355ee1859d39ddd1b30946b9f69a98ac.ppt
- Количество слайдов: 46
 Automated High-Resolution Protein Structure Determination using Residual Dipolar Couplings Anna Yershova Department of Computer Science Duke University February 5, 2010 1 Feb 5 2010, NC State University Automated Protein Structure Determination using RDCs
Automated High-Resolution Protein Structure Determination using Residual Dipolar Couplings Anna Yershova Department of Computer Science Duke University February 5, 2010 1 Feb 5 2010, NC State University Automated Protein Structure Determination using RDCs
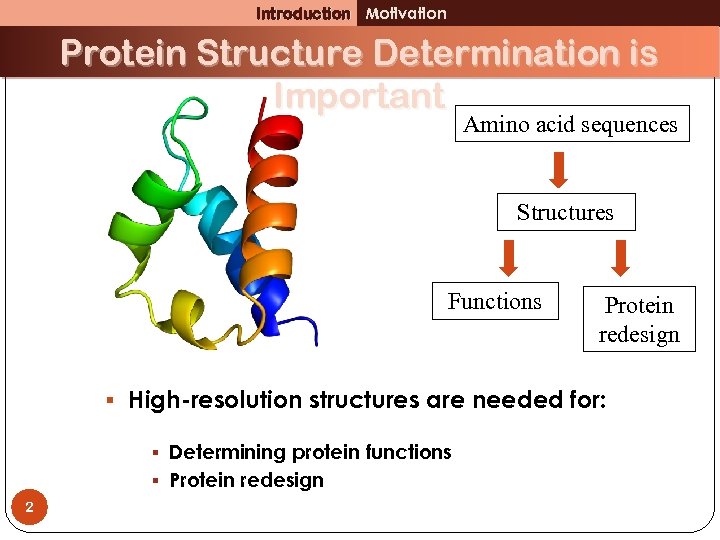 Introduction Motivation Protein Structure Determination is Important Amino acid sequences Structures Functions Protein redesign § High-resolution structures are needed for: § Determining protein functions § Protein redesign 2
Introduction Motivation Protein Structure Determination is Important Amino acid sequences Structures Functions Protein redesign § High-resolution structures are needed for: § Determining protein functions § Protein redesign 2
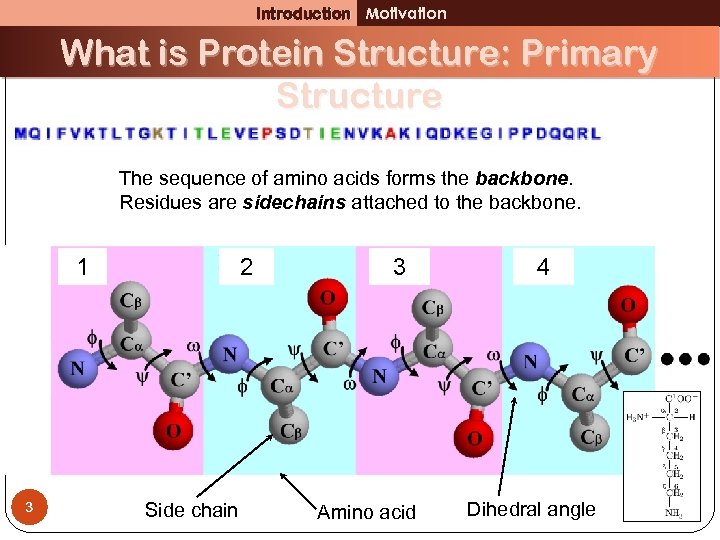 Introduction Motivation What is Protein Structure: Primary Structure The sequence of amino acids forms the backbone. Residues are sidechains attached to the backbone. 1 3 2 Side chain 3 Amino acid 4 Dihedral angle
Introduction Motivation What is Protein Structure: Primary Structure The sequence of amino acids forms the backbone. Residues are sidechains attached to the backbone. 1 3 2 Side chain 3 Amino acid 4 Dihedral angle
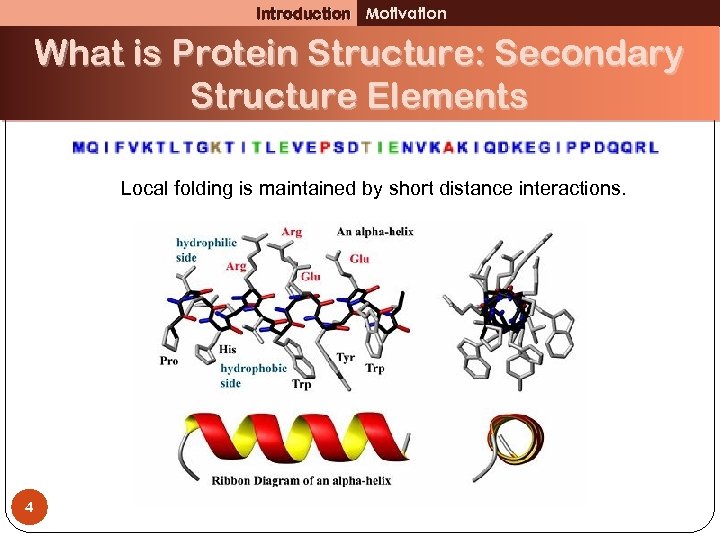 Introduction Motivation What is Protein Structure: Secondary Structure Elements Local folding is maintained by short distance interactions. 4
Introduction Motivation What is Protein Structure: Secondary Structure Elements Local folding is maintained by short distance interactions. 4
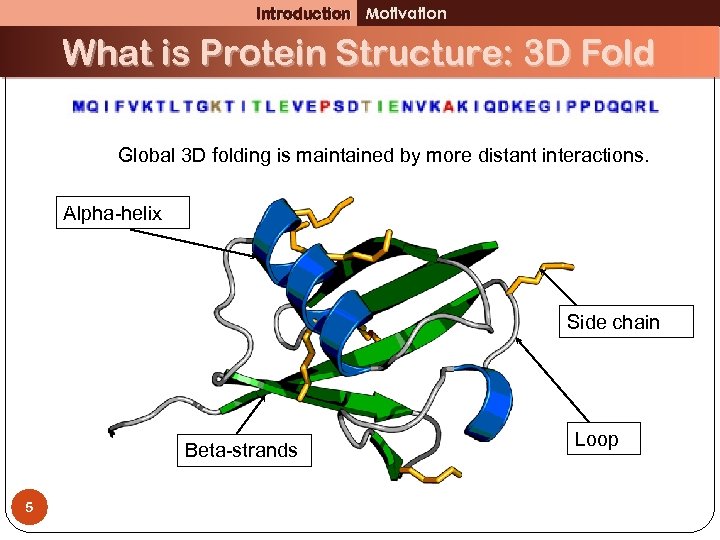 Introduction Motivation What is Protein Structure: 3 D Fold Global 3 D folding is maintained by more distant interactions. Alpha-helix Side chain Beta-strands 5 Loop
Introduction Motivation What is Protein Structure: 3 D Fold Global 3 D folding is maintained by more distant interactions. Alpha-helix Side chain Beta-strands 5 Loop
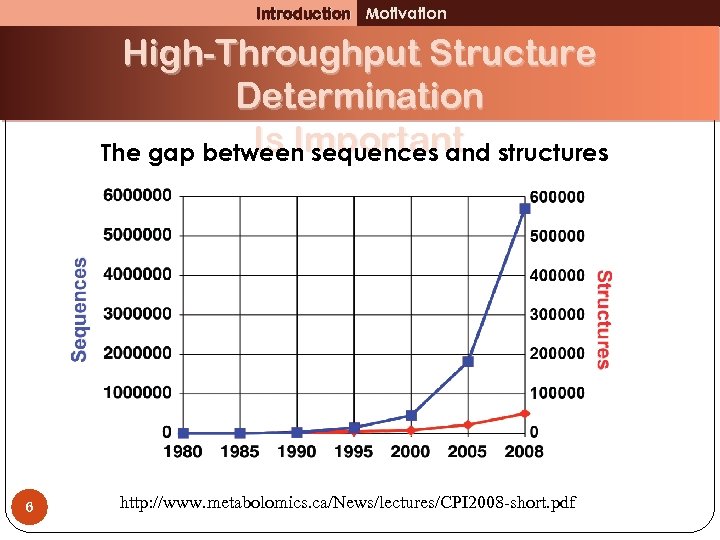 Introduction Motivation High-Throughput Structure Determination Is Important The gap between sequences and structures 6 http: //www. metabolomics. ca/News/lectures/CPI 2008 -short. pdf
Introduction Motivation High-Throughput Structure Determination Is Important The gap between sequences and structures 6 http: //www. metabolomics. ca/News/lectures/CPI 2008 -short. pdf
