66f64c6efdda62948119162a06745072.ppt
- Количество слайдов: 38

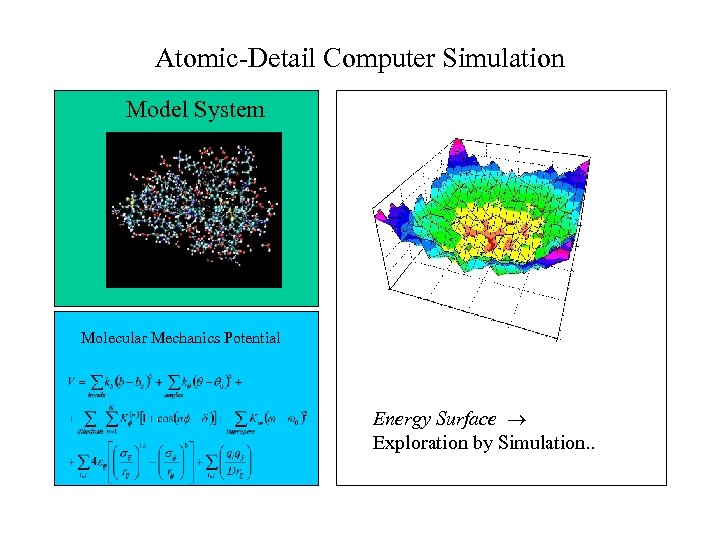 Atomic-Detail Computer Simulation Model System Molecular Mechanics Potential Energy Surface Exploration by Simulation. .
Atomic-Detail Computer Simulation Model System Molecular Mechanics Potential Energy Surface Exploration by Simulation. .
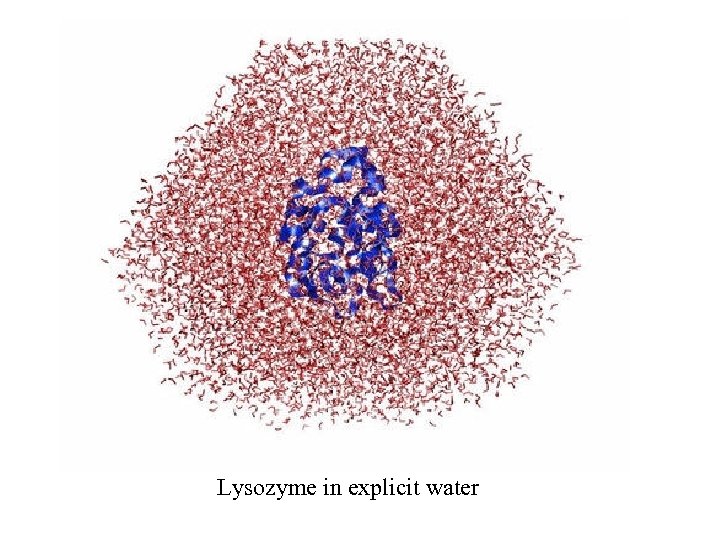 Lysozyme in explicit water
Lysozyme in explicit water
 Model System • set of atoms • explicit/implicit solvent • periodic boundary conditions Potential Function • empirical • chemically intuitive • quick to calculate Tradeoff: simplicity (timescale) versus accuracy
Model System • set of atoms • explicit/implicit solvent • periodic boundary conditions Potential Function • empirical • chemically intuitive • quick to calculate Tradeoff: simplicity (timescale) versus accuracy
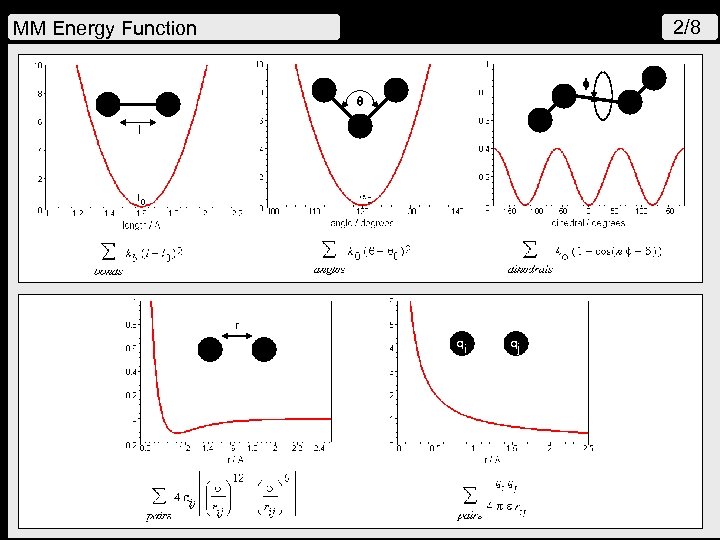 2/8 MM Energy Function q l r qi qj
2/8 MM Energy Function q l r qi qj
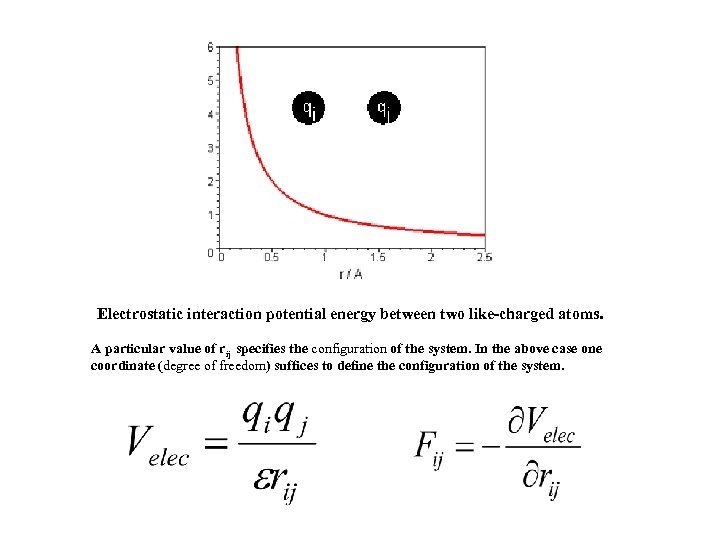 Electrostatic interaction potential energy between two like-charged atoms. A particular value of rij specifies the configuration of the system. In the above case one coordinate (degree of freedom) suffices to define the configuration of the system.
Electrostatic interaction potential energy between two like-charged atoms. A particular value of rij specifies the configuration of the system. In the above case one coordinate (degree of freedom) suffices to define the configuration of the system.
