41a651553ac304a13ba25fe6f520aba1.ppt
- Количество слайдов: 62
 Ariadne Genomics technology: Extraction from the literature and network analysis Dr. Anton Yuryev Ariadne Genomics Inc. © 2006 Ariadne Genomics. All Rights Reserved.
Ariadne Genomics technology: Extraction from the literature and network analysis Dr. Anton Yuryev Ariadne Genomics Inc. © 2006 Ariadne Genomics. All Rights Reserved.
 Pathway Studio product line • • • Pathway Studio desktop Pathway Studio workgroup Pathway Studio enterprise Main functionality: 1) Data mining and pathway building 2) Analysis of high-throughput data 3) Text-mining, fact extraction and database building © 2006 Ariadne Genomics. All Rights Reserved. 2
Pathway Studio product line • • • Pathway Studio desktop Pathway Studio workgroup Pathway Studio enterprise Main functionality: 1) Data mining and pathway building 2) Analysis of high-throughput data 3) Text-mining, fact extraction and database building © 2006 Ariadne Genomics. All Rights Reserved. 2
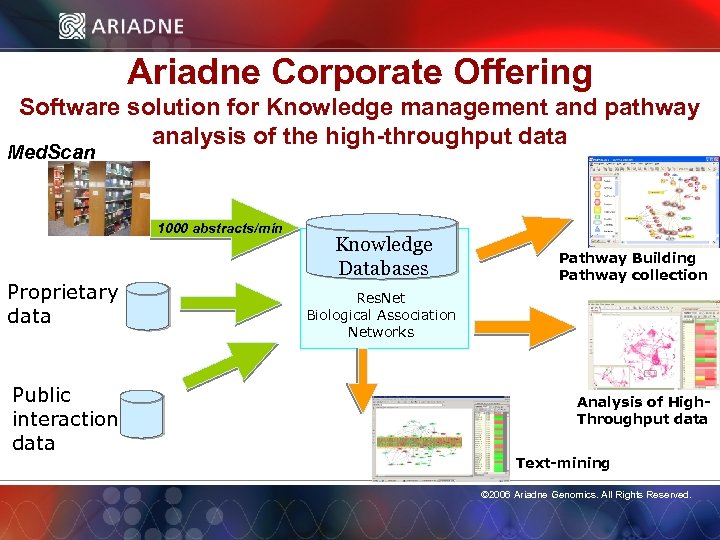 Ariadne Corporate Offering Software solution for Knowledge management and pathway analysis of the high-throughput data Med. Scan 1000 abstracts/min Proprietary data Public interaction data Knowledge Databases Pathway Building Pathway collection Res. Net Biological Association Networks Analysis of High. Throughput data © 2006 Ariadne Genomics. All Rights Reserved. Text-mining © 2006 Ariadne Genomics. All Rights Reserved. 3
Ariadne Corporate Offering Software solution for Knowledge management and pathway analysis of the high-throughput data Med. Scan 1000 abstracts/min Proprietary data Public interaction data Knowledge Databases Pathway Building Pathway collection Res. Net Biological Association Networks Analysis of High. Throughput data © 2006 Ariadne Genomics. All Rights Reserved. Text-mining © 2006 Ariadne Genomics. All Rights Reserved. 3
 Ariadne Database Construction • Automatic fact extraction by Med. Scan from organism-specific subset of Pub. Med and full-text journals • Import of Ariadne proprietary curated data – Curated physical interaction – 712 signaling line pathways • Import of publicly available curated interaction data: Entrez Gene, BIND, HPRD, KEGG, Gene Ontology • Import of publicly available high-throughput interaction data (Y 2 K, Massspec etc) • Import of user proprietary data: – Proprietary or publicly available experimental data in PSI, Bio. Pax or Tabdelimited formats – Data mined by Med. Scan tool from literature sources not included with database • User manual curation © 2006 Ariadne Genomics. All Rights Reserved. 4
Ariadne Database Construction • Automatic fact extraction by Med. Scan from organism-specific subset of Pub. Med and full-text journals • Import of Ariadne proprietary curated data – Curated physical interaction – 712 signaling line pathways • Import of publicly available curated interaction data: Entrez Gene, BIND, HPRD, KEGG, Gene Ontology • Import of publicly available high-throughput interaction data (Y 2 K, Massspec etc) • Import of user proprietary data: – Proprietary or publicly available experimental data in PSI, Bio. Pax or Tabdelimited formats – Data mined by Med. Scan tool from literature sources not included with database • User manual curation © 2006 Ariadne Genomics. All Rights Reserved. 4