 Introduction Motivation Current Approaches for Structure Determination §X-ray crystallography § Difficulty: growing good quality crystals §Nuclear Magnetic Resonance (NMR) spectroscopy § Difficulty: lengthy (expensive) time in processing and analyzing experimental data Both require expressing and purifying proteins. 7
Introduction Motivation Current Approaches for Structure Determination §X-ray crystallography § Difficulty: growing good quality crystals §Nuclear Magnetic Resonance (NMR) spectroscopy § Difficulty: lengthy (expensive) time in processing and analyzing experimental data Both require expressing and purifying proteins. 7
 Introduction Motivation Bruce Donald’s Lab Michael Zeng Chittu Tripathy Lincong Wang Pei Zhou Bruce Donald Cheng-Yu Chen John Mac. Master 8
Introduction Motivation Bruce Donald’s Lab Michael Zeng Chittu Tripathy Lincong Wang Pei Zhou Bruce Donald Cheng-Yu Chen John Mac. Master 8
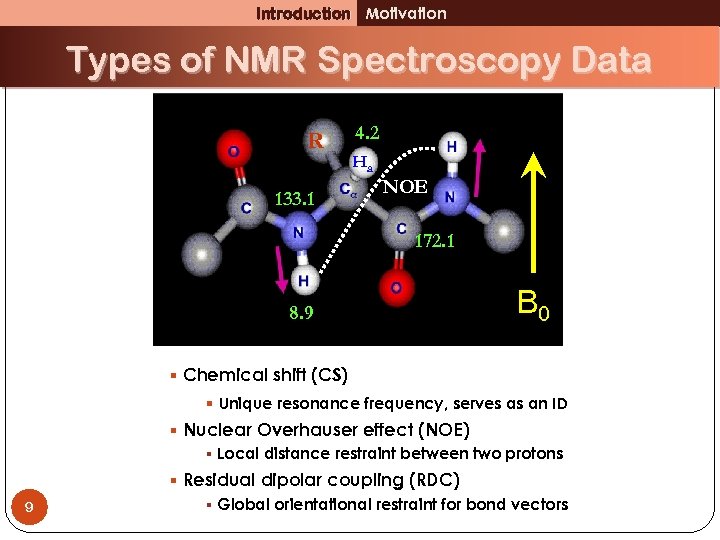 Introduction Motivation Types of NMR Spectroscopy Data R 133. 1 4. 2 Ha NOE 172. 1 8. 9 B 0 § Chemical shift (CS) § Unique resonance frequency, serves as an ID § Nuclear Overhauser effect (NOE) § Local distance restraint between two protons 9 § Residual dipolar coupling (RDC) § Global orientational restraint for bond vectors
Introduction Motivation Types of NMR Spectroscopy Data R 133. 1 4. 2 Ha NOE 172. 1 8. 9 B 0 § Chemical shift (CS) § Unique resonance frequency, serves as an ID § Nuclear Overhauser effect (NOE) § Local distance restraint between two protons 9 § Residual dipolar coupling (RDC) § Global orientational restraint for bond vectors
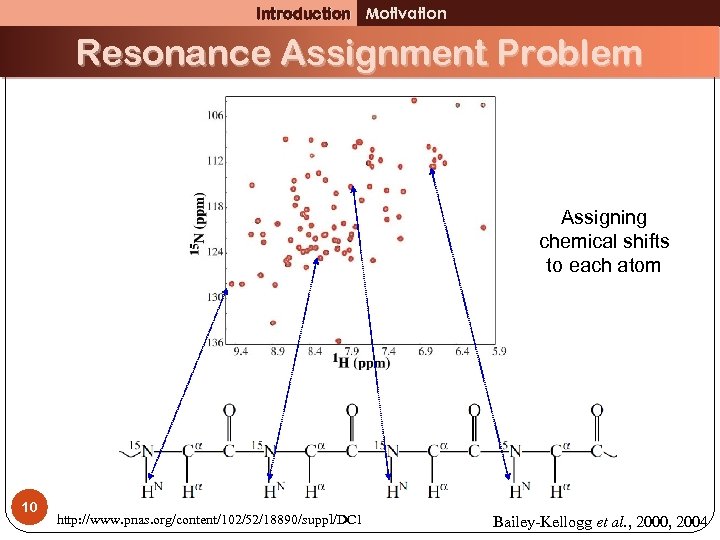 Introduction Motivation Resonance Assignment Problem Assigning chemical shifts to each atom 10 http: //www. pnas. org/content/102/52/18890/suppl/DC 1 Bailey-Kellogg et al. , 2000, 2004
Introduction Motivation Resonance Assignment Problem Assigning chemical shifts to each atom 10 http: //www. pnas. org/content/102/52/18890/suppl/DC 1 Bailey-Kellogg et al. , 2000, 2004
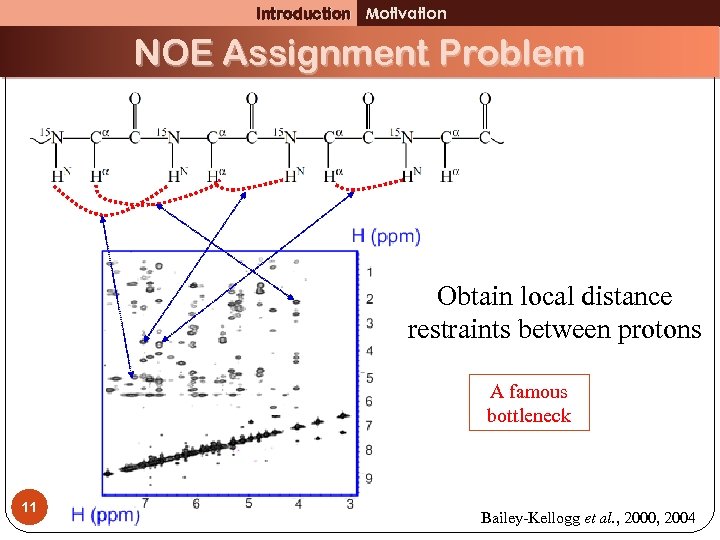 Introduction Motivation NOE Assignment Problem Obtain local distance restraints between protons A famous bottleneck 11 Bailey-Kellogg et al. , 2000, 2004
Introduction Motivation NOE Assignment Problem Obtain local distance restraints between protons A famous bottleneck 11 Bailey-Kellogg et al. , 2000, 2004
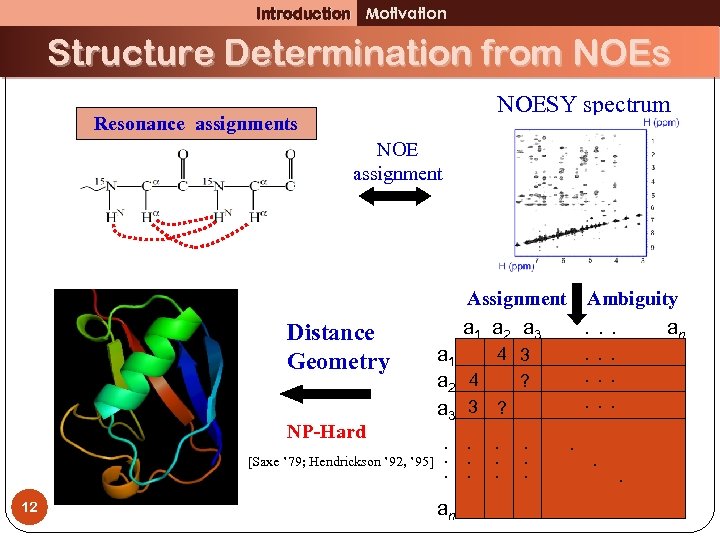 Introduction Motivation Structure Determination from NOEs NOESY spectrum Resonance assignments NOE assignment Distance Geometry NP-Hard [Saxe ’ 79; Hendrickson ’ 92, ’ 95] 12 Assignment a 1 a 2 a 3 Ambiguity. . . an . . 4 3 a 1 a 2 4 ? a 3 3 ? . . . an . . .
Introduction Motivation Structure Determination from NOEs NOESY spectrum Resonance assignments NOE assignment Distance Geometry NP-Hard [Saxe ’ 79; Hendrickson ’ 92, ’ 95] 12 Assignment a 1 a 2 a 3 Ambiguity. . . an . . 4 3 a 1 a 2 4 ? a 3 3 ? . . . an . . .