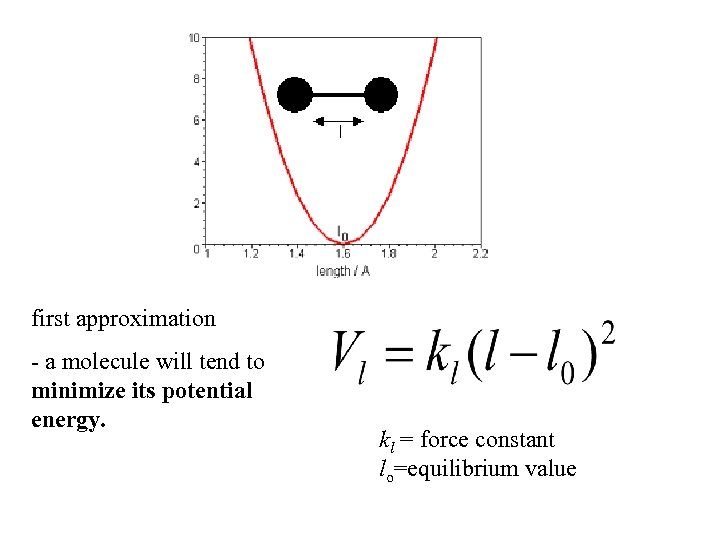 first approximation - a molecule will tend to minimize its potential energy. kl = force constant lo=equilibrium value
first approximation - a molecule will tend to minimize its potential energy. kl = force constant lo=equilibrium value

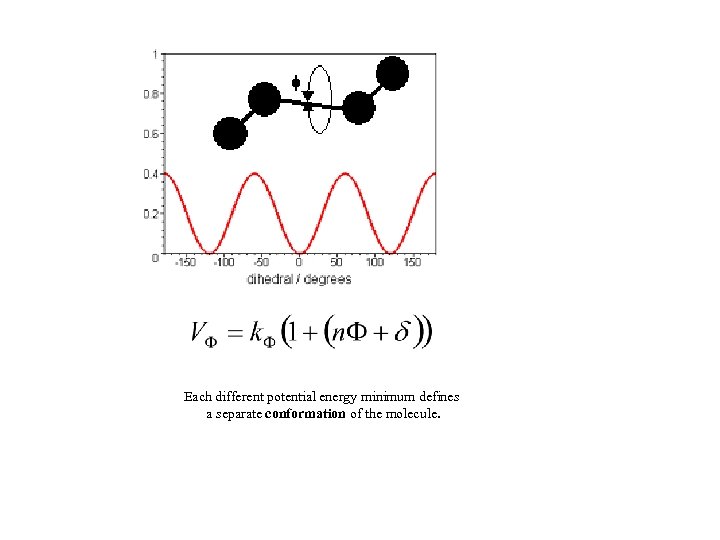 Each different potential energy minimum defines a separate conformation of the molecule.
Each different potential energy minimum defines a separate conformation of the molecule.

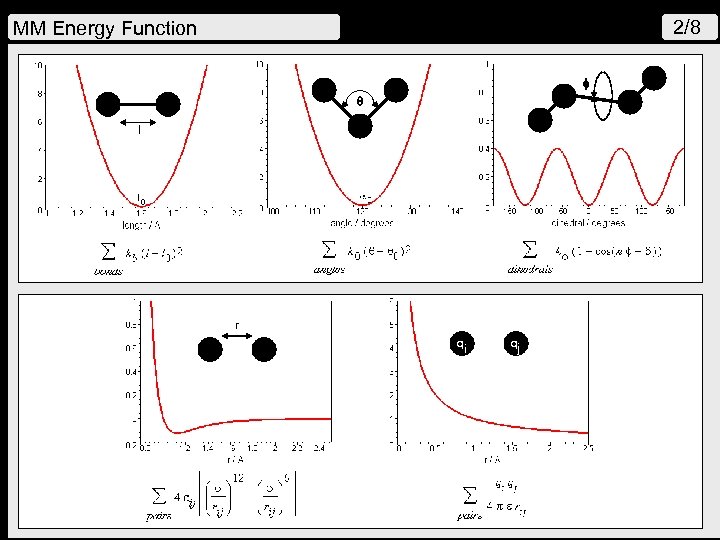 2/8 MM Energy Function q l r qi qj
2/8 MM Energy Function q l r qi qj
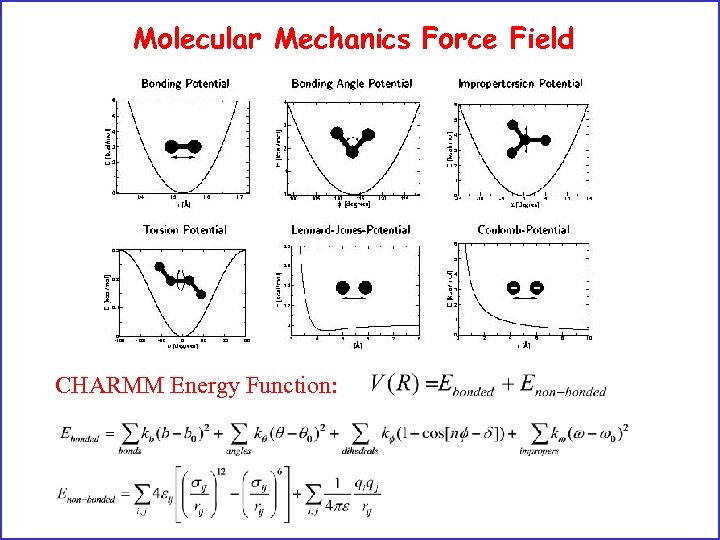 Molecular Mechanics Force Field CHARMM Energy Function:
Molecular Mechanics Force Field CHARMM Energy Function:
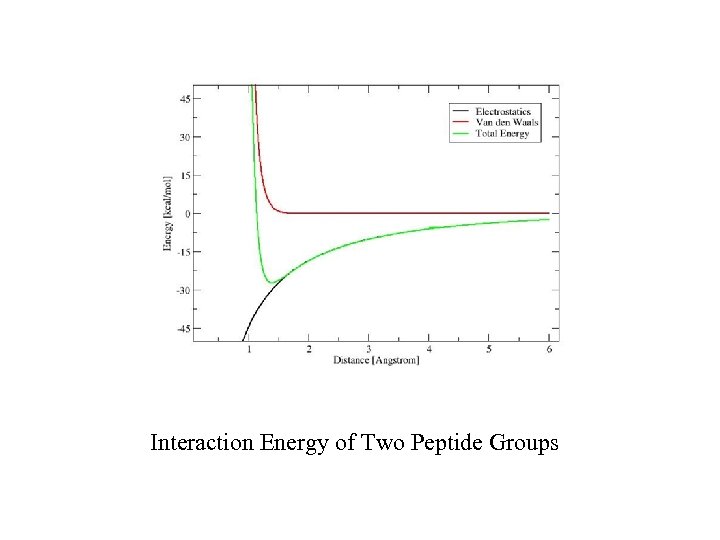 Interaction Energy of Two Peptide Groups
Interaction Energy of Two Peptide Groups
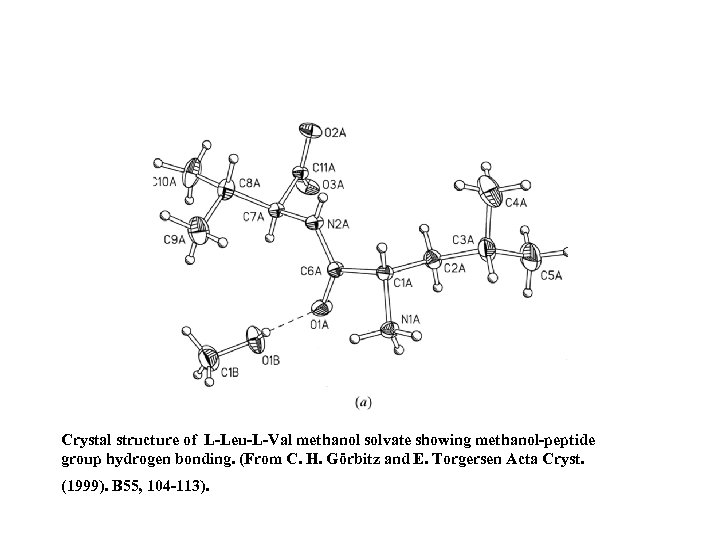 Crystal structure of L-Leu-L-Val methanol solvate showing methanol-peptide group hydrogen bonding. (From C. H. Görbitz and E. Torgersen Acta Cryst. (1999). B 55, 104 -113).
