8406f81fe4aa6690517624eb6f3e3a33.ppt
- Количество слайдов: 16
 Attention Deficit Hyperactivity Disorder ADHD
Attention Deficit Hyperactivity Disorder ADHD
 ADHD • Childhood-onset, severe impairing inattention, overactivity, impulsiveness • Clinical variability • Highly heritable especially when severe antisocial symptoms present
ADHD • Childhood-onset, severe impairing inattention, overactivity, impulsiveness • Clinical variability • Highly heritable especially when severe antisocial symptoms present
 Current work • Identifying large, rare CNVs • Identifying common variants-GWAS of 800 • Identifying biological pathways that underlie ADHD • What are the links between biology, genes and clinical variability in ADHD? • How do associated genes exert risk effects on the clinical outcome?
Current work • Identifying large, rare CNVs • Identifying common variants-GWAS of 800 • Identifying biological pathways that underlie ADHD • What are the links between biology, genes and clinical variability in ADHD? • How do associated genes exert risk effects on the clinical outcome?
 In-house results
In-house results
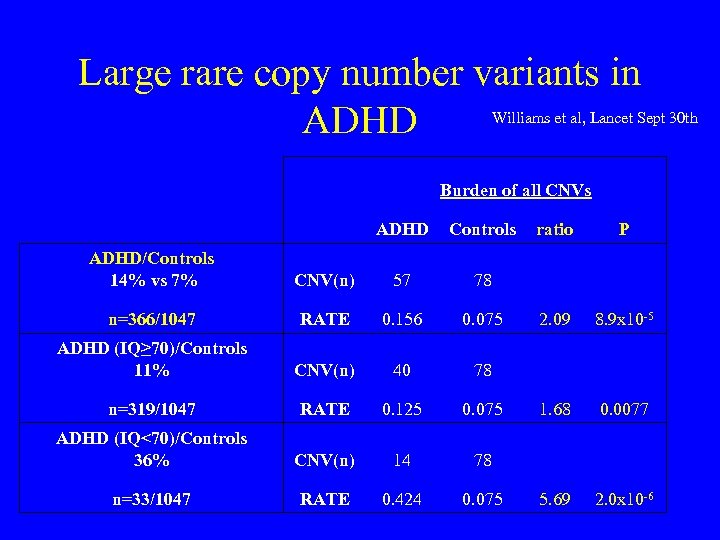 Large rare copy number variants in ADHD Williams et al, Lancet Sept 30 th Burden of all CNVs ADHD Controls ratio P ADHD/Controls 14% vs 7% CNV(n) 57 78 n=366/1047 RATE 0. 156 0. 075 2. 09 8. 9 x 10 -5 ADHD (IQ≥ 70)/Controls 11% CNV(n) 40 78 n=319/1047 RATE 0. 125 0. 075 1. 68 0. 0077 ADHD (IQ<70)/Controls 36% CNV(n) 14 78 n=33/1047 RATE 0. 424 0. 075 5. 69 2. 0 x 10 -6
Large rare copy number variants in ADHD Williams et al, Lancet Sept 30 th Burden of all CNVs ADHD Controls ratio P ADHD/Controls 14% vs 7% CNV(n) 57 78 n=366/1047 RATE 0. 156 0. 075 2. 09 8. 9 x 10 -5 ADHD (IQ≥ 70)/Controls 11% CNV(n) 40 78 n=319/1047 RATE 0. 125 0. 075 1. 68 0. 0077 ADHD (IQ<70)/Controls 36% CNV(n) 14 78 n=33/1047 RATE 0. 424 0. 075 5. 69 2. 0 x 10 -6
 Large rare CNVs in ADHD • Significant overlap of regions implicated in autism and schizophrenia • 16 p 13. 11 duplications OR 13. 9 p=0. 0008 (95% CI 2. 3 -82. 2)
Large rare CNVs in ADHD • Significant overlap of regions implicated in autism and schizophrenia • 16 p 13. 11 duplications OR 13. 9 p=0. 0008 (95% CI 2. 3 -82. 2)