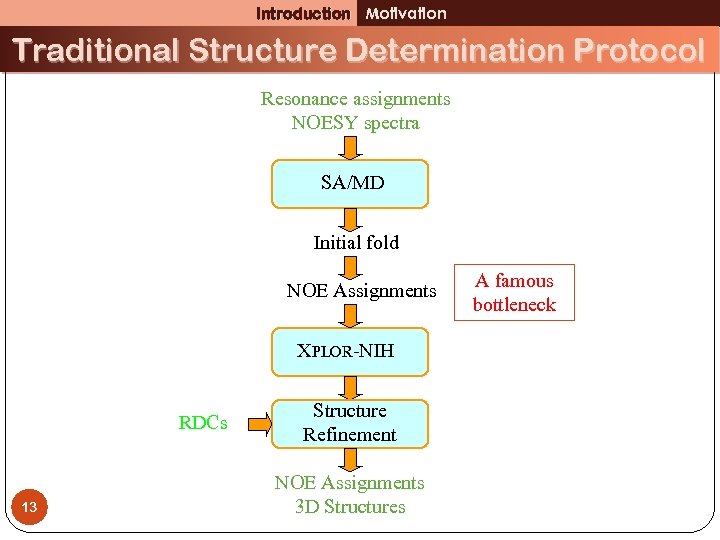 Introduction Motivation Traditional Structure Determination Protocol Resonance assignments NOESY spectra SA/MD Initial fold NOE Assignments XPLOR-NIH RDCs 13 Structure Refinement NOE Assignments 3 D Structures A famous bottleneck
Introduction Motivation Traditional Structure Determination Protocol Resonance assignments NOESY spectra SA/MD Initial fold NOE Assignments XPLOR-NIH RDCs 13 Structure Refinement NOE Assignments 3 D Structures A famous bottleneck
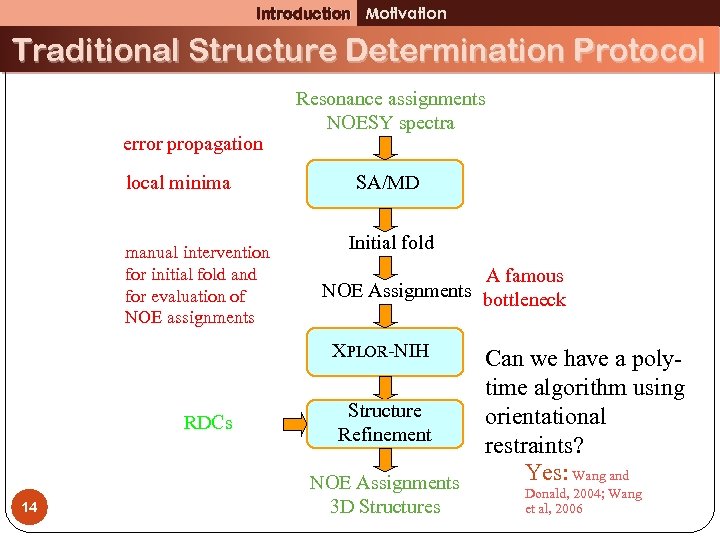 Introduction Motivation Traditional Structure Determination Protocol error propagation local minima manual intervention for initial fold and for evaluation of NOE assignments Resonance assignments NOESY spectra SA/MD Initial fold A famous NOE Assignments bottleneck XPLOR-NIH RDCs 14 Structure Refinement NOE Assignments 3 D Structures Can we have a polytime algorithm using orientational restraints? Yes: Wang and Donald, 2004; Wang et al, 2006
Introduction Motivation Traditional Structure Determination Protocol error propagation local minima manual intervention for initial fold and for evaluation of NOE assignments Resonance assignments NOESY spectra SA/MD Initial fold A famous NOE Assignments bottleneck XPLOR-NIH RDCs 14 Structure Refinement NOE Assignments 3 D Structures Can we have a polytime algorithm using orientational restraints? Yes: Wang and Donald, 2004; Wang et al, 2006
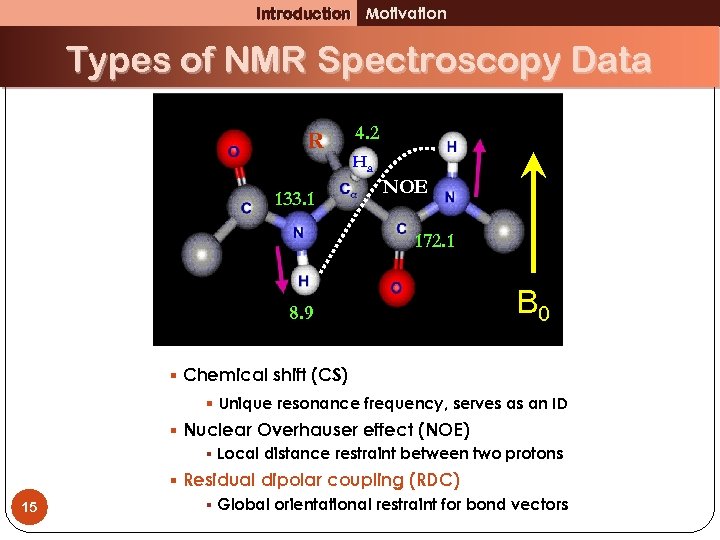 Introduction Motivation Types of NMR Spectroscopy Data R 133. 1 4. 2 Ha NOE 172. 1 8. 9 B 0 § Chemical shift (CS) § Unique resonance frequency, serves as an ID § Nuclear Overhauser effect (NOE) § Local distance restraint between two protons 15 § Residual dipolar coupling (RDC) § Global orientational restraint for bond vectors
Introduction Motivation Types of NMR Spectroscopy Data R 133. 1 4. 2 Ha NOE 172. 1 8. 9 B 0 § Chemical shift (CS) § Unique resonance frequency, serves as an ID § Nuclear Overhauser effect (NOE) § Local distance restraint between two protons 15 § Residual dipolar coupling (RDC) § Global orientational restraint for bond vectors
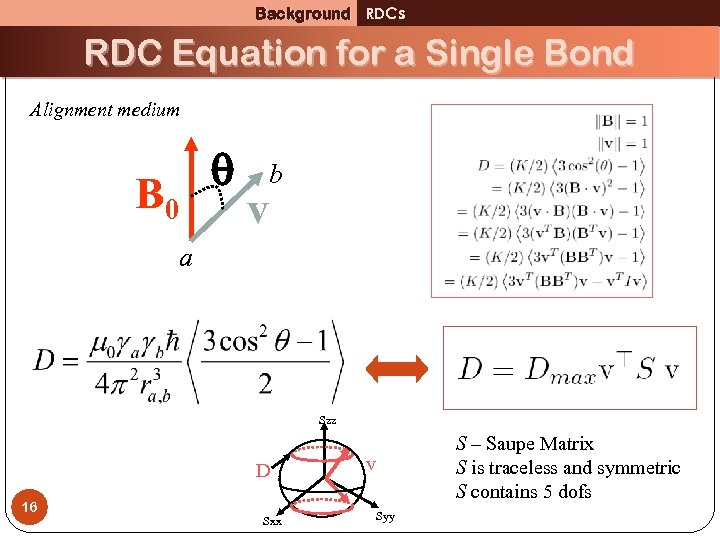 Background RDCs RDC Equation for a Single Bond Alignment medium B 0 v b a Szz D 16 Sxx v Syy S – Saupe Matrix S is traceless and symmetric S contains 5 dofs
Background RDCs RDC Equation for a Single Bond Alignment medium B 0 v b a Szz D 16 Sxx v Syy S – Saupe Matrix S is traceless and symmetric S contains 5 dofs
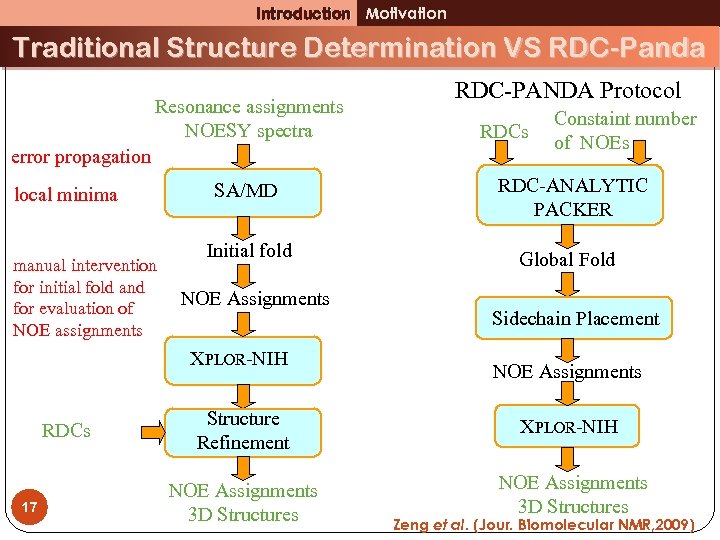 Introduction Motivation Traditional Structure Determination VS RDC-Panda Resonance assignments NOESY spectra error propagation local minima manual intervention for initial fold and for evaluation of NOE assignments 17 RDCs Constaint number of NOEs SA/MD RDC-ANALYTIC PACKER Initial fold Global Fold NOE Assignments XPLOR-NIH RDCs RDC-PANDA Protocol Sidechain Placement NOE Assignments Structure Refinement XPLOR-NIH NOE Assignments 3 D Structures Zeng et al. (Jour. Biomolecular NMR, 2009)