Crystal structure of L-Leu-L-Val methanol solvate showing methanol-peptide group hydrogen bonding. (From C. H. Görbitz and E. Torgersen Acta Cryst. (1999). B 55, 104 -113).
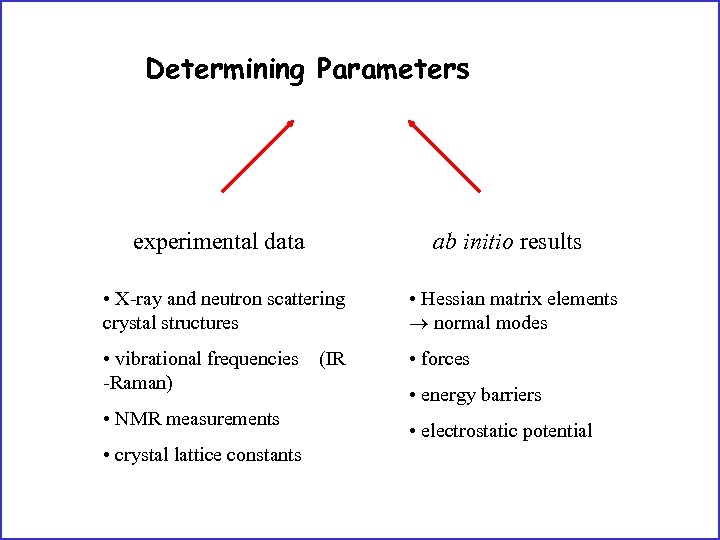 Determining Parameters experimental data ab initio results • X-ray and neutron scattering crystal structures • Hessian matrix elements normal modes • vibrational frequencies (IR -Raman) • forces • NMR measurements • crystal lattice constants • energy barriers • electrostatic potential
Determining Parameters experimental data ab initio results • X-ray and neutron scattering crystal structures • Hessian matrix elements normal modes • vibrational frequencies (IR -Raman) • forces • NMR measurements • crystal lattice constants • energy barriers • electrostatic potential
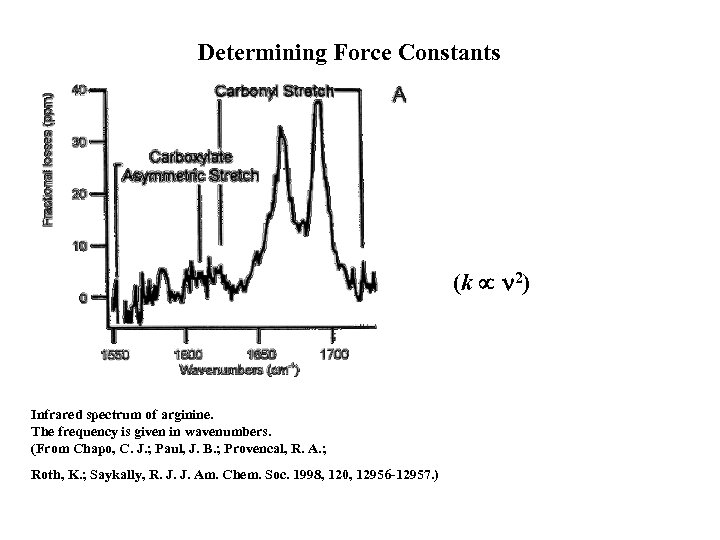 Determining Force Constants (k 2) Infrared spectrum of arginine. The frequency is given in wavenumbers. (From Chapo, C. J. ; Paul, J. B. ; Provencal, R. A. ; Roth, K. ; Saykally, R. J. J. Am. Chem. Soc. 1998, 120, 12956 -12957. )
Determining Force Constants (k 2) Infrared spectrum of arginine. The frequency is given in wavenumbers. (From Chapo, C. J. ; Paul, J. B. ; Provencal, R. A. ; Roth, K. ; Saykally, R. J. J. Am. Chem. Soc. 1998, 120, 12956 -12957. )
 Basics of Quantum Chemistry. Schrödinger equation: H =E where E is the energy of the system, H is the Hamiltonian operator, H=T+V. V=Vnn+Vne+Vee. Born-Oppenheimer Approximation Potential Energy Surface.
Basics of Quantum Chemistry. Schrödinger equation: H =E where E is the energy of the system, H is the Hamiltonian operator, H=T+V. V=Vnn+Vne+Vee. Born-Oppenheimer Approximation Potential Energy Surface.