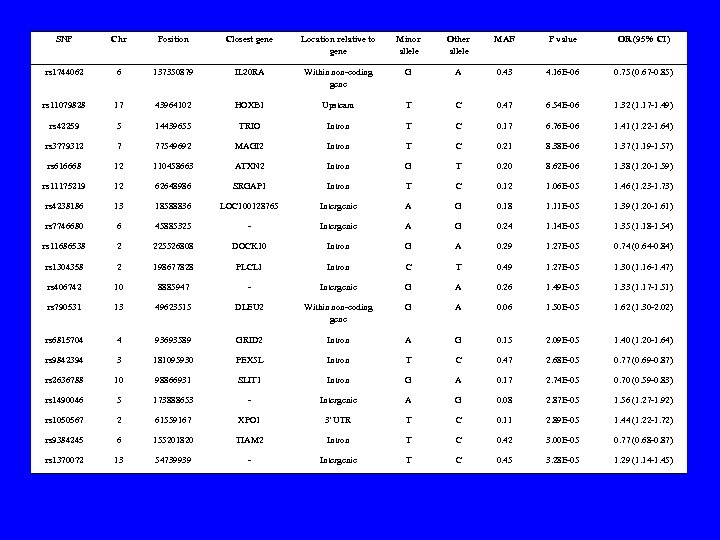 SNP Chr Position Closest gene Location relative to gene Minor allele Other allele MAF P value OR (95% CI) rs 1744062 6 137350879 IL 20 RA Within non-coding gene G A 0. 43 4. 16 E-06 0. 75 (0. 67 -0. 85) rs 11079828 17 43964102 HOXB 1 Upsteam T C 0. 47 6. 54 E-06 1. 32 (1. 17 -1. 49) rs 42259 5 14439655 TRIO Intron T C 0. 17 6. 76 E-06 1. 41 (1. 22 -1. 64) rs 3779312 7 77549692 MAGI 2 Intron T C 0. 21 8. 38 E-06 1. 37 (1. 19 -1. 57) rs 616668 12 110458663 ATXN 2 Intron G T 0. 20 8. 62 E-06 1. 38 (1. 20 -1. 59) rs 11175219 12 62648986 SRGAP 1 Intron T C 0. 12 1. 06 E-05 1. 46 (1. 23 -1. 73) rs 4238186 13 18588836 LOC 100128765 Intergenic A G 0. 18 1. 11 E-05 1. 39 (1. 20 -1. 61) rs 7746680 6 45885325 - Intergenic A G 0. 24 1. 14 E-05 1. 35 (1. 18 -1. 54) rs 11686538 2 225526808 DOCK 10 Intron G A 0. 29 1. 27 E-05 0. 74 (0. 64 -0. 84) rs 1304358 2 198677828 PLCL 1 Intron C T 0. 49 1. 27 E-05 1. 30 (1. 16 -1. 47) rs 406742 10 8885947 - Intergenic G A 0. 26 1. 49 E-05 1. 33 (1. 17 -1. 51) rs 790531 13 49623515 DLEU 2 Within non-coding gene G A 0. 06 1. 50 E-05 1. 62 (1. 30 -2. 02) rs 6815704 4 93693589 GRID 2 Intron A G 0. 15 2. 09 E-05 1. 40 (1. 20 -1. 64) rs 9842394 3 181095930 PEX 5 L Intron T C 0. 47 2. 68 E-05 0. 77 (0. 69 -0. 87) rs 2636788 10 98866931 SLIT 1 Intron G A 0. 17 2. 74 E-05 0. 70 (0. 59 -0. 83) rs 1490046 5 173888653 - Intergenic A G 0. 08 2. 87 E-05 1. 56 (1. 27 -1. 92) rs 1050567 2 61559167 XPO 1 3' UTR T C 0. 11 2. 89 E-05 1. 44 (1. 22 -1. 72) rs 9384245 6 155201820 TIAM 2 Intron T C 0. 42 3. 00 E-05 0. 77 (0. 68 -0. 87) rs 1370072 13 54739939 - Intergenic T C 0. 45 3. 28 E-05 1. 29 (1. 14 -1. 45)
SNP Chr Position Closest gene Location relative to gene Minor allele Other allele MAF P value OR (95% CI) rs 1744062 6 137350879 IL 20 RA Within non-coding gene G A 0. 43 4. 16 E-06 0. 75 (0. 67 -0. 85) rs 11079828 17 43964102 HOXB 1 Upsteam T C 0. 47 6. 54 E-06 1. 32 (1. 17 -1. 49) rs 42259 5 14439655 TRIO Intron T C 0. 17 6. 76 E-06 1. 41 (1. 22 -1. 64) rs 3779312 7 77549692 MAGI 2 Intron T C 0. 21 8. 38 E-06 1. 37 (1. 19 -1. 57) rs 616668 12 110458663 ATXN 2 Intron G T 0. 20 8. 62 E-06 1. 38 (1. 20 -1. 