Introduction Motivation Traditional Structure Determination VS RDC-Panda Resonance assignments NOESY spectra error propagation local minima manual intervention for initial fold and for evaluation of NOE assignments 17 RDCs Constaint number of NOEs SA/MD RDC-ANALYTIC PACKER Initial fold Global Fold NOE Assignments XPLOR-NIH RDCs RDC-PANDA Protocol Sidechain Placement NOE Assignments Structure Refinement XPLOR-NIH NOE Assignments 3 D Structures Zeng et al. (Jour. Biomolecular NMR, 2009)
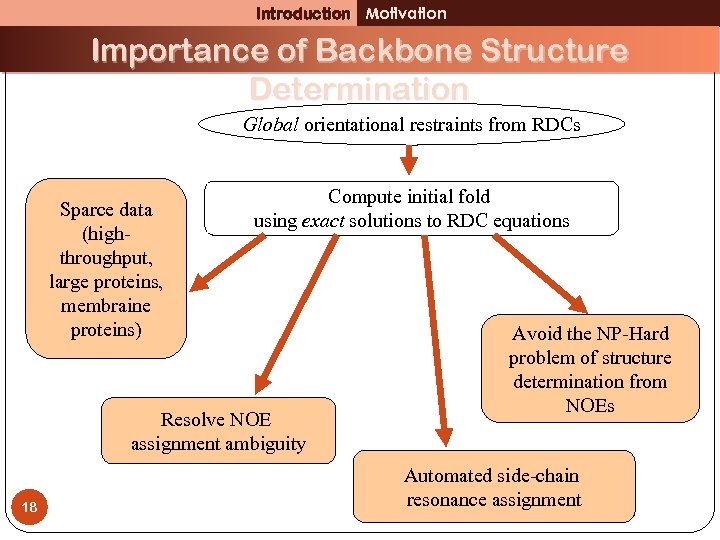 Introduction Motivation Importance of Backbone Structure Determination Global orientational restraints from RDCs Sparce data (highthroughput, large proteins, membraine proteins) Compute initial fold using exact solutions to RDC equations Resolve NOE assignment ambiguity 18 Avoid the NP-Hard problem of structure determination from NOEs Automated side-chain resonance assignment
Introduction Motivation Importance of Backbone Structure Determination Global orientational restraints from RDCs Sparce data (highthroughput, large proteins, membraine proteins) Compute initial fold using exact solutions to RDC equations Resolve NOE assignment ambiguity 18 Avoid the NP-Hard problem of structure determination from NOEs Automated side-chain resonance assignment
 Introduction Motivation Current Limitations of RDC-Panda Because it requires only 2 RDCs per residue: § Only SSE elements can be reliably determined, NOEs are needed to determine structure of loops § Difficulty in handling missing data 19
Introduction Motivation Current Limitations of RDC-Panda Because it requires only 2 RDCs per residue: § Only SSE elements can be reliably determined, NOEs are needed to determine structure of loops § Difficulty in handling missing data 19
 Introduction Motivation My Current Project § Improve current protein structure determination techniques from our lab § Design new algorithms for protein backbone structure determination using orientational restraints from RDCs 20
Introduction Motivation My Current Project § Improve current protein structure determination techniques from our lab § Design new algorithms for protein backbone structure determination using orientational restraints from RDCs 20
 Introduction Motivation Literature Overview Distance geometry based structure determination Braun, 1987 Crippen and Havel, 1988 More and Wu, 1999 Heuristic based structure determination Brünger, 1992 Nilges et al. , 1997 Güntert, 2003 Rieping et al. , 2005 RDC-based structure determination Tolman et al. , 1995 Tjandra and Bax, 1997 Hus et al. , 2001 Tian et al. , 2001 Prestegard et al. , 2004 Wang and Donald (CSB 2004) Wang and Donald (Jour. Biomolecular NMR, 21 2004) Wang, Mettu and Donald (JCB 2005) Donald and Martin (Progress in NMR Spectroscopy, 2009 ) Ruan et al. , 2008 Zeng et al. (Jour. Biomolecular NMR, 2009) • Heuristic based automated NOE assignment – Mumenthaler et al. , 1997 – Nilges et al. , 1997, 2003 – Herrmann et al. , 2002 – Schwieters et al. , 2003 – Kuszewski et al. , 2004 – Huang et al. , 2006 • Automated NOE assignment starting with initial fold computed from RDCs – Wang and Donald (CSB 2005) – Zeng et al. (CSB 2008) – Zeng et al. (Jour. Biomolecular NMR, 2009) • Automated side-chain resonance assignment – – Li and Sanctuary, 1996, 1997 Marin et al. , 2004 Masse et al. , 2006 Zeng et al. (In submission, 2009)
