3cf3e897fb27818bcf1cf98c6a623643.ppt
- Количество слайдов: 28

Array. Express A public database for microarray based gene expression data http: //www. ebi. ac. uk/microarray/ European Bioinformatics Institute EMBL-EBI Alvis Brazma, Helen Parkinson, Ugis Sarkans, Mohammadreza Shojatalab, Jaak Vilo + team MGED IV, Boston, February 2002
Array. Express Tuesday, February 12 th, 2002 Opened to public • • Standards: MIAME-compliant Data model: MAGE-OM Data input: MAGE-ML, web Data output: HTML, MAGE-ML, TAB-delimited, link to Expression Profiler • Data curation: Team of curators • Data sets: Yeast, human
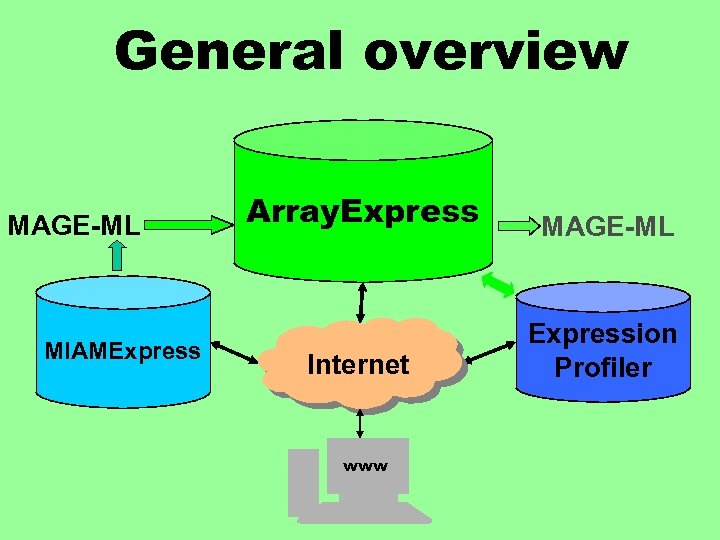
General overview MAGE-ML MIAMExpress Array. Express MAGE-ML Internet Expression Profiler www
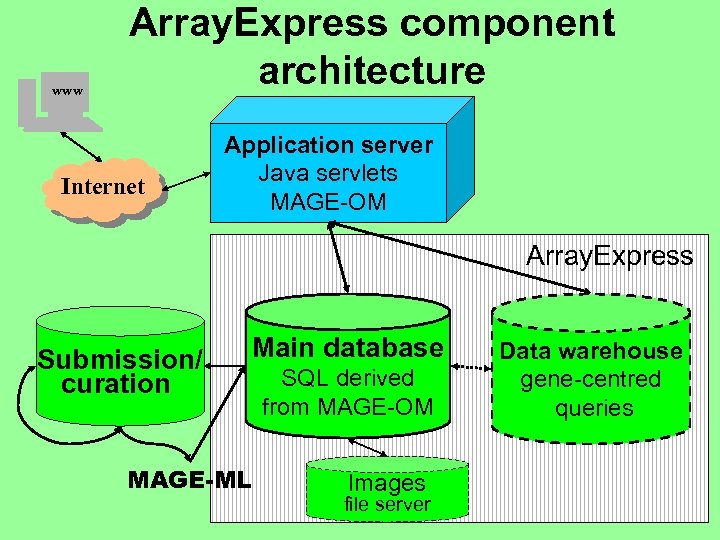
www Array. Express component architecture Internet Application server Java servlets MAGE-OM Array. Express Submission/ curation Main database MAGE-ML SQL derived from MAGE-OM Images file server Data warehouse gene-centred queries
Array. Express - features • MIAME-compliant, MAGE-ML, MAGE-OM • Can deal with: • raw quantitation data • processed data • data transformations • Independent of: • experimental platforms • image analysis methods • data normalization methods
Array. Express: details • • Database schema derived from MAGE-OM Standard SQL, we use Oracle Data loader for MAGE-ML - generated Web interface (first release 12. 2. 2002) • Queries by experiment, array, sample • Browsing • Object model-based query mechanism, automatic mapping to SQL
Simplified Array. Express model
MIAMExpress • • • Data annotation and submission tool MIAME based web interface Experiment, Array, Protocol submissions Uses CV/ontology wherever possible Creates MAGE-ML files for loading into Array. Express • Based on My. SQL, Perl, CGI, Apache