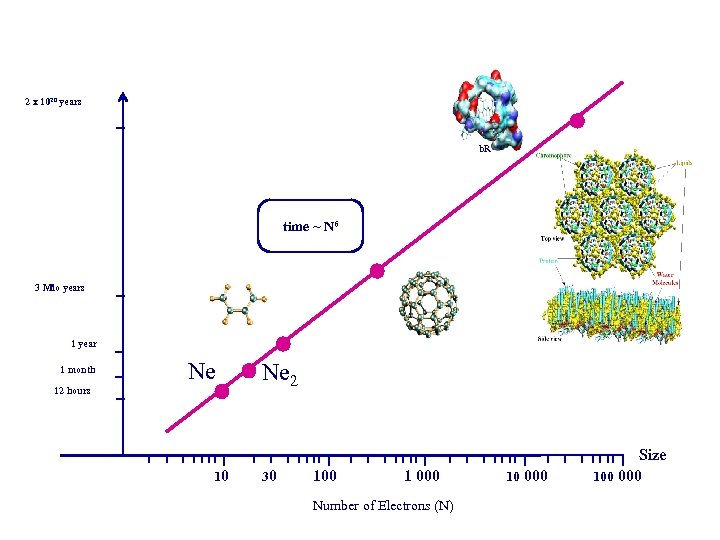 2 x 1020 years b. R time ~ N 6 3 Mio years 1 year 1 month Ne 12 hours 10 Ne 2 30 100 1 000 Number of Electrons (N) 10 000 Size 100 000
2 x 1020 years b. R time ~ N 6 3 Mio years 1 year 1 month Ne 12 hours 10 Ne 2 30 100 1 000 Number of Electrons (N) 10 000 Size 100 000
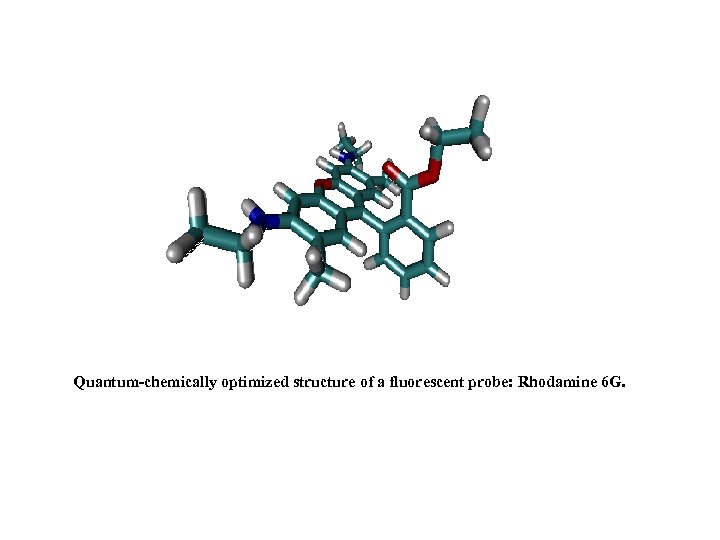 Quantum-chemically optimized structure of a fluorescent probe: Rhodamine 6 G.
Quantum-chemically optimized structure of a fluorescent probe: Rhodamine 6 G.
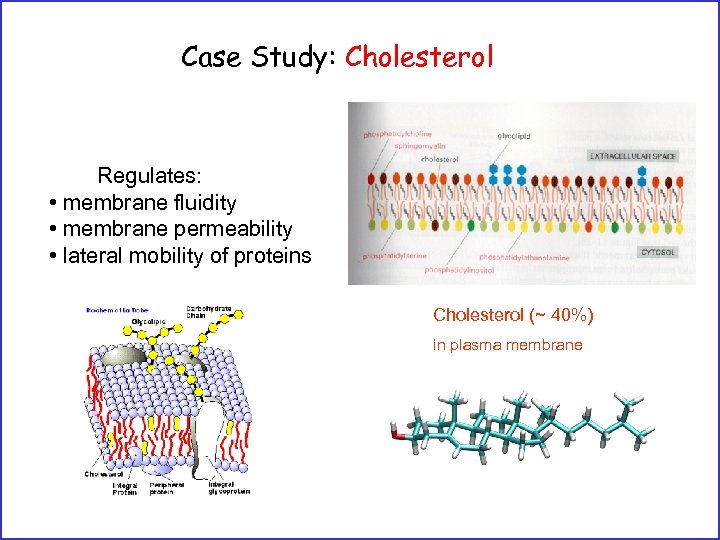 Case Study: Cholesterol Regulates: • membrane fluidity • membrane permeability • lateral mobility of proteins Cholesterol (~ 40%) in plasma membrane
Case Study: Cholesterol Regulates: • membrane fluidity • membrane permeability • lateral mobility of proteins Cholesterol (~ 40%) in plasma membrane
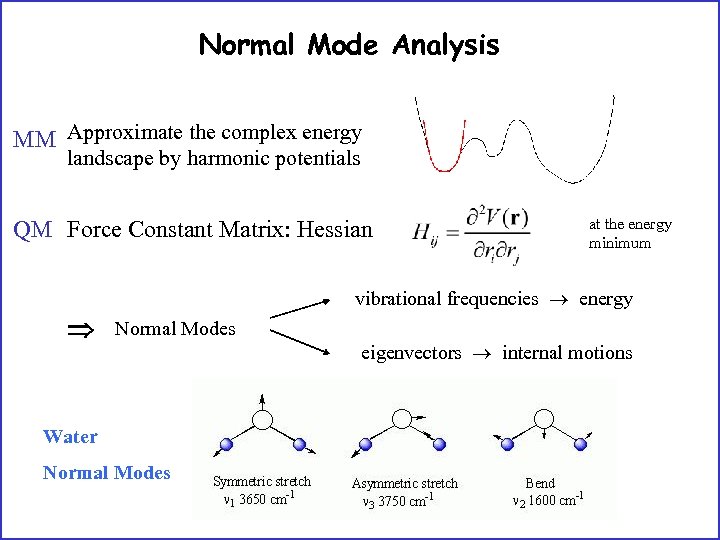 Normal Mode Analysis MM Approximate the complex energy landscape by harmonic potentials QM Force Constant Matrix: Hessian at the energy minimum vibrational frequencies energy Normal Modes Water Normal Modes eigenvectors internal motions
Normal Mode Analysis MM Approximate the complex energy landscape by harmonic potentials QM Force Constant Matrix: Hessian at the energy minimum vibrational frequencies energy Normal Modes Water Normal Modes eigenvectors internal motions
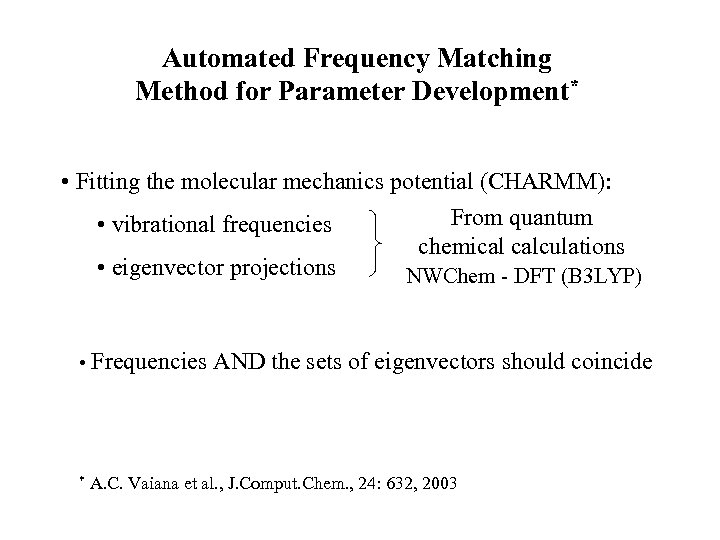 Automated Frequency Matching Method for Parameter Development* • Fitting the molecular mechanics potential (CHARMM): From quantum • vibrational frequencies chemical calculations • eigenvector projections NWChem - DFT (B 3 LYP) • Frequencies AND the sets of eigenvectors should coincide * A. C. Vaiana et al. , J. Comput. Chem. , 24: 632, 2003
Automated Frequency Matching Method for Parameter Development* • Fitting the molecular mechanics potential (CHARMM): From quantum • vibrational frequencies chemical calculations • eigenvector projections NWChem - DFT (B 3 LYP) • Frequencies AND the sets of eigenvectors should coincide * A. C. Vaiana et al. , J. Comput. Chem. , 24: 632, 2003