Introduction Motivation Literature Overview Distance geometry based structure determination Braun, 1987 Crippen and Havel, 1988 More and Wu, 1999 Heuristic based structure determination Brünger, 1992 Nilges et al. , 1997 Güntert, 2003 Rieping et al. , 2005 RDC-based structure determination Tolman et al. , 1995 Tjandra and Bax, 1997 Hus et al. , 2001 Tian et al. , 2001 Prestegard et al. , 2004 Wang and Donald (CSB 2004) Wang and Donald (Jour. Biomolecular NMR, 21 2004) Wang, Mettu and Donald (JCB 2005) Donald and Martin (Progress in NMR Spectroscopy, 2009 ) Ruan et al. , 2008 Zeng et al. (Jour. Biomolecular NMR, 2009) • Heuristic based automated NOE assignment – Mumenthaler et al. , 1997 – Nilges et al. , 1997, 2003 – Herrmann et al. , 2002 – Schwieters et al. , 2003 – Kuszewski et al. , 2004 – Huang et al. , 2006 • Automated NOE assignment starting with initial fold computed from RDCs – Wang and Donald (CSB 2005) – Zeng et al. (CSB 2008) – Zeng et al. (Jour. Biomolecular NMR, 2009) • Automated side-chain resonance assignment – – Li and Sanctuary, 1996, 1997 Marin et al. , 2004 Masse et al. , 2006 Zeng et al. (In submission, 2009)
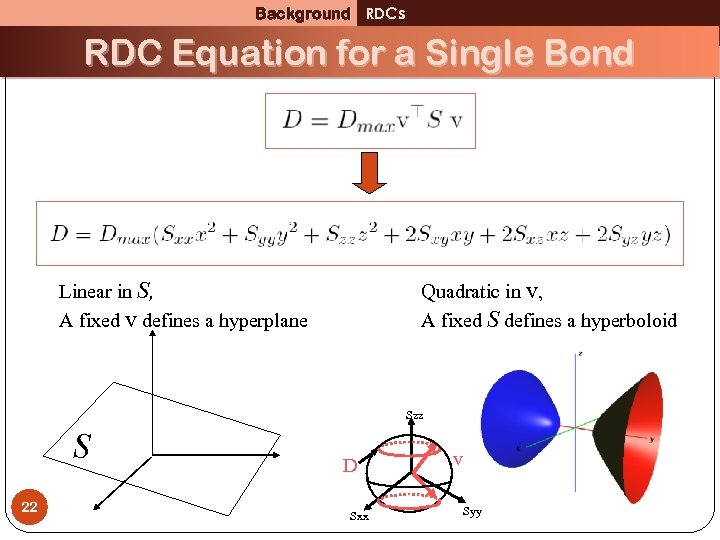 Background RDCs RDC Equation for a Single Bond Linear in S, A fixed v defines a hyperplane Quadratic in v, A fixed S defines a hyperboloid Szz S 22 D Sxx v Syy
Background RDCs RDC Equation for a Single Bond Linear in S, A fixed v defines a hyperplane Quadratic in v, A fixed S defines a hyperboloid Szz S 22 D Sxx v Syy
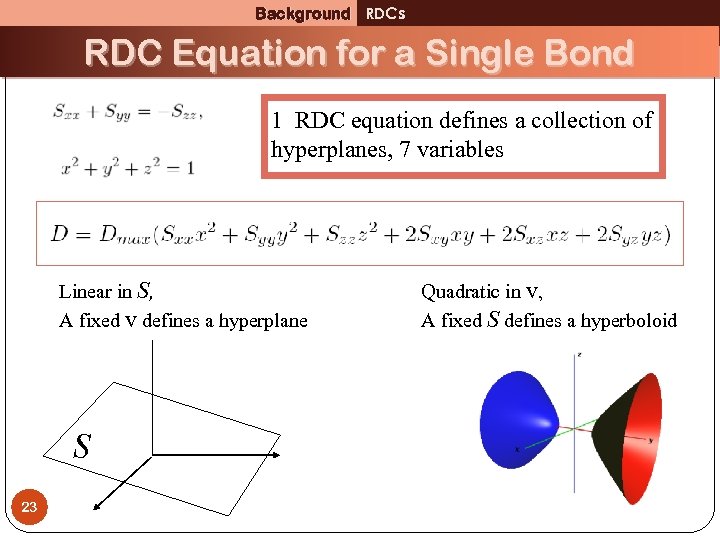 Background RDCs RDC Equation for a Single Bond 1 RDC equation defines a collection of hyperplanes, 7 variables Linear in S, A fixed v defines a hyperplane S 23 Quadratic in v, A fixed S defines a hyperboloid
Background RDCs RDC Equation for a Single Bond 1 RDC equation defines a collection of hyperplanes, 7 variables Linear in S, A fixed v defines a hyperplane S 23 Quadratic in v, A fixed S defines a hyperboloid
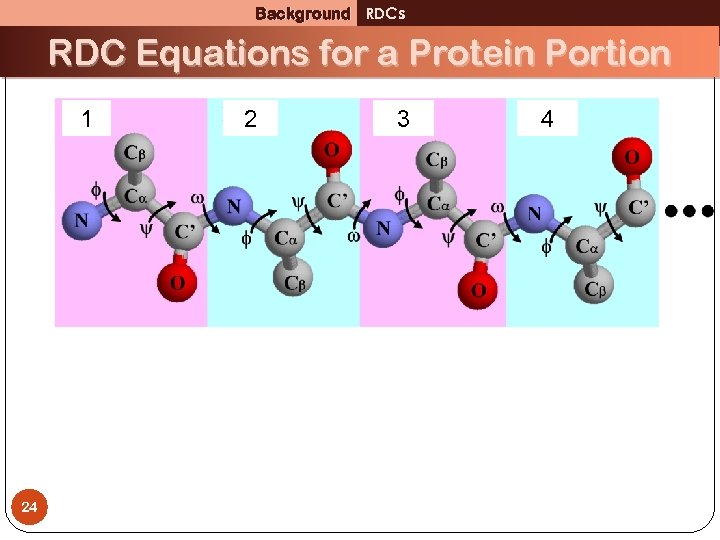 Background RDCs RDC Equations for a Protein Portion 1 24 2 3 4
Background RDCs RDC Equations for a Protein Portion 1 24 2 3 4
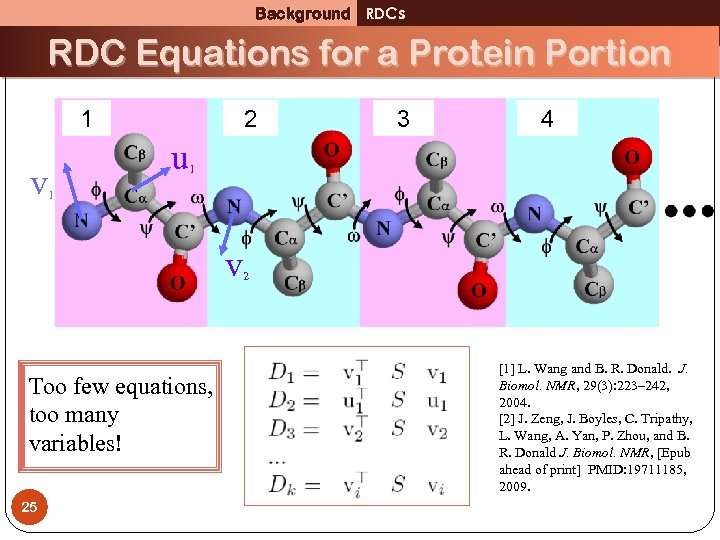 Background RDCs RDC Equations for a Protein Portion 1 v 2 3 4 u 1 1 v 2 Too few equations, too many variables! 25 [1] L. Wang and B. R. Donald. J. Biomol. NMR, 29(3): 223– 242, 2004. [2] J. Zeng, J. Boyles, C. Tripathy, L. Wang, A. Yan, P. Zhou, and B. R. Donald J. Biomol. NMR, [Epub ahead of print] PMID: 19711185, 2009.
Background RDCs RDC Equations for a Protein Portion 1 v 2 3 4 u 1 1 v 2 Too few equations, too many variables! 25 [1] L. Wang and B. R. Donald. J. Biomol. NMR, 29(3): 223– 242, 2004. [2] J. Zeng, J. Boyles, C. Tripathy, L. Wang, A. Yan, P. Zhou, and B. R. Donald J. Biomol. NMR, [Epub ahead of print] PMID: 19711185, 2009.
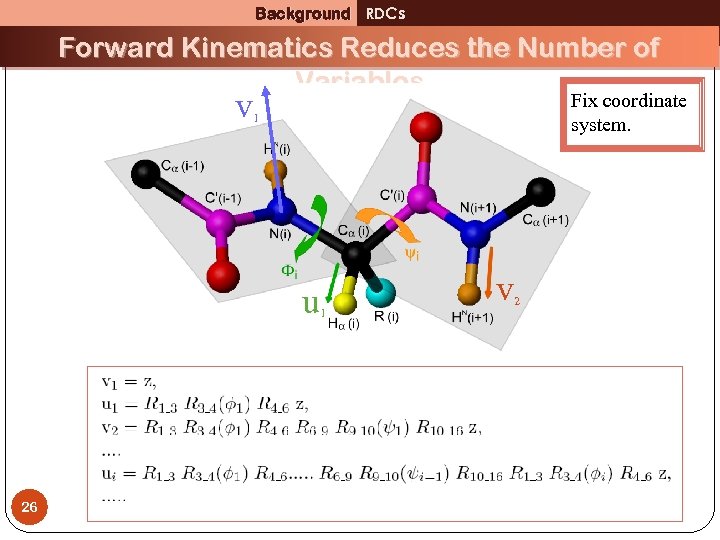 Background RDCs Forward Kinematics Reduces the Number of Variables v Fix coordinate system. 1 u 1 26 v 2
Background RDCs Forward Kinematics Reduces the Number of Variables v Fix coordinate system. 1 u 1 26 v 2
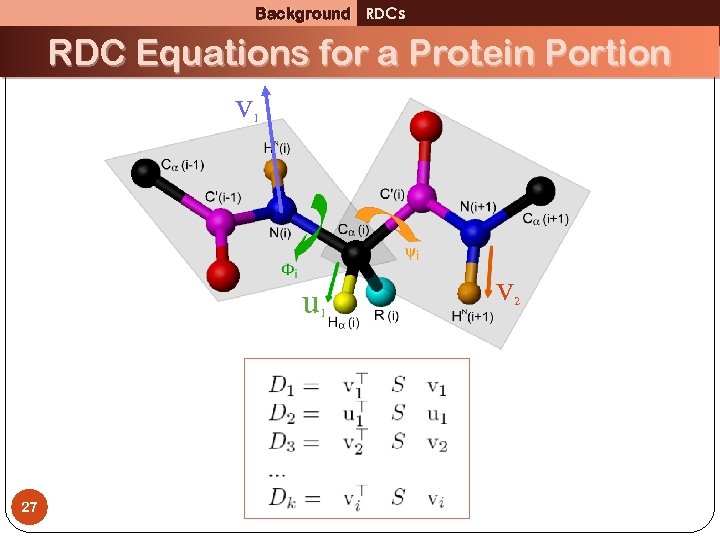 Background RDCs RDC Equations for a Protein Portion v 1 u 1 27 v 2
Background RDCs RDC Equations for a Protein Portion v 1 u 1 27 v 2
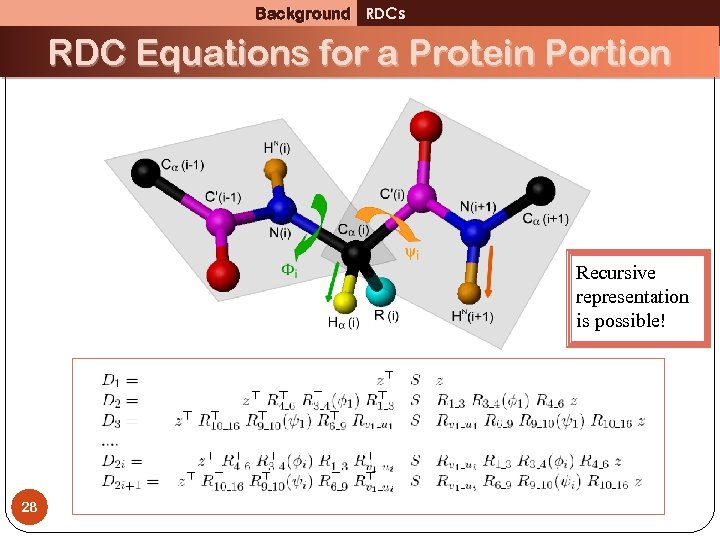 Background RDCs RDC Equations for a Protein Portion Recursive representation is possible! 28