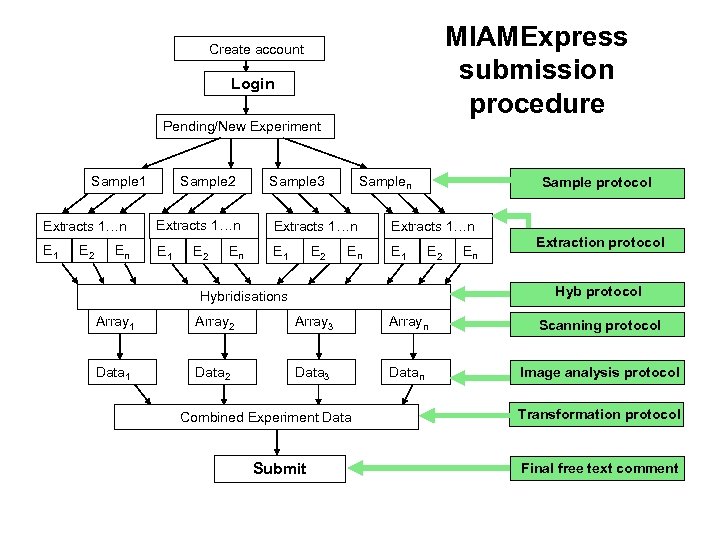
MIAMExpress submission procedure Create account Login Pending/New Experiment Sample 1 Extracts 1…n E 1 E 2 En Sample 2 Extracts 1…n E 1 E 2 En Sample 3 Extracts 1…n E 1 E 2 En Sample protocol Extracts 1…n E 1 E 2 En Extraction protocol Hybridisations Array 1 Array 2 Array 3 Arrayn Scanning protocol Data 1 Data 2 Data 3 Datan Image analysis protocol Combined Experiment Data Submit Transformation protocol Final free text comment
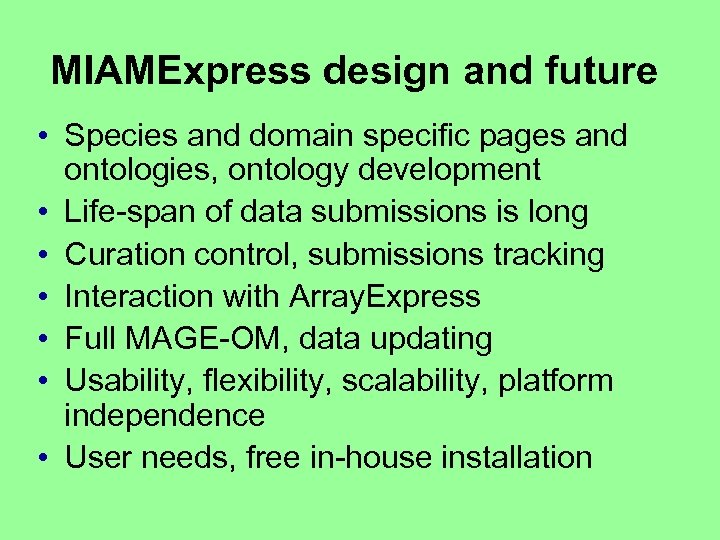
MIAMExpress design and future • Species and domain specific pages and ontologies, ontology development • Life-span of data submissions is long • Curation control, submissions tracking • Interaction with Array. Express • Full MAGE-OM, data updating • Usability, flexibility, scalability, platform independence • User needs, free in-house installation