 Additional Commercial datasets • > 130 KEGG metabolic pathways • >70 STKE pathways (AAAS) • >10, 000 ERGO pathways for 587 organisms (Integrated genomics) • >100, 000 protein interactions from Hynet (Prolexys) • >600 disease pathways Path. Art (Jubilant) © 2006 Ariadne Genomics. All Rights Reserved. 5
Additional Commercial datasets • > 130 KEGG metabolic pathways • >70 STKE pathways (AAAS) • >10, 000 ERGO pathways for 587 organisms (Integrated genomics) • >100, 000 protein interactions from Hynet (Prolexys) • >600 disease pathways Path. Art (Jubilant) © 2006 Ariadne Genomics. All Rights Reserved. 5
 Pathway Studio Enterprise distinctions • Web-client for instant pathway publishing – Connection between multiple geographical sites • 3 -tier architecture with Java API to connect third party applications and algorithms • Med. Scan Enterprise license: – open Med. Scan dictionaries and pattern rules files for customization – distribution of Med. Scan data across entire company • GSEA, NEA and network clustering algorithms for analysis of high-throughout data © 2006 Ariadne Genomics. All Rights Reserved. 6
Pathway Studio Enterprise distinctions • Web-client for instant pathway publishing – Connection between multiple geographical sites • 3 -tier architecture with Java API to connect third party applications and algorithms • Med. Scan Enterprise license: – open Med. Scan dictionaries and pattern rules files for customization – distribution of Med. Scan data across entire company • GSEA, NEA and network clustering algorithms for analysis of high-throughout data © 2006 Ariadne Genomics. All Rights Reserved. 6
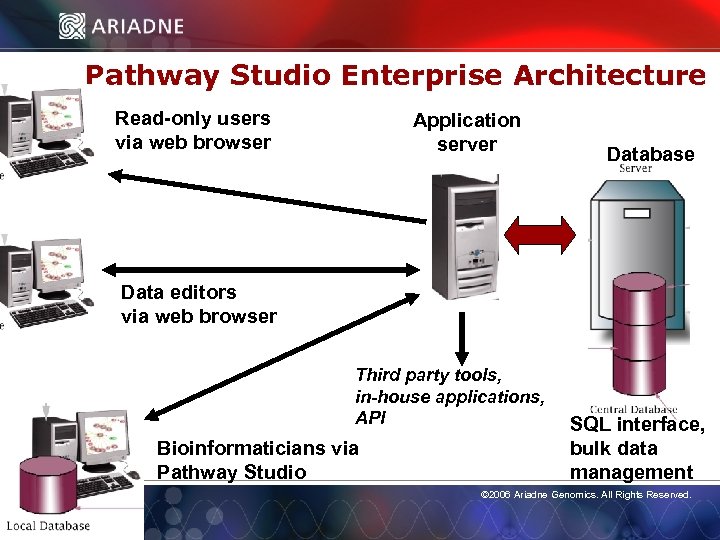 Pathway Studio Enterprise Architecture Read-only users via web browser Application server Database Data editors via web browser Third party tools, in-house applications, API Bioinformaticians via Pathway Studio SQL interface, bulk data © 2006 Ariadne Genomics. All Rights Reserved. management © 2006 Ariadne Genomics. All Rights Reserved. 7
Pathway Studio Enterprise Architecture Read-only users via web browser Application server Database Data editors via web browser Third party tools, in-house applications, API Bioinformaticians via Pathway Studio SQL interface, bulk data © 2006 Ariadne Genomics. All Rights Reserved. management © 2006 Ariadne Genomics. All Rights Reserved. 7
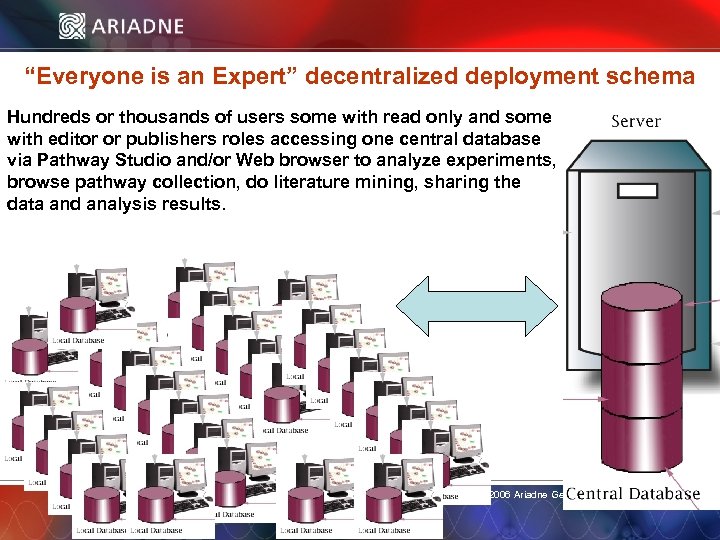 “Everyone is an Expert” decentralized deployment schema Hundreds or thousands of users some with read only and some with editor or publishers roles accessing one central database via Pathway Studio and/or Web browser to analyze experiments, browse pathway collection, do literature mining, sharing the data and analysis results. © 2006 Ariadne Genomics. All Rights Reserved. 8