59) rs 11175219 12 62648986 SRGAP 1 Intron T C 0. 12 1. 06 E-05 1. 46 (1. 23 -1. 73) rs 4238186 13 18588836 LOC 100128765 Intergenic A G 0. 18 1. 11 E-05 1. 39 (1. 20 -1. 61) rs 7746680 6 45885325 - Intergenic A G 0. 24 1. 14 E-05 1. 35 (1. 18 -1. 54) rs 11686538 2 225526808 DOCK 10 Intron G A 0. 29 1. 27 E-05 0. 74 (0. 64 -0. 84) rs 1304358 2 198677828 PLCL 1 Intron C T 0. 49 1. 27 E-05 1. 30 (1. 16 -1. 47) rs 406742 10 8885947 - Intergenic G A 0. 26 1. 49 E-05 1. 33 (1. 17 -1. 51) rs 790531 13 49623515 DLEU 2 Within non-coding gene G A 0. 06 1. 50 E-05 1. 62 (1. 30 -2. 02) rs 6815704 4 93693589 GRID 2 Intron A G 0. 15 2. 09 E-05 1. 40 (1. 20 -1. 64) rs 9842394 3 181095930 PEX 5 L Intron T C 0. 47 2. 68 E-05 0. 77 (0. 69 -0. 87) rs 2636788 10 98866931 SLIT 1 Intron G A 0. 17 2. 74 E-05 0. 70 (0. 59 -0. 83) rs 1490046 5 173888653 - Intergenic A G 0. 08 2. 87 E-05 1. 56 (1. 27 -1. 92) rs 1050567 2 61559167 XPO 1 3' UTR T C 0. 11 2. 89 E-05 1. 44 (1. 22 -1. 72) rs 9384245 6 155201820 TIAM 2 Intron T C 0. 42 3. 00 E-05 0. 77 (0. 68 -0. 87) rs 1370072 13 54739939 - Intergenic T C 0. 45 3. 28 E-05 1. 29 (1. 14 -1. 45)
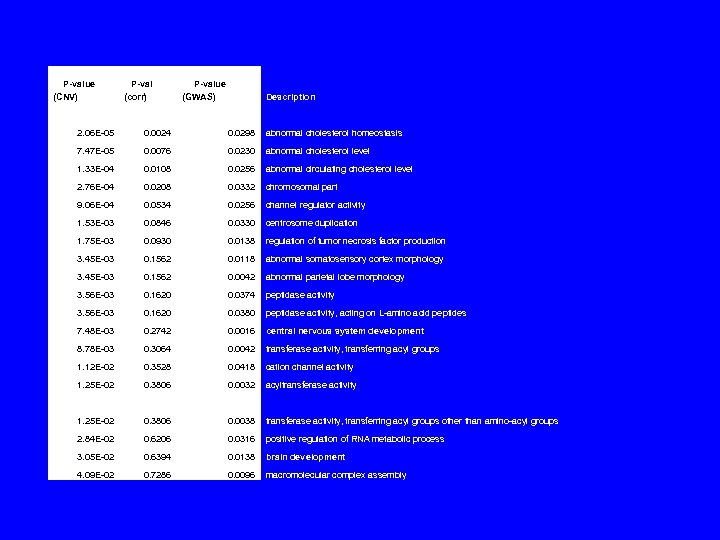 P-value (CNV) P-val (corr) P-value (GWAS) Description 2. 06 E-05 0. 0024 0. 0298 abnormal cholesterol homeostasis 7. 47 E-05 0. 0076 0. 0230 abnormal cholesterol level 1. 33 E-04 0. 0108 0. 0256 abnormal circulating cholesterol level 2. 76 E-04 0. 0208 0. 0332 chromosomal part 9. 06 E-04 0. 0534 0. 0256 channel regulator activity 1. 53 E-03 0. 0846 0. 0330 centrosome duplication 1. 75 E-03 0. 0930 0. 0138 regulation of tumor necrosis factor production 3. 45 E-03 0. 1562 0. 0118 abnormal somatosensory cortex morphology 3. 45 E-03 0. 1562 0. 0042 abnormal parietal lobe morphology 3. 56 E-03 0. 1620 0. 0374 peptidase activity 3. 56 E-03 0. 1620 0. 0380 peptidase activity, acting on L-amino acid peptides 7. 48 E-03 0. 2742 0. 0016 central nervous system development 8. 78 E-03 0. 3064 0. 0042 transferase activity, transferring acyl groups 1. 12 E-02 0. 3528 0. 0418 cation channel activity 1. 25 E-02 0. 3806 0. 0032 acyltransferase activity 1. 25 E-02 0. 3806 0. 0038 transferase activity, transferring acyl groups other than amino-acyl groups 2. 84 E-02 0. 6206 0. 0316 positive regulation of RNA metabolic process 3. 05 E-02 0. 6394 0. 0138 brain development 4. 09 E-02 0. 7286 0. 0096 macromolecular complex assembly