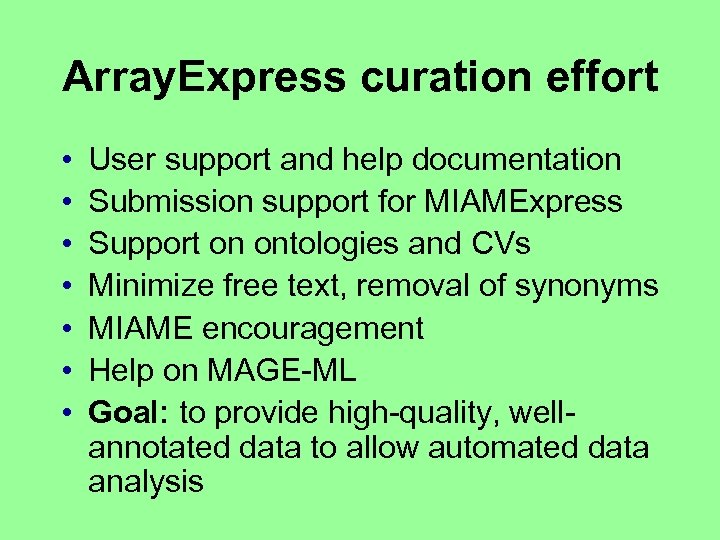
Array. Express curation effort • • User support and help documentation Submission support for MIAMExpress Support on ontologies and CVs Minimize free text, removal of synonyms MIAME encouragement Help on MAGE-ML Goal: to provide high-quality, wellannotated data to allow automated data analysis
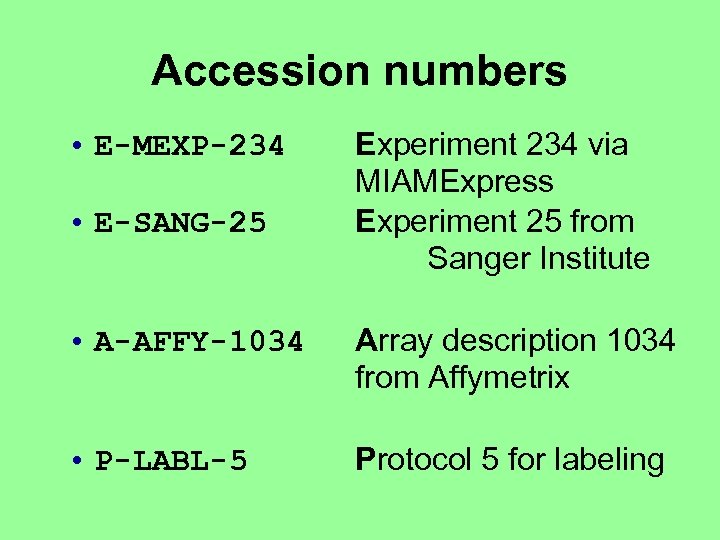
Accession numbers • E-MEXP-234 • E-SANG-25 Experiment 234 via MIAMExpress Experiment 25 from Sanger Institute • A-AFFY-1034 Array description 1034 from Affymetrix • P-LABL-5 Protocol 5 for labeling
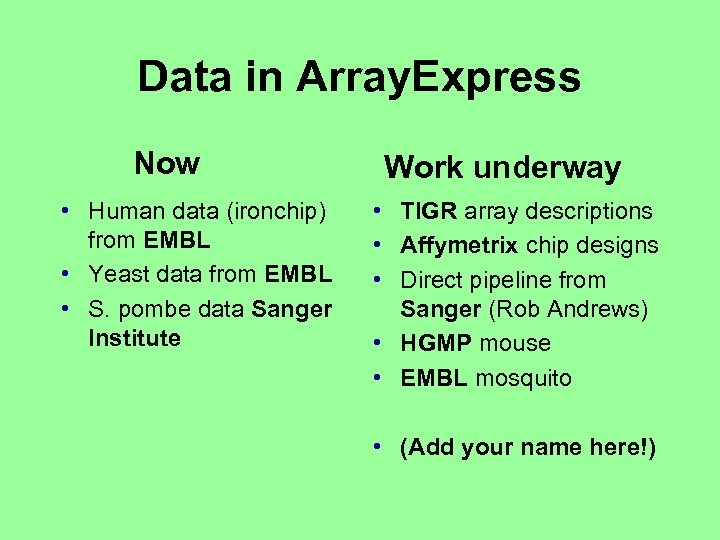
Data in Array. Express Now • Human data (ironchip) from EMBL • Yeast data from EMBL • S. pombe data Sanger Institute Work underway • TIGR array descriptions • Affymetrix chip designs • Direct pipeline from Sanger (Rob Andrews) • HGMP mouse • EMBL mosquito • (Add your name here!)