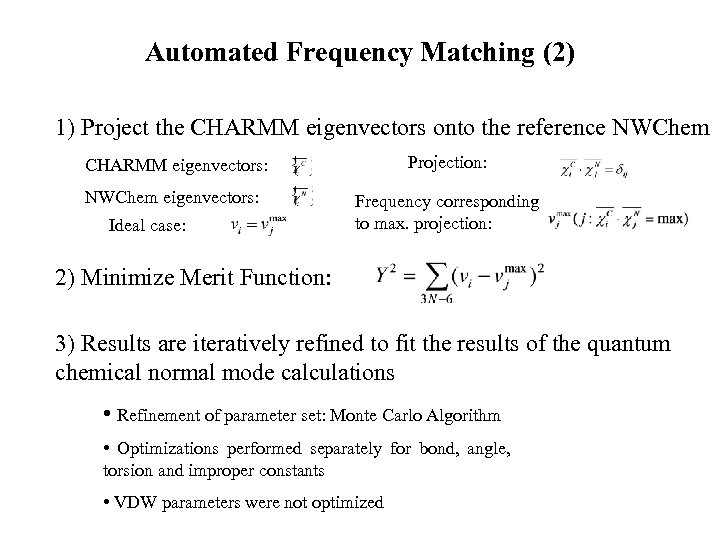 Automated Frequency Matching (2) 1) Project the CHARMM eigenvectors onto the reference NWChem CHARMM eigenvectors: Projection: NWChem eigenvectors: Frequency corresponding to max. projection: Ideal case: 2) Minimize Merit Function: 3) Results are iteratively refined to fit the results of the quantum chemical normal mode calculations • Refinement of parameter set: Monte Carlo Algorithm • Optimizations performed separately for bond, angle, torsion and improper constants • VDW parameters were not optimized
Automated Frequency Matching (2) 1) Project the CHARMM eigenvectors onto the reference NWChem CHARMM eigenvectors: Projection: NWChem eigenvectors: Frequency corresponding to max. projection: Ideal case: 2) Minimize Merit Function: 3) Results are iteratively refined to fit the results of the quantum chemical normal mode calculations • Refinement of parameter set: Monte Carlo Algorithm • Optimizations performed separately for bond, angle, torsion and improper constants • VDW parameters were not optimized
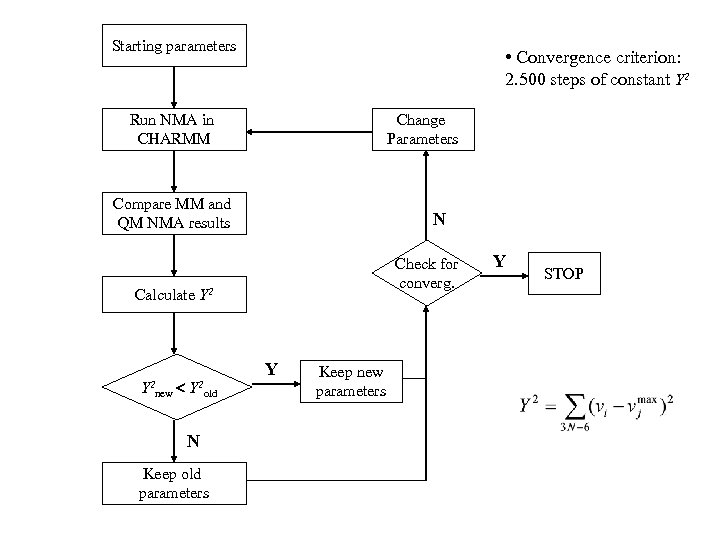 Starting parameters • Convergence criterion: 2. 500 steps of constant Y 2 Run NMA in CHARMM Change Parameters Compare MM and QM NMA results N Check for converg. Calculate Y 2 new Y 2 old N Keep old parameters Y Keep new parameters Y STOP
Starting parameters • Convergence criterion: 2. 500 steps of constant Y 2 Run NMA in CHARMM Change Parameters Compare MM and QM NMA results N Check for converg. Calculate Y 2 new Y 2 old N Keep old parameters Y Keep new parameters Y STOP
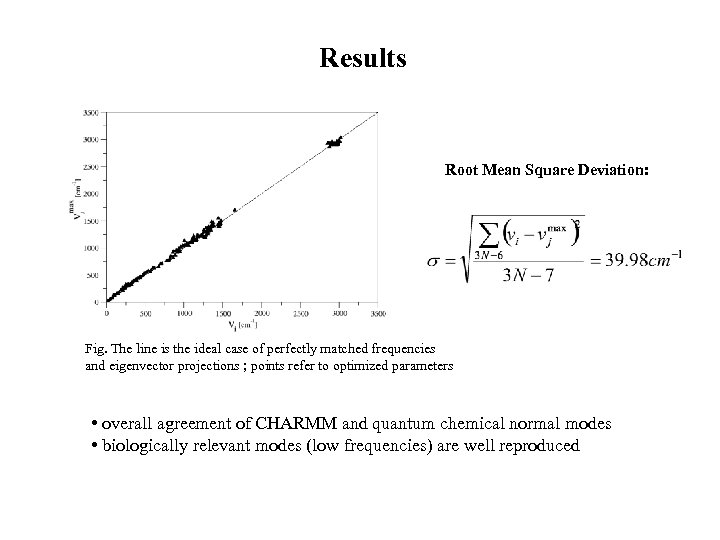 Results Root Mean Square Deviation: Fig. The line is the ideal case of perfectly matched frequencies and eigenvector projections ; points refer to optimized parameters • overall agreement of CHARMM and quantum chemical normal modes • biologically relevant modes (low frequencies) are well reproduced
Results Root Mean Square Deviation: Fig. The line is the ideal case of perfectly matched frequencies and eigenvector projections ; points refer to optimized parameters • overall agreement of CHARMM and quantum chemical normal modes • biologically relevant modes (low frequencies) are well reproduced