“Everyone is an Expert” decentralized deployment schema Hundreds or thousands of users some with read only and some with editor or publishers roles accessing one central database via Pathway Studio and/or Web browser to analyze experiments, browse pathway collection, do literature mining, sharing the data and analysis results. © 2006 Ariadne Genomics. All Rights Reserved. 8
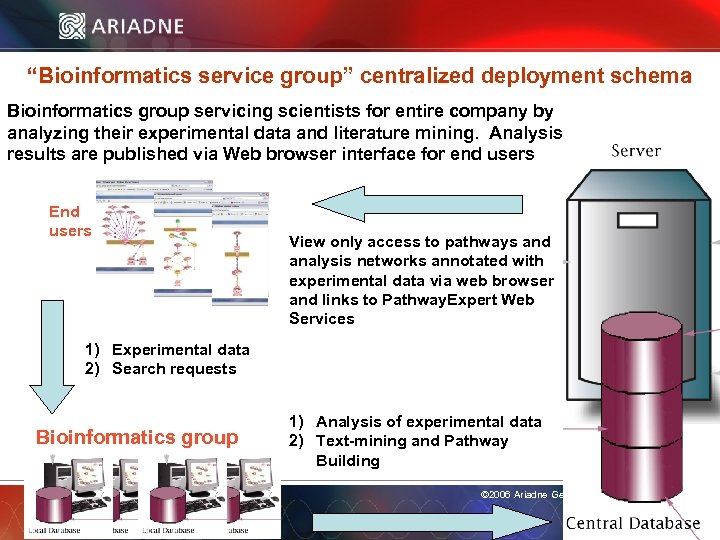 “Bioinformatics service group” centralized deployment schema Bioinformatics group servicing scientists for entire company by analyzing their experimental data and literature mining. Analysis results are published via Web browser interface for end users End users View only access to pathways and analysis networks annotated with experimental data via web browser and links to Pathway. Expert Web Services 1) Experimental data 2) Search requests Bioinformatics group 1) Analysis of experimental data 2) Text-mining and Pathway © 2006 Ariadne Genomics. All Rights Reserved. Building © 2006 Ariadne Genomics. All Rights Reserved. 9
“Bioinformatics service group” centralized deployment schema Bioinformatics group servicing scientists for entire company by analyzing their experimental data and literature mining. Analysis results are published via Web browser interface for end users End users View only access to pathways and analysis networks annotated with experimental data via web browser and links to Pathway. Expert Web Services 1) Experimental data 2) Search requests Bioinformatics group 1) Analysis of experimental data 2) Text-mining and Pathway © 2006 Ariadne Genomics. All Rights Reserved. Building © 2006 Ariadne Genomics. All Rights Reserved. 9
 “Disease area” decentralized clusters deployment schema Disease area groups have bioinformatics, biologists and chemists working as a team with focus on one disease Cardiovascular group Digestive disorders group Cancer group CNS group © 2006 Ariadne Genomics. All Rights Reserved. 10
“Disease area” decentralized clusters deployment schema Disease area groups have bioinformatics, biologists and chemists working as a team with focus on one disease Cardiovascular group Digestive disorders group Cancer group CNS group © 2006 Ariadne Genomics. All Rights Reserved. 10
 Plan of the talk 1) Text-mining, fact extraction and database building - Stay current with the literature Build focused literature networks Build focus databases 2) Data mining and pathway building - Understand molecular mechanisms of disease and processes Maintain pathway collection Build focus databases 3) Analysis of high-throughput data - Functional ontology analysis Network analysis © 2006 Ariadne Genomics. All Rights Reserved. 11
Plan of the talk 1) Text-mining, fact extraction and database building - Stay current with the literature Build focused literature networks Build focus databases 2) Data mining and pathway building - Understand molecular mechanisms of disease and processes Maintain pathway collection Build focus databases 3) Analysis of high-throughput data - Functional ontology analysis Network analysis © 2006 Ariadne Genomics. All Rights Reserved. 11
 Introduction to Med. Scan technology © 2006 Ariadne Genomics. All Rights Reserved.
Introduction to Med. Scan technology © 2006 Ariadne Genomics. All Rights Reserved.