Data browsing and queries


Experiment info

Sample info
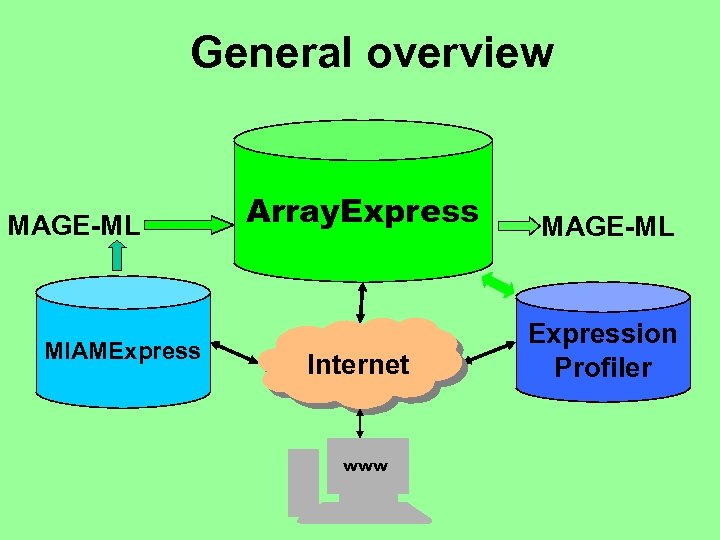
General overview MAGE-ML MIAMExpress Array. Express MAGE-ML Internet Expression Profiler www
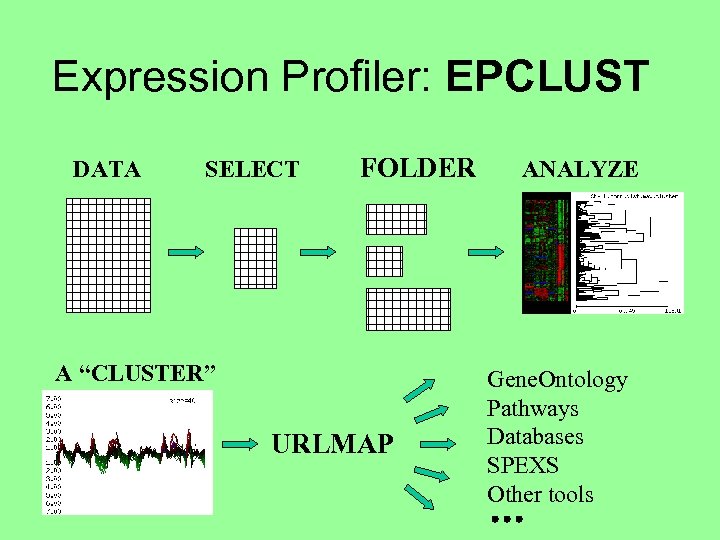
Expression Profiler: EPCLUST DATA SELECT FOLDER A “CLUSTER” URLMAP ANALYZE Gene. Ontology Pathways Databases SPEXS Other tools
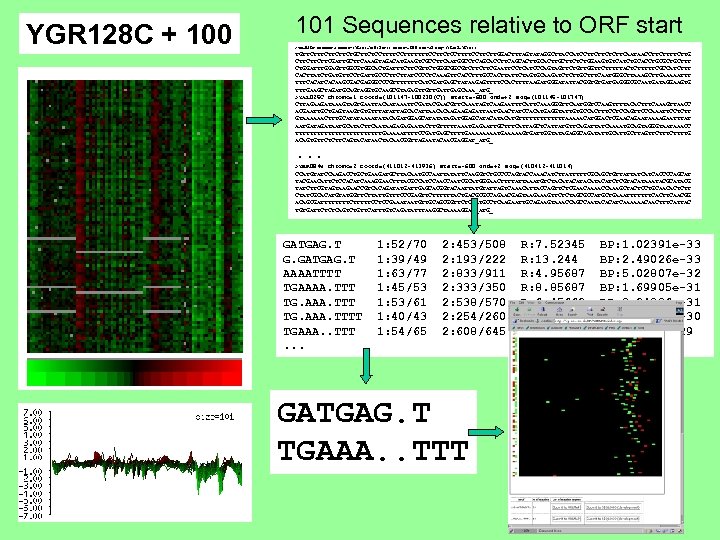
YGR 128 C + 100 101 Sequences relative to ORF start >YAL 036 C chromo=1 coord=(76154 -75048(C)) start=-600 end=+2 seq=(76152 -76754) TGTTCTTCTTCTGCTTCTCCTTTTTTTCCTTCTCCTTTTCCTTCTTGGACTTTAGTATAGGCTTACCATCCTTCTTCAATAACCTTCTTG CTTCTTCTTCGATTGCTTCAAAGTAGACATGAAGTCGCCTTCAATGGCCTCAGCACCTTCAGCACTTGCTTCTCTGGAAGTGTCATCTGCACCTGCGCTGCTTT CTGGATTTGGAGTTGGCGTGGCACTGATTTCTTCGTTCTGGGCGGCGTCTTCTTCGAATTCCTCATCCCAGTAGTTCTGTTGGTTCTTTTTACTCTTTTTCGCCATCTTT CACTTATCTGATGTTCCTGATTGCCCTTCTTATCCCCTCAAAGTTCACCTTTGCCACTTATTCTAGTGCAAGATCTCTTGCTTTCAATGGGCTTAAAGCTTGAAAAATTT TTTCACAAGCGACGAGGGCCCGTTTTTTTCATCGATGAGCTATAAGAGTTTTCCACTTTTAAGATGGGATATTACGGTGTGATGAGGGCGCAATGATAGGAAGTG TTTGAAGCTAGATGCAGTAGGTGCAAGCGTAGAGTTGTTGAGCAAA_ATG_ >YAL 025 C chromo=1 coord=(101147 -100230(C)) start=-600 end=+2 seq=(101145 -101747) CTTAGAAGATAAAGTAGTGAATTACAATAAATTCGATACGAACGTTCAAATAGTCAAGAATTTCAAAGGGTTCAATGGTCCAAGTTTTACACTTTCAAAGTTAACC ACGAATTGCTGAGTAAGTGTGTTTATATTAGCACATTAACACAAGAAGAGATTAATGAACTATCCACATGAGGTATTGTGCCACTTTCCTCCAGTTCCCAAATTCCTCTT