P-value (CNV) P-val (corr) P-value (GWAS) Description 2. 06 E-05 0. 0024 0. 0298 abnormal cholesterol homeostasis 7. 47 E-05 0. 0076 0. 0230 abnormal cholesterol level 1. 33 E-04 0. 0108 0. 0256 abnormal circulating cholesterol level 2. 76 E-04 0. 0208 0. 0332 chromosomal part 9. 06 E-04 0. 0534 0. 0256 channel regulator activity 1. 53 E-03 0. 0846 0. 0330 centrosome duplication 1. 75 E-03 0. 0930 0. 0138 regulation of tumor necrosis factor production 3. 45 E-03 0. 1562 0. 0118 abnormal somatosensory cortex morphology 3. 45 E-03 0. 1562 0. 0042 abnormal parietal lobe morphology 3. 56 E-03 0. 1620 0. 0374 peptidase activity 3. 56 E-03 0. 1620 0. 0380 peptidase activity, acting on L-amino acid peptides 7. 48 E-03 0. 2742 0. 0016 central nervous system development 8. 78 E-03 0. 3064 0. 0042 transferase activity, transferring acyl groups 1. 12 E-02 0. 3528 0. 0418 cation channel activity 1. 25 E-02 0. 3806 0. 0032 acyltransferase activity 1. 25 E-02 0. 3806 0. 0038 transferase activity, transferring acyl groups other than amino-acyl groups 2. 84 E-02 0. 6206 0. 0316 positive regulation of RNA metabolic process 3. 05 E-02 0. 6394 0. 0138 brain development 4. 09 E-02 0. 7286 0. 0096 macromolecular complex assembly
 Questions? • Does clinical variability in presentation index biological heterogeneity? • Can we pick out biologically homogeneous subtypes? • How do our GWAS and CNV findings relate to animal models of ADHD and imaging studies showing affected brain regions?
Questions? • Does clinical variability in presentation index biological heterogeneity? • Can we pick out biologically homogeneous subtypes? • How do our GWAS and CNV findings relate to animal models of ADHD and imaging studies showing affected brain regions?
 COMT influencing antisocial behaviour in ADHD
COMT influencing antisocial behaviour in ADHD
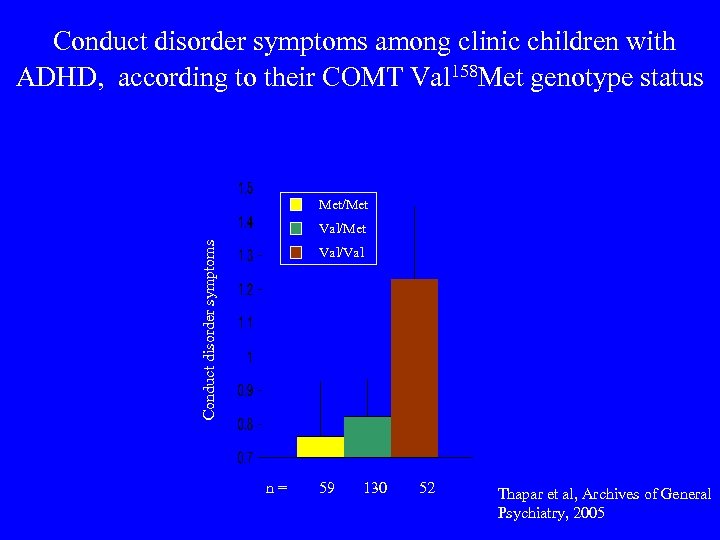 Conduct disorder symptoms among clinic children with ADHD, according to their COMT Val 158 Met genotype status Met/Met Conduct disorder symptoms Val/Met Val/Val n = 59 130 52 Thapar et al, Archives of General Psychiatry, 2005