Background RDCs RDC Equations for a Protein Portion Recursive representation is possible! 28
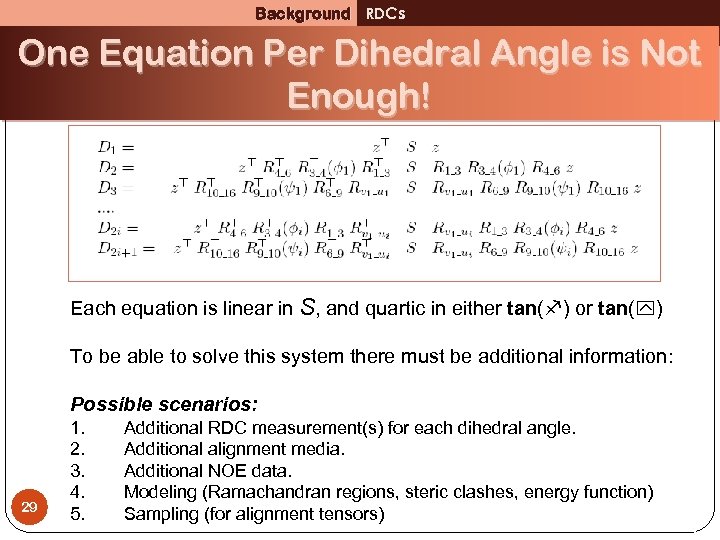 Background RDCs One Equation Per Dihedral Angle is Not Enough! Each equation is linear in S, and quartic in either tan( ) or tan( ) To be able to solve this system there must be additional information: Possible scenarios: 29 1. 2. 3. 4. 5. Additional RDC measurement(s) for each dihedral angle. Additional alignment media. Additional NOE data. Modeling (Ramachandran regions, steric clashes, energy function) Sampling (for alignment tensors)
Background RDCs One Equation Per Dihedral Angle is Not Enough! Each equation is linear in S, and quartic in either tan( ) or tan( ) To be able to solve this system there must be additional information: Possible scenarios: 29 1. 2. 3. 4. 5. Additional RDC measurement(s) for each dihedral angle. Additional alignment media. Additional NOE data. Modeling (Ramachandran regions, steric clashes, energy function) Sampling (for alignment tensors)
 Background RDC-Panda The RDC-PANDA Structure Determination Package Current requirements • • 2 RDCs per residue to obtain SSE structures Sparse NOEs to pack the SSEs Current bottlenecks • • 30 Missing data (even in long SSEs) Long loops Sampling for computing alignment tensor(s) Sampling for the orientation of the first pp [1] L. Wang and B. R. Donald. J. Biomol. NMR, 29(3): 223– 242, 2004. [2] J. Zeng, J. Boyles, C. Tripathy, L. Wang, A. Yan, P. Zhou, and B. R. Donald J. Biomol. NMR, [Epub ahead of print] PMID: 19711185, 2009.
Background RDC-Panda The RDC-PANDA Structure Determination Package Current requirements • • 2 RDCs per residue to obtain SSE structures Sparse NOEs to pack the SSEs Current bottlenecks • • 30 Missing data (even in long SSEs) Long loops Sampling for computing alignment tensor(s) Sampling for the orientation of the first pp [1] L. Wang and B. R. Donald. J. Biomol. NMR, 29(3): 223– 242, 2004. [2] J. Zeng, J. Boyles, C. Tripathy, L. Wang, A. Yan, P. Zhou, and B. R. Donald J. Biomol. NMR, [Epub ahead of print] PMID: 19711185, 2009.
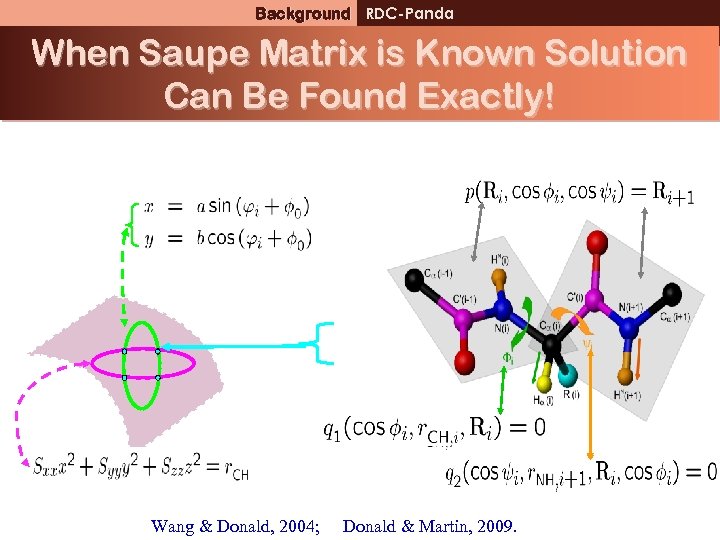 Ellipse equations for CHBackground RDC-Panda bond vector When Saupe Matrix is Known Solution Can Be Found Exactly! Wang & Donald, 2004; Donald & Martin, 2009.
Ellipse equations for CHBackground RDC-Panda bond vector When Saupe Matrix is Known Solution Can Be Found Exactly! Wang & Donald, 2004; Donald & Martin, 2009.
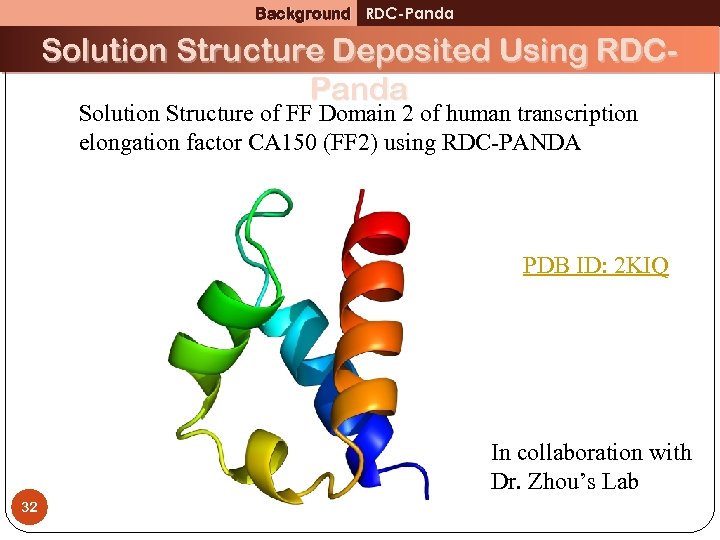 Background RDC-Panda Solution Structure Deposited Using RDCPanda Solution Structure of FF Domain 2 of human transcription elongation factor CA 150 (FF 2) using RDC-PANDA PDB ID: 2 KIQ In collaboration with Dr. Zhou’s Lab 32
Background RDC-Panda Solution Structure Deposited Using RDCPanda Solution Structure of FF Domain 2 of human transcription elongation factor CA 150 (FF 2) using RDC-PANDA PDB ID: 2 KIQ In collaboration with Dr. Zhou’s Lab 32
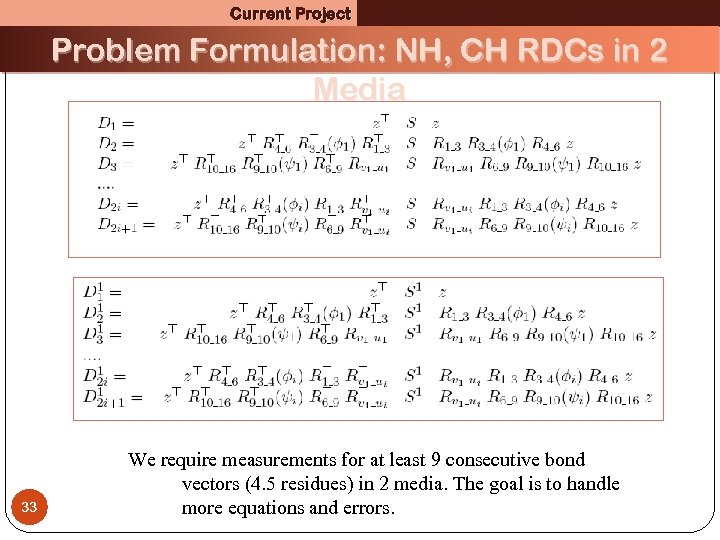 Current Project Problem Formulation: NH, CH RDCs in 2 Media 33 We require measurements for at least 9 consecutive bond vectors (4. 5 residues) in 2 media. The goal is to handle more equations and errors.