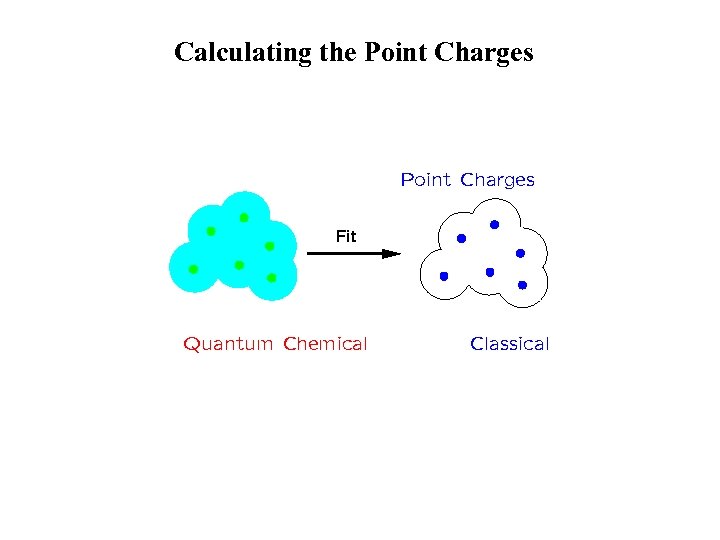 Calculating the Point Charges
Calculating the Point Charges
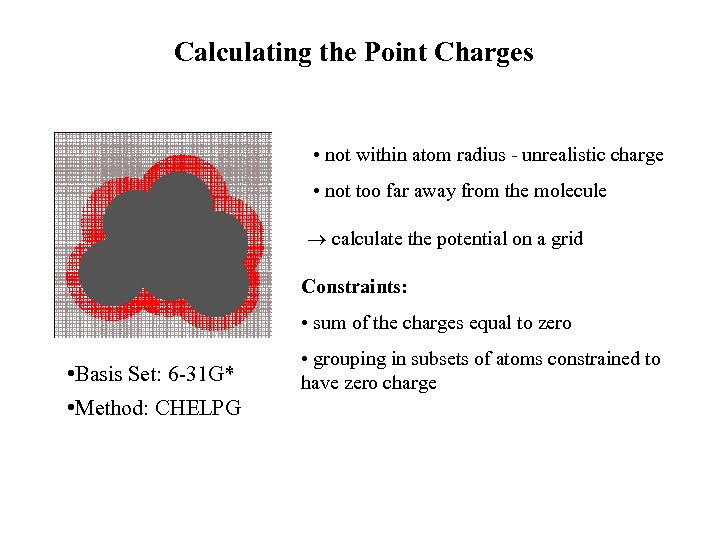 Calculating the Point Charges • not within atom radius - unrealistic charge • not too far away from the molecule calculate the potential on a grid Constraints: • sum of the charges equal to zero • Basis Set: 6 -31 G* • Method: CHELPG • grouping in subsets of atoms constrained to have zero charge
Calculating the Point Charges • not within atom radius - unrealistic charge • not too far away from the molecule calculate the potential on a grid Constraints: • sum of the charges equal to zero • Basis Set: 6 -31 G* • Method: CHELPG • grouping in subsets of atoms constrained to have zero charge
 The electrostatic potential (r) at a point r is defined as the work done to bring a unit positive charge from infinity to the point. The electrostatic interaction energy between a point charge q located at r and the molecule equals q (r). Electrostatic potential mapped onto the electron density surface for 2 -bromo-2 -chloro-1, 1, 1 -trifluoroethane (halothane). (From: Pei Tang, Igor Zubryzcki, Yan Xu J comp chem. 22 436 (2001)).
The electrostatic potential (r) at a point r is defined as the work done to bring a unit positive charge from infinity to the point. The electrostatic interaction energy between a point charge q located at r and the molecule equals q (r). Electrostatic potential mapped onto the electron density surface for 2 -bromo-2 -chloro-1, 1, 1 -trifluoroethane (halothane). (From: Pei Tang, Igor Zubryzcki, Yan Xu J comp chem. 22 436 (2001)).
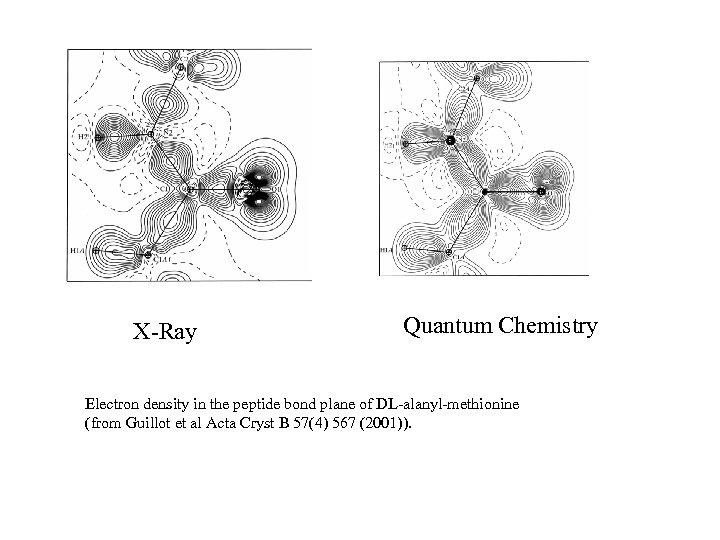 X-Ray Quantum Chemistry Electron density in the peptide bond plane of DL-alanyl-methionine (from Guillot et al Acta Cryst B 57(4) 567 (2001)).
X-Ray Quantum Chemistry Electron density in the peptide bond plane of DL-alanyl-methionine (from Guillot et al Acta Cryst B 57(4) 567 (2001)).