Conduct disorder symptoms among clinic children with ADHD, according to their COMT Val 158 Met genotype status Met/Met Conduct disorder symptoms Val/Met Val/Val n = 59 130 52 Thapar et al, Archives of General Psychiatry, 2005
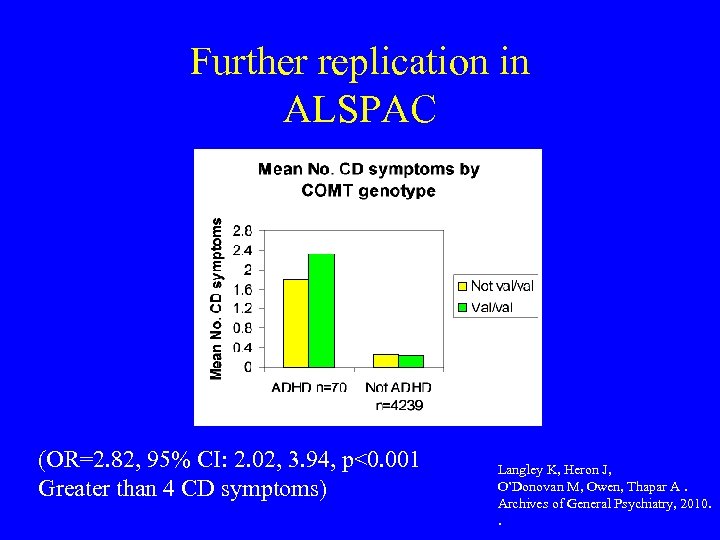 Further replication in ALSPAC (OR=2. 82, 95% CI: 2. 02, 3. 94, p<0. 001 Greater than 4 CD symptoms) Langley K, Heron J, O’Donovan M, Owen, Thapar A. Archives of General Psychiatry, 2010. .
Further replication in ALSPAC (OR=2. 82, 95% CI: 2. 02, 3. 94, p<0. 001 Greater than 4 CD symptoms) Langley K, Heron J, O’Donovan M, Owen, Thapar A. Archives of General Psychiatry, 2010. .
 Mechanisms? COMT Val/val Antisocial behaviour in ADHD
Mechanisms? COMT Val/val Antisocial behaviour in ADHD
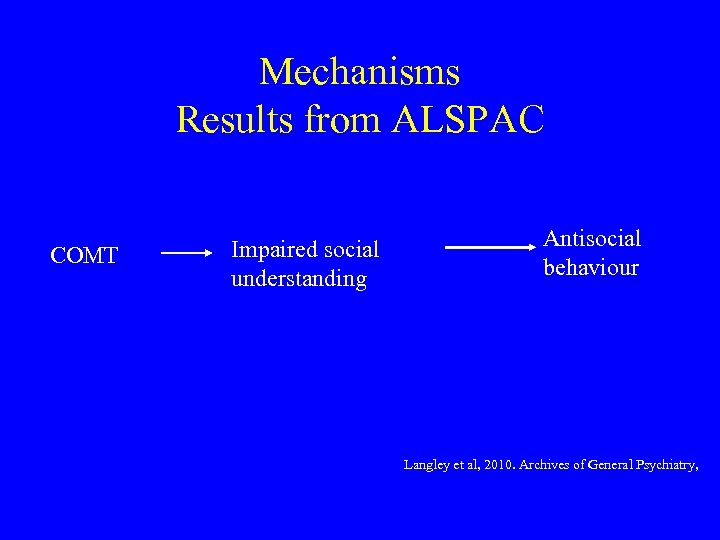 Mechanisms Results from ALSPAC COMT Impaired social understanding Antisocial behaviour Langley et al, 2010. Archives of General Psychiatry,
Mechanisms Results from ALSPAC COMT Impaired social understanding Antisocial behaviour Langley et al, 2010. Archives of General Psychiatry,
 What next? • Detailed cognitive and psychophysiological testing of ADHD children in experimental lab based on COMT Val 158 Met genotype • What should we be looking at (gene/mutation-wise) that might be important in relation to COMT function and related pathways?
What next? • Detailed cognitive and psychophysiological testing of ADHD children in experimental lab based on COMT Val 158 Met genotype • What should we be looking at (gene/mutation-wise) that might be important in relation to COMT function and related pathways?
 …. Final add on • 3 wave study of psychiatric problems in adolescents who are at high genetic risk, developing a risk prediction tool • Developing a risk prediction tool that is being incorporated into GP software (commercial software company) • Computing/risk prediction expertise?
…. Final add on • 3 wave study of psychiatric problems in adolescents who are at high genetic risk, developing a risk prediction tool • Developing a risk prediction tool that is being incorporated into GP software (commercial software company) • Computing/risk prediction expertise?


