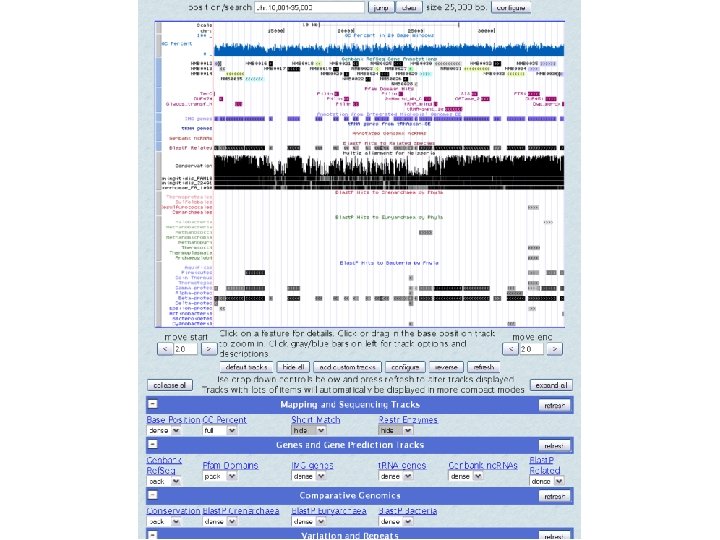
UCSC browser MC 58
Psuedomonas aeruginosa - search
Psuedo tools
Psuedo tools
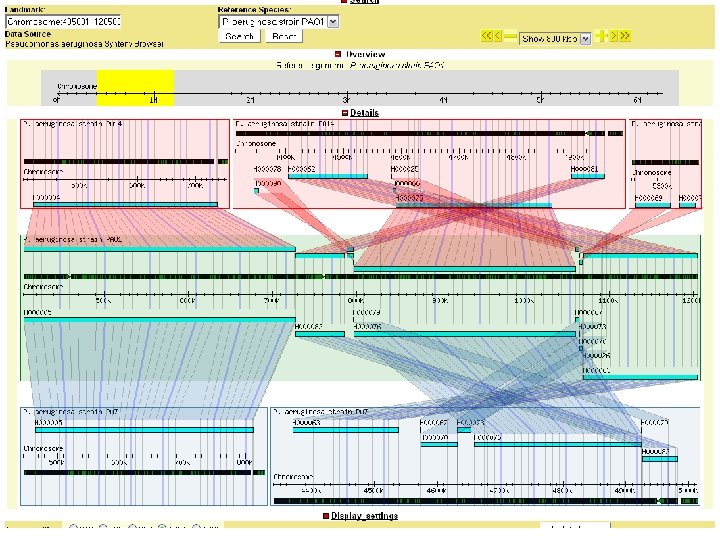
Psuedo synteny
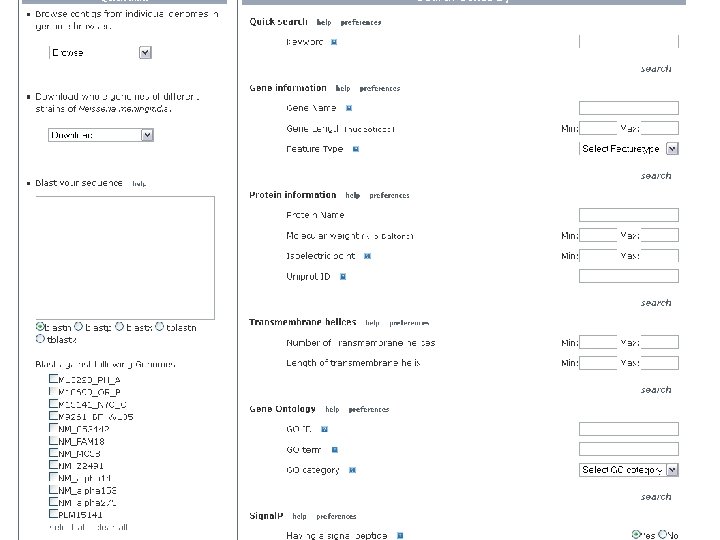
NBase
Database models • How are the databases for these web applications implemented? – Flat files - plain text tables • examples include GBrowse, CGView – RDBMS • May be standard (like Chado, an extensive model organism db schema) • May be created in-house, which is probably the case with many of these multiple genome online databases