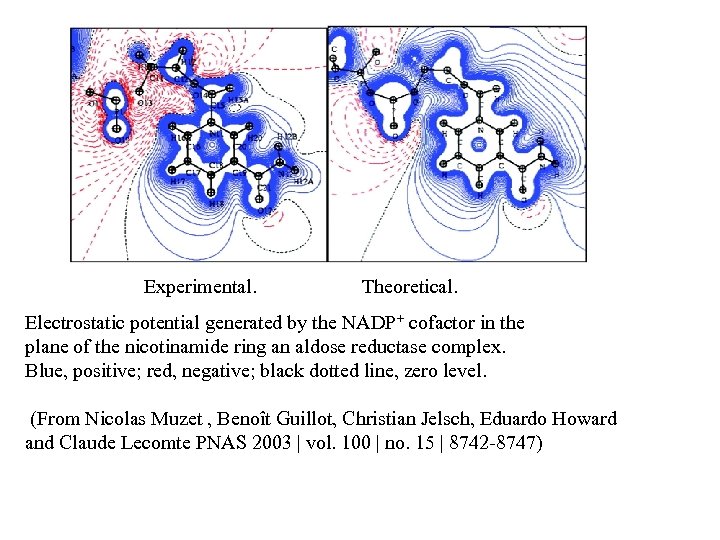 Experimental. Theoretical. Electrostatic potential generated by the NADP+ cofactor in the plane of the nicotinamide ring an aldose reductase complex. Blue, positive; red, negative; black dotted line, zero level. (From Nicolas Muzet , Benoît Guillot, Christian Jelsch, Eduardo Howard and Claude Lecomte PNAS 2003 | vol. 100 | no. 15 | 8742 -8747)
Experimental. Theoretical. Electrostatic potential generated by the NADP+ cofactor in the plane of the nicotinamide ring an aldose reductase complex. Blue, positive; red, negative; black dotted line, zero level. (From Nicolas Muzet , Benoît Guillot, Christian Jelsch, Eduardo Howard and Claude Lecomte PNAS 2003 | vol. 100 | no. 15 | 8742 -8747)
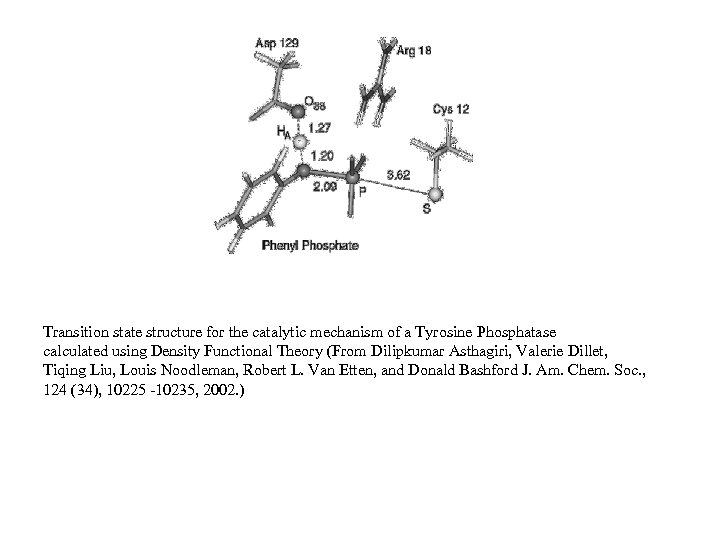 Transition state structure for the catalytic mechanism of a Tyrosine Phosphatase calculated using Density Functional Theory (From Dilipkumar Asthagiri, Valerie Dillet, Tiqing Liu, Louis Noodleman, Robert L. Van Etten, and Donald Bashford J. Am. Chem. Soc. , 124 (34), 10225 -10235, 2002. )
Transition state structure for the catalytic mechanism of a Tyrosine Phosphatase calculated using Density Functional Theory (From Dilipkumar Asthagiri, Valerie Dillet, Tiqing Liu, Louis Noodleman, Robert L. Van Etten, and Donald Bashford J. Am. Chem. Soc. , 124 (34), 10225 -10235, 2002. )
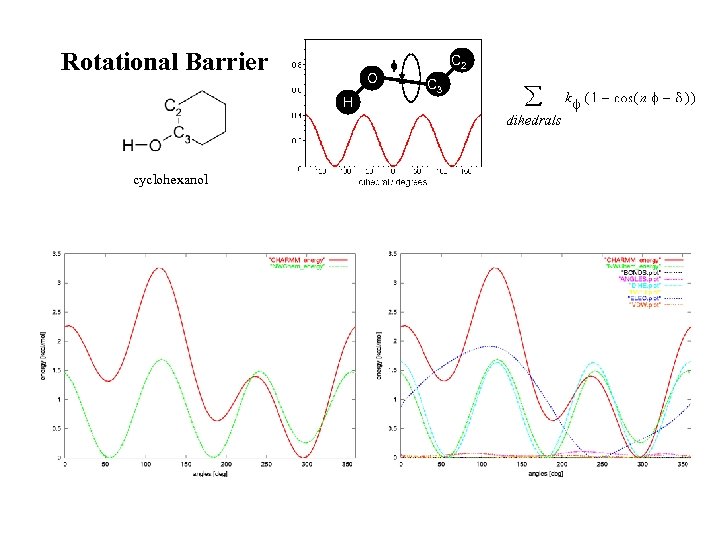 Rotational Barrier O H cyclohexanol C 2 C 3
Rotational Barrier O H cyclohexanol C 2 C 3
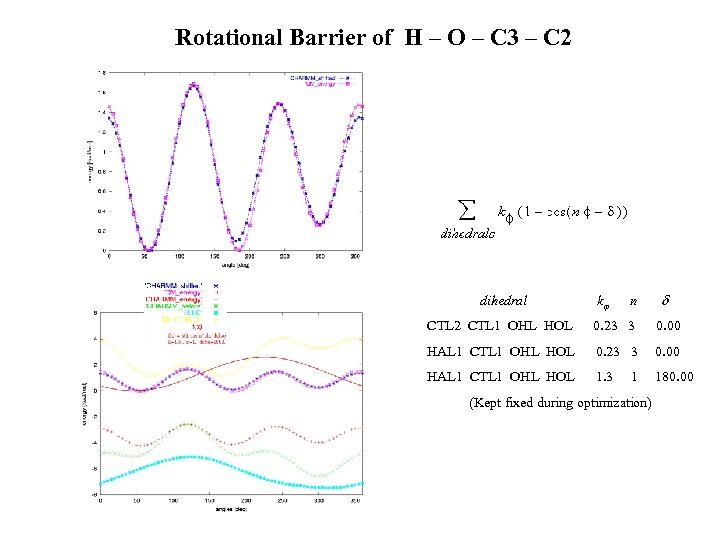 Rotational Barrier of H – O – C 3 – C 2 dihedral k n CTL 2 CTL 1 OHL HOL 0. 23 3 0. 00 HAL 1 CTL 1 OHL HOL 1. 3 180. 00 1 (Kept fixed during optimization)
Rotational Barrier of H – O – C 3 – C 2 dihedral k n CTL 2 CTL 1 OHL HOL 0. 23 3 0. 00 HAL 1 CTL 1 OHL HOL 1. 3 180. 00 1 (Kept fixed during optimization)

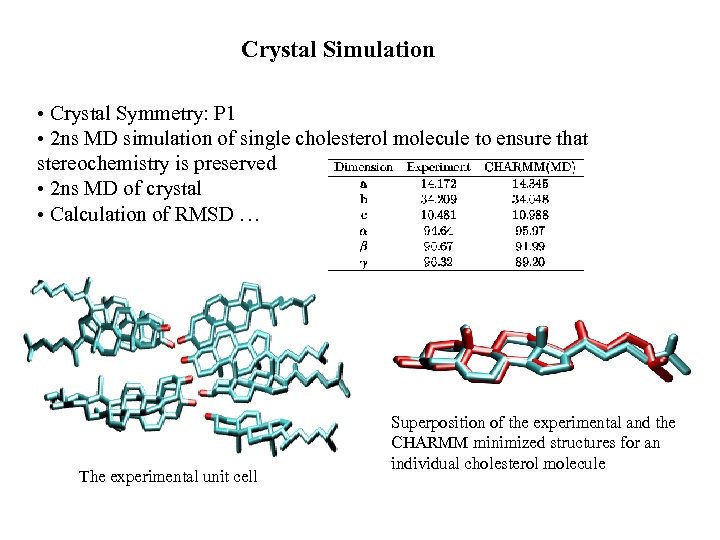 Crystal Simulation • Crystal Symmetry: P 1 • 2 ns MD simulation of single cholesterol molecule to ensure that stereochemistry is preserved • 2 ns MD of crystal • Calculation of RMSD … The experimental unit cell Superposition of the experimental and the CHARMM minimized structures for an individual cholesterol molecule