Current Project Problem Formulation: NH, CH RDCs in 2 Media 33 We require measurements for at least 9 consecutive bond vectors (4. 5 residues) in 2 media. The goal is to handle more equations and errors.
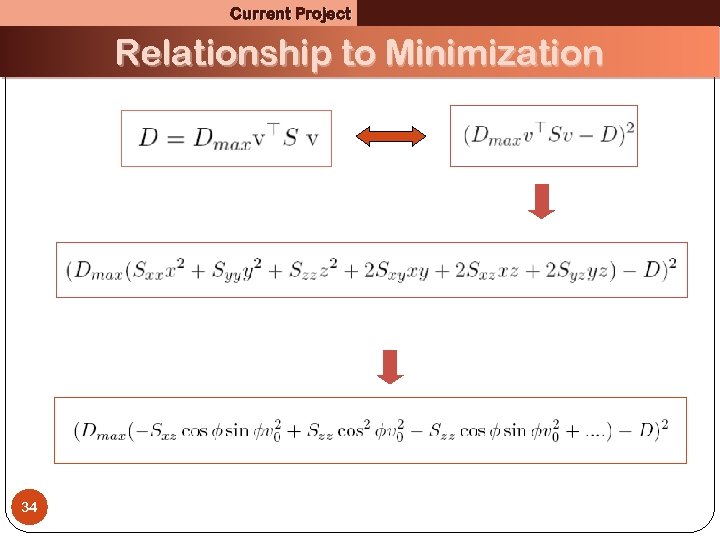 Current Project Relationship to Minimization 34
Current Project Relationship to Minimization 34
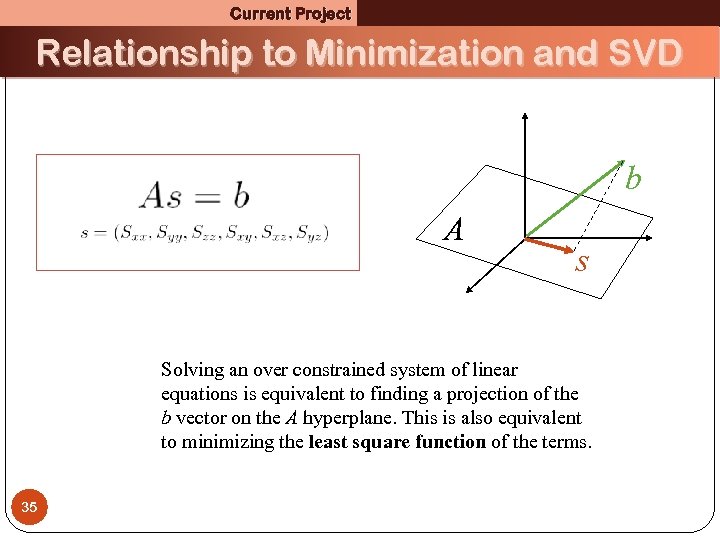 Current Project Relationship to Minimization and SVD b A s Solving an over constrained system of linear equations is equivalent to finding a projection of the b vector on the A hyperplane. This is also equivalent to minimizing the least square function of the terms. 35
Current Project Relationship to Minimization and SVD b A s Solving an over constrained system of linear equations is equivalent to finding a projection of the b vector on the A hyperplane. This is also equivalent to minimizing the least square function of the terms. 35
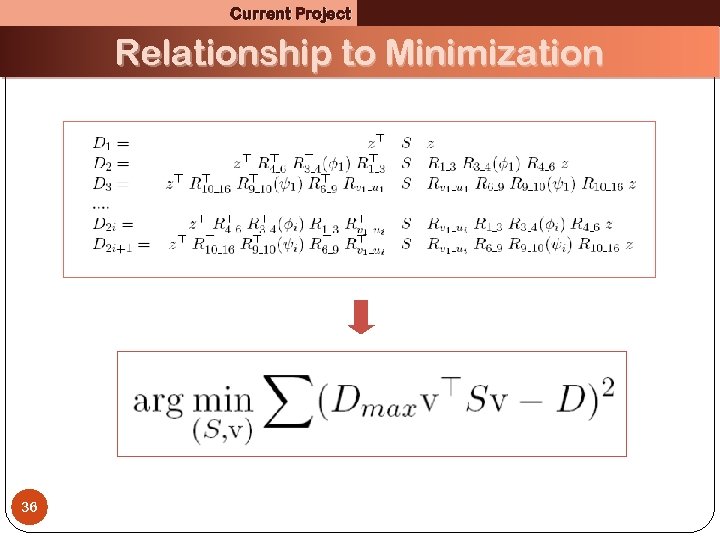 Current Project Relationship to Minimization 36
Current Project Relationship to Minimization 36
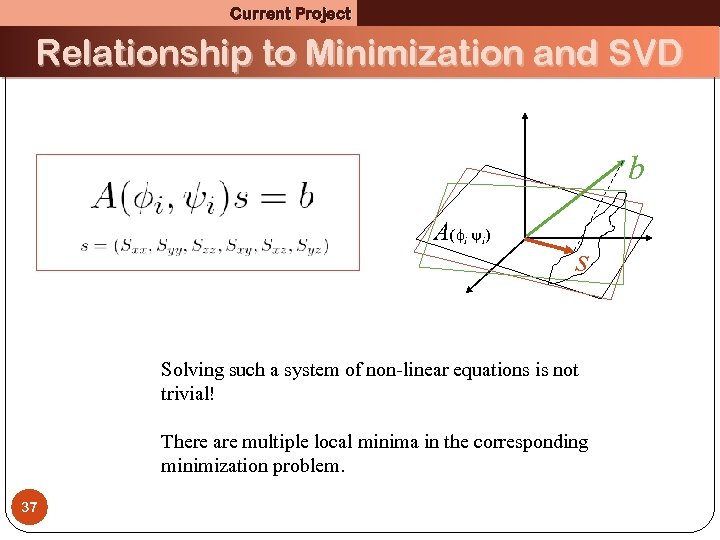 Current Project Relationship to Minimization and SVD b A( i i) s Solving such a system of non-linear equations is not trivial! There are multiple local minima in the corresponding minimization problem. 37
Current Project Relationship to Minimization and SVD b A( i i) s Solving such a system of non-linear equations is not trivial! There are multiple local minima in the corresponding minimization problem. 37
 Current Project Advantages If the minimization problem is solved then • Computation of packed SSEs and loops is possible without additional NOE data. • Saupe matrices for each of the alignment medium can be computed without sampling. • Robust handling of missing values 38
Current Project Advantages If the minimization problem is solved then • Computation of packed SSEs and loops is possible without additional NOE data. • Saupe matrices for each of the alignment medium can be computed without sampling. • Robust handling of missing values 38
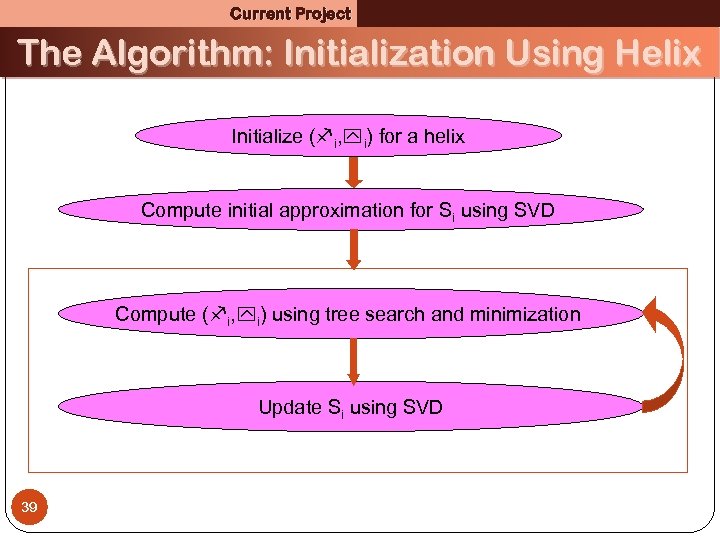 Current Project The Algorithm: Initialization Using Helix Initialize ( i, i) for a helix Compute initial approximation for Si using SVD Compute ( i, i) using tree search and minimization Update Si using SVD 39
Current Project The Algorithm: Initialization Using Helix Initialize ( i, i) for a helix Compute initial approximation for Si using SVD Compute ( i, i) using tree search and minimization Update Si using SVD 39
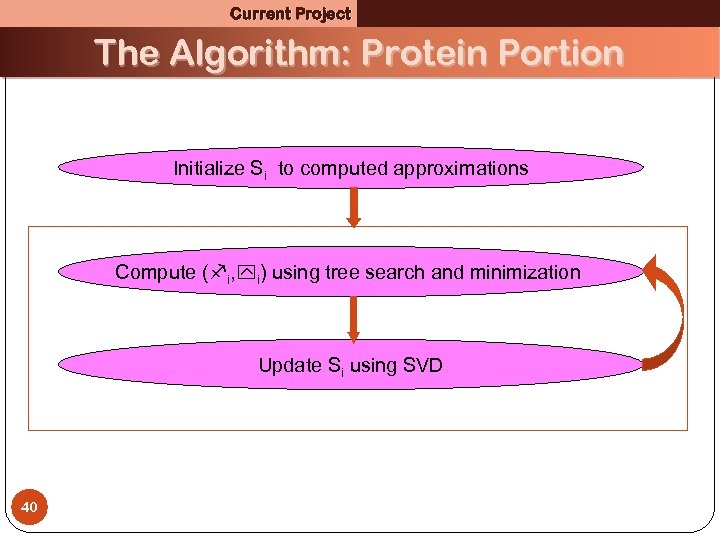 Current Project The Algorithm: Protein Portion Initialize Si to computed approximations Compute ( i, i) using tree search and minimization Update Si using SVD 40
Current Project The Algorithm: Protein Portion Initialize Si to computed approximations Compute ( i, i) using tree search and minimization Update Si using SVD 40
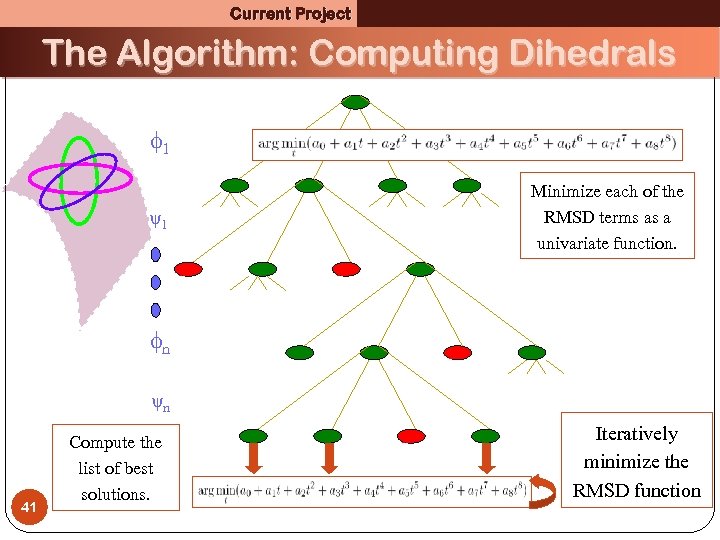 Current Project The Algorithm: Computing Dihedrals 1 Minimize each of the RMSD terms as a univariate function. ψ1 x x n x ψn 41 Compute the list of best solutions. x Iteratively minimize the RMSD function
Current Project The Algorithm: Computing Dihedrals 1 Minimize each of the RMSD terms as a univariate function. ψ1 x x n x ψn 41 Compute the list of best solutions. x Iteratively minimize the RMSD function