NBase has a hybrid schema • The next few slides show it • Bio: : DB: : GFF which is the GBrowse default
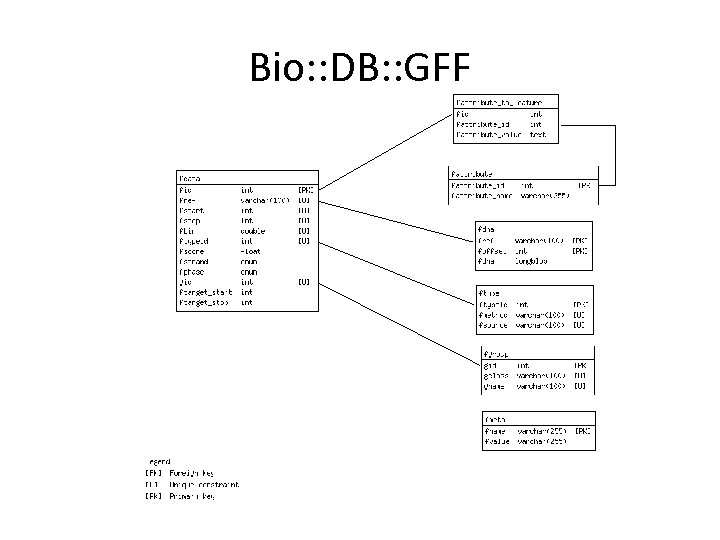
Bio: : DB: : GFF
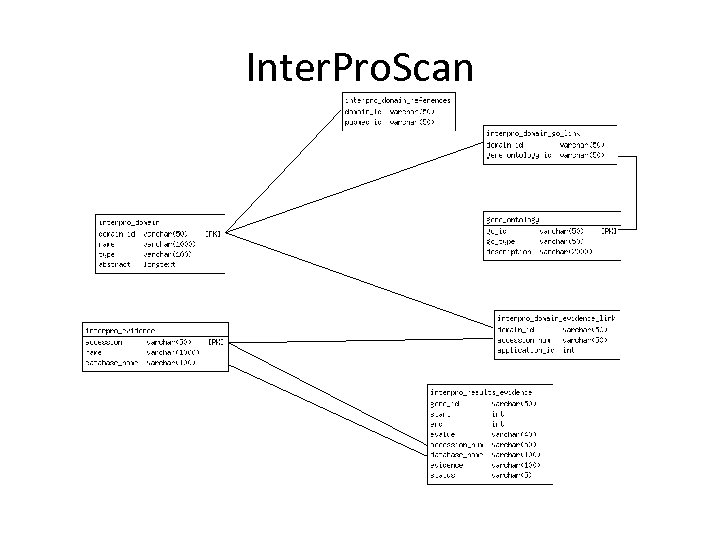
Inter. Pro. Scan
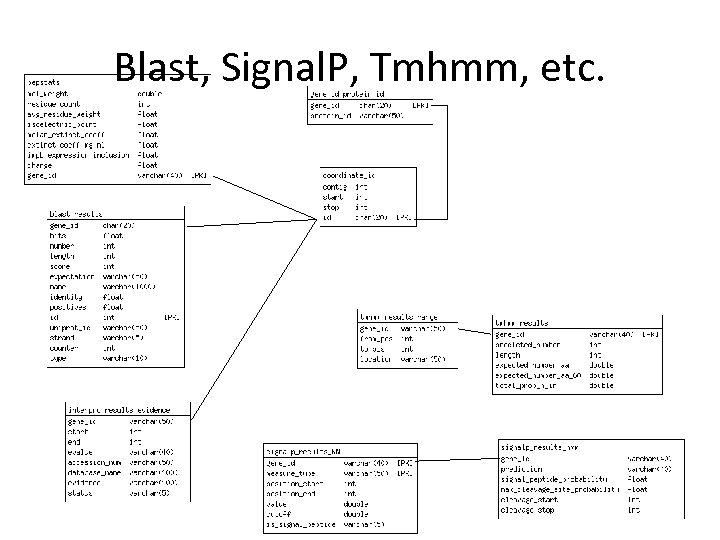
Blast, Signal. P, Tmhmm, etc.

Chado
Survey Conclusions • Genome browsers generally provide – Search forms for querying the database, including BLAST – Detailed profile for each genome feature – hyperlinks to more detailed information in external databases such as KEGG, . . – Graphical viewer – Analysis tools – Genome data downloads

Survey Conclusions – Backend implementation, the database design of these genome browsers, is kind of a mystery, we don't have access to their schemas unless they are using a standard one like Chado, GFF, Bio. SQL – They might be using a home-built or modified schema customized for their purposes
8ab5bb59eab87d245159f78e9a9cd37c.ppt