Crystal Simulation • Crystal Symmetry: P 1 • 2 ns MD simulation of single cholesterol molecule to ensure that stereochemistry is preserved • 2 ns MD of crystal • Calculation of RMSD … The experimental unit cell Superposition of the experimental and the CHARMM minimized structures for an individual cholesterol molecule
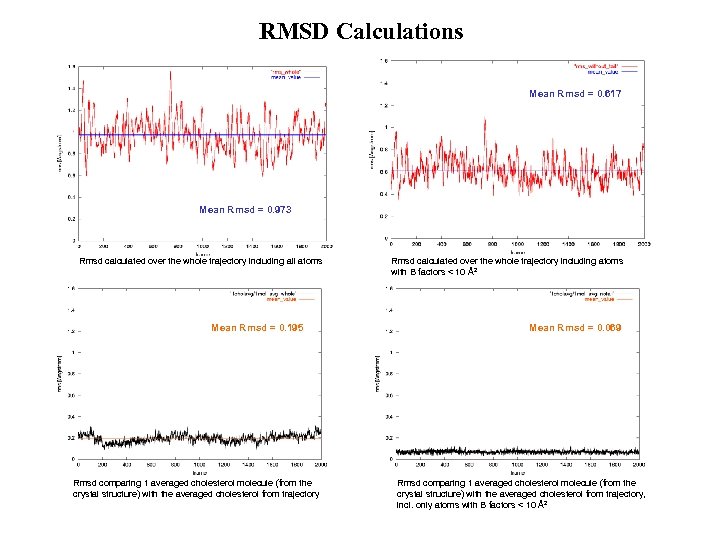 RMSD Calculations Mean Rmsd = 0. 617 Mean Rmsd = 0. 973 Rmsd calculated over the whole trajectory including all atoms Mean Rmsd = 0. 195 Rmsd comparing 1 averaged cholesterol molecule (from the crystal structure) with the averaged cholesterol from trajectory Rmsd calculated over the whole trajectory including atoms with B factors < 10 Å2 Mean Rmsd = 0. 069 Rmsd comparing 1 averaged cholesterol molecule (from the crystal structure) with the averaged cholesterol from trajectory, incl. only atoms with B factors < 10 Å2
RMSD Calculations Mean Rmsd = 0. 617 Mean Rmsd = 0. 973 Rmsd calculated over the whole trajectory including all atoms Mean Rmsd = 0. 195 Rmsd comparing 1 averaged cholesterol molecule (from the crystal structure) with the averaged cholesterol from trajectory Rmsd calculated over the whole trajectory including atoms with B factors < 10 Å2 Mean Rmsd = 0. 069 Rmsd comparing 1 averaged cholesterol molecule (from the crystal structure) with the averaged cholesterol from trajectory, incl. only atoms with B factors < 10 Å2

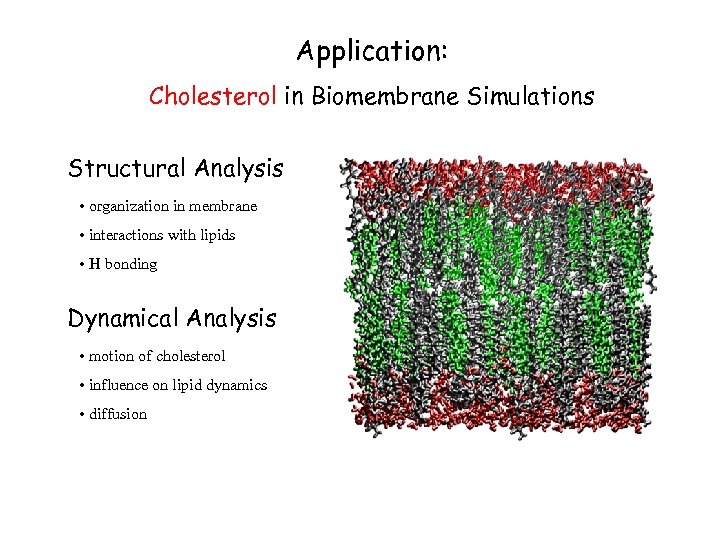 Application: Cholesterol in Biomembrane Simulations Structural Analysis • organization in membrane • interactions with lipids • H bonding Dynamical Analysis • motion of cholesterol • influence on lipid dynamics • diffusion
Application: Cholesterol in Biomembrane Simulations Structural Analysis • organization in membrane • interactions with lipids • H bonding Dynamical Analysis • motion of cholesterol • influence on lipid dynamics • diffusion