GTAAAAAACTTTGCATATAAAATATACAGATGGAGCATATATAGATGGAGCATACATGTTTTTTTAAAAACATGGACTCGAACAGAATAAAAGAATTTAT AATGATAATGCATACTTCAATAAGAGAGAATACTTGTTTTTAAATGAGAATTGCTTTCATTAGCTCATTATGTTCAGATTATCAAAATGCAGTAGGGTAATAAACC TTTTTTTTTTTTGAAAAATTTTCCGATGAGCTTTTGAAAATGAAAAAGTGATTGGTATAGAGGCAGATATTGCTTAGTTCTTTTG ACAGTGTTCTCTTCAGTACATAACTACAACGGTTAGAATACAACGAGGAT_ATG_ . . . >YBR 084 W chromo=2 coord=(411012 -413936) start=-600 end=+2 seq=(410412 -411014) CCATGTATCCAAGACCTGCTGAAGATGCTTACAATGCCAATTATATTCAAGGTCTGCCCCAGTACCAAACATCTTATTTTTCGCAGCTGTTATTATCATCACCCCAGCAT TACGAACATTCTCCACATCAAAGGAACTTTACGCCATCCAATCGCATGGGAACTTTTATTAAATGTCTACATACATCTCGTACATAAATACGCATACG TATCTTCGTAGTAAGAACCGTCACAGATATGATTGAGCACGGTACAATTATGTATTAGTCAAACATTACCAGTTCTCGAACAAAACCAAAGCTACTCCTGCAACACTCTT CTATCGCACATGTATGGTTCTTATTGTTTCCCGAGTTCTTTTTTACTGACGCGCCAGAACGAGTAAGAAAGTTCTCTAGCGCCATGCTGAAATTTTTTTCACTTCAACGG ACAGCGATTTTCTTTTTCCTCCGAAATAATGTTGCAGCGGTTCTCGATGCCTCAAGAATTGCAGAAGTAAACCAGCCAATACACATCAAAAAACAACTTTCATTAC TGTGATTCTCTCAGTCTGTTCATTTGTCAGATATTTAAGGCTAAAAGGAA_ATG_ GATGAG. T G. GATGAG. T AAAATTTT TGAAAA. TTT TG. AAA. TTTT TGAAA. . TTT. . . 1: 52/70 1: 39/49 1: 63/77 1: 45/53 1: 53/61 1: 40/43 1: 54/65 2: 453/508 2: 193/222 2: 833/911 2: 333/350 2: 538/570 2: 254/260 2: 608/645 GATGAG. T TGAAA. . TTT R: 7. 52345 R: 13. 244 R: 4. 95687 R: 8. 85687 R: 6. 45662 R: 10. 3214 R: 5. 82106 BP: 1. 02391 e-33 BP: 2. 49026 e-33 BP: 5. 02807 e-32 BP: 1. 69905 e-31 BP: 3. 24836 e-31 BP: 3. 84624 e-30 BP: 1. 0887 e-29