 Current Project Advantages • The algorithm is converging, since every step minimizes RMSD function • If the data was “perfect” then the solution to the minimization problem would be the roots of the polynomials in the RMSD terms, and the algorithm would find ALL of them. • The minima of the RMSD terms give a good collection of initial structures for finding local and global minima • Robust handling of missing values 42
Current Project Advantages • The algorithm is converging, since every step minimizes RMSD function • If the data was “perfect” then the solution to the minimization problem would be the roots of the polynomials in the RMSD terms, and the algorithm would find ALL of them. • The minima of the RMSD terms give a good collection of initial structures for finding local and global minima • Robust handling of missing values 42
![Preliminary Results: Ubiquitin Helix 43 Conformation of the portion [25 -31] of the helix Preliminary Results: Ubiquitin Helix 43 Conformation of the portion [25 -31] of the helix](https://present5.com/presentation/355ee1859d39ddd1b30946b9f69a98ac/image-43.jpg) Preliminary Results: Ubiquitin Helix 43 Conformation of the portion [25 -31] of the helix for human ubiquitin computed using NH and CH RDCs in two media (red) has been superimposed on the same portion from high-resolution X-ray structure (PDB Id: 1 UBQ) (green). The backbone RMSD is 0. 58 Å. Protein RMSD (Hz) Alignment Tensor (Syy, Szz) Ubq : 25 -31 C H : 0. 32 (23. 66, 16. 48) NH: 0. 24 (53. 25, 7. 65)
Preliminary Results: Ubiquitin Helix 43 Conformation of the portion [25 -31] of the helix for human ubiquitin computed using NH and CH RDCs in two media (red) has been superimposed on the same portion from high-resolution X-ray structure (PDB Id: 1 UBQ) (green). The backbone RMSD is 0. 58 Å. Protein RMSD (Hz) Alignment Tensor (Syy, Szz) Ubq : 25 -31 C H : 0. 32 (23. 66, 16. 48) NH: 0. 24 (53. 25, 7. 65)
![Preliminary Results: Ubiquitin Strand 44 Conformation of the portion [2 -7] of the beta-strand Preliminary Results: Ubiquitin Strand 44 Conformation of the portion [2 -7] of the beta-strand](https://present5.com/presentation/355ee1859d39ddd1b30946b9f69a98ac/image-44.jpg) Preliminary Results: Ubiquitin Strand 44 Conformation of the portion [2 -7] of the beta-strand for human ubiquitin computed using NH and CH RDCs in two media has been superimposed on the same portion from high-resolution X-ray structure (PDB Id: 1 UBQ). The backbone RMSD is 1. 151 Å. Protein RMSD (Hz) Alignment Tensor (Syy, Szz) Ubq: beta 2 -7 C H : (53. 32, 4. 83) NH: (48. 03, 14. 32)
Preliminary Results: Ubiquitin Strand 44 Conformation of the portion [2 -7] of the beta-strand for human ubiquitin computed using NH and CH RDCs in two media has been superimposed on the same portion from high-resolution X-ray structure (PDB Id: 1 UBQ). The backbone RMSD is 1. 151 Å. Protein RMSD (Hz) Alignment Tensor (Syy, Szz) Ubq: beta 2 -7 C H : (53. 32, 4. 83) NH: (48. 03, 14. 32)
 Conclusions • Complete and exhaustive search over the space of all structures minimizing the RDC fit function seems feasible due to understanding the structure of the solution. • Possible and exiting extensions to more/different data Funding: NIH Thank you! 45
Conclusions • Complete and exhaustive search over the space of all structures minimizing the RDC fit function seems feasible due to understanding the structure of the solution. • Possible and exiting extensions to more/different data Funding: NIH Thank you! 45
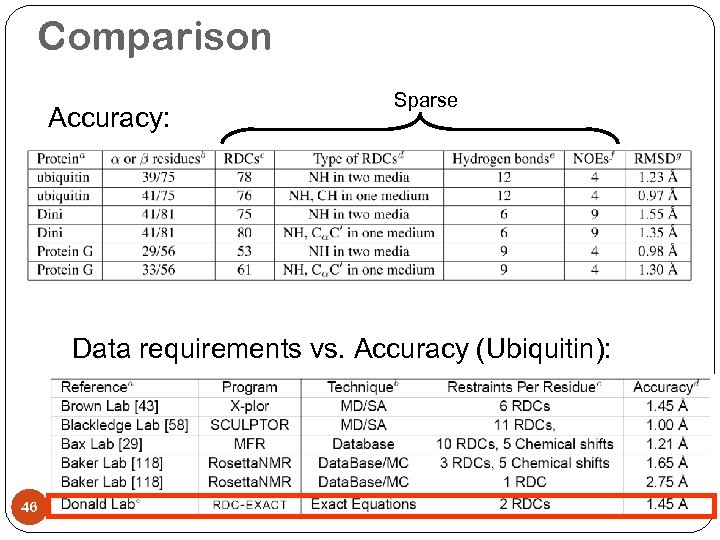 Comparison Accuracy: Sparse Data requirements vs. Accuracy (Ubiquitin): 46
Comparison Accuracy: Sparse Data requirements vs. Accuracy (Ubiquitin): 46