1 mismatch GATGAG. T TGAAA. . TTT GATGAG. T W/30 TGAAA. . TTT Upstream sequence (600 bp)

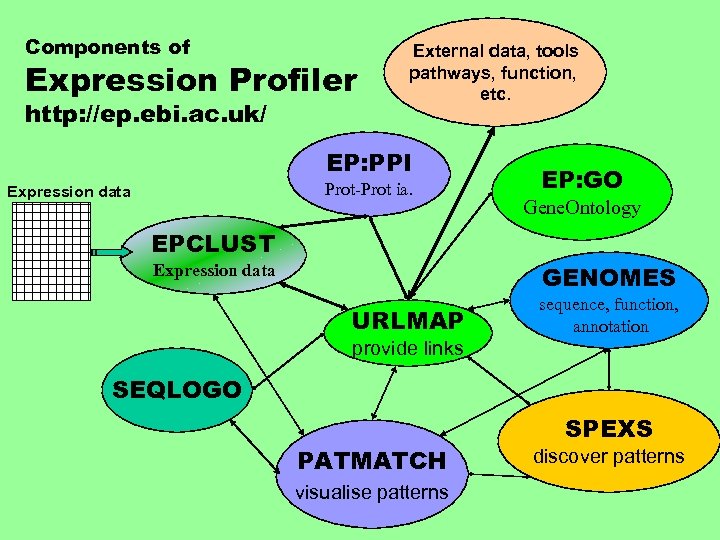
Components of Expression Profiler http: //ep. ebi. ac. uk/ External data, tools pathways, function, etc. EP: PPI Prot-Prot ia. Expression data EP: GO Gene. Ontology EPCLUST Expression data GENOMES URLMAP sequence, function, annotation provide links SEQLOGO PATMATCH visualise patterns SPEXS discover patterns

Ackowledgments: the team (3) 1999 November MGED 1 in Hinxton, EBI Alvis Brazma Alan Robinson Jaak Vilo

Ackowledgments: the team (5) Alvis Brazma, Alan Robinson 2000 August Database Ugis Sarkans Expression Profiler Research, students Jaak Vilo Thomas Schlitt

Ackowledgments: the team (9) 2001 June Alvis Brazma Database Curation Ugis Sarkans Helen Parkinson MIAMExpress Mohammadreza Shojatalab Expression Profiler Research, students Jaak Vilo Patrick Kemmeren Thomas Schlitt Katja Kivinen Johan Rung

Ackowledgments: the team (19) 2002 February Alvis Brazma Database Curation Ugis Sarkans Helen Parkinson Ahmet Oezcimen Susanna Sansone Gonzalo Garcia Philippe Rocca-Serra Ele Holloway MIAMExpress Mohammadreza Shojatalab Niran Abeygunawardena Expression Profiler Research, students Jaak Vilo Patrick Kemmeren Misha Kapushesky Thomas Schlitt Katja Kivinen Johan Rung Koichi Tazaki Lev Soinov Anastasia Samsonova
3cf3e897fb27818bcf1cf98c6a623643.ppt