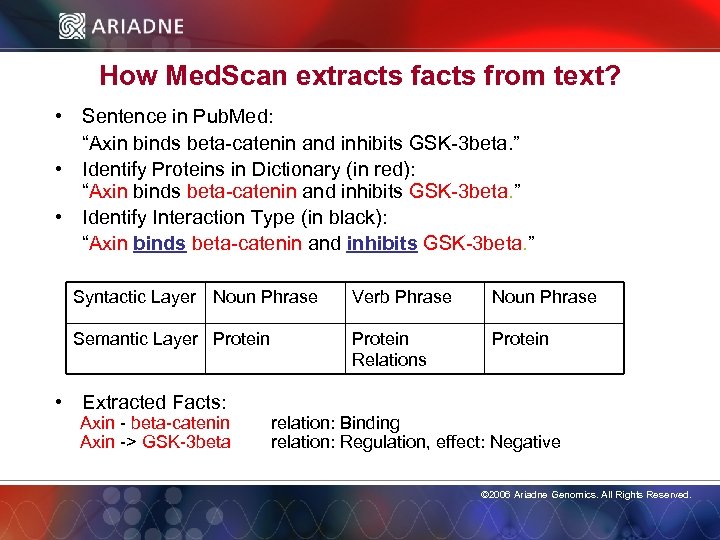 How Med. Scan extracts from text? • Sentence in Pub. Med: “Axin binds beta-catenin and inhibits GSK-3 beta. ” • Identify Proteins in Dictionary (in red): “Axin binds beta-catenin and inhibits GSK-3 beta. ” • Identify Interaction Type (in black): “Axin binds beta-catenin and inhibits GSK-3 beta. ” Syntactic Layer Noun Phrase Verb Phrase Noun Phrase Semantic Layer Protein Relations Protein • Extracted Facts: Axin - beta-catenin Axin -> GSK-3 beta relation: Binding relation: Regulation, effect: Negative © 2006 Ariadne Genomics. All Rights Reserved. 13
How Med. Scan extracts from text? • Sentence in Pub. Med: “Axin binds beta-catenin and inhibits GSK-3 beta. ” • Identify Proteins in Dictionary (in red): “Axin binds beta-catenin and inhibits GSK-3 beta. ” • Identify Interaction Type (in black): “Axin binds beta-catenin and inhibits GSK-3 beta. ” Syntactic Layer Noun Phrase Verb Phrase Noun Phrase Semantic Layer Protein Relations Protein • Extracted Facts: Axin - beta-catenin Axin -> GSK-3 beta relation: Binding relation: Regulation, effect: Negative © 2006 Ariadne Genomics. All Rights Reserved. 13
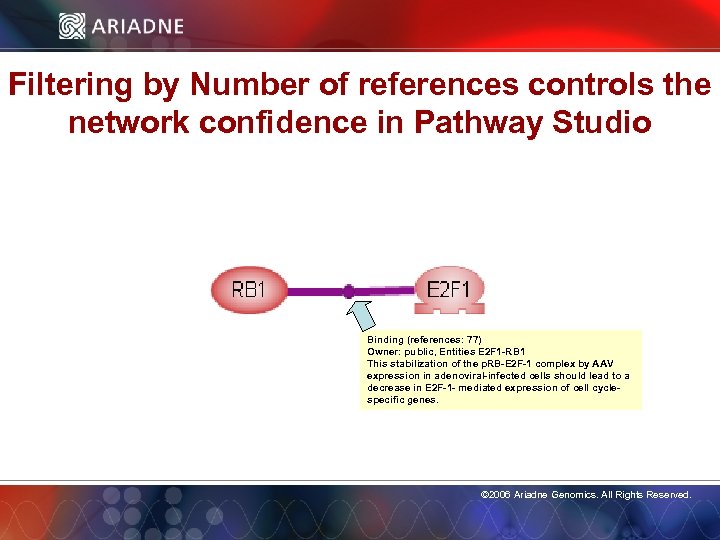 Filtering by Number of references controls the network confidence in Pathway Studio Binding (references: 77) Owner: public, Entities E 2 F 1 -RB 1 This stabilization of the p. RB-E 2 F-1 complex by AAV expression in adenoviral-infected cells should lead to a decrease in E 2 F-1 - mediated expression of cell cyclespecific genes. © 2006 Ariadne Genomics. All Rights Reserved. 14
Filtering by Number of references controls the network confidence in Pathway Studio Binding (references: 77) Owner: public, Entities E 2 F 1 -RB 1 This stabilization of the p. RB-E 2 F-1 complex by AAV expression in adenoviral-infected cells should lead to a decrease in E 2 F-1 - mediated expression of cell cyclespecific genes. © 2006 Ariadne Genomics. All Rights Reserved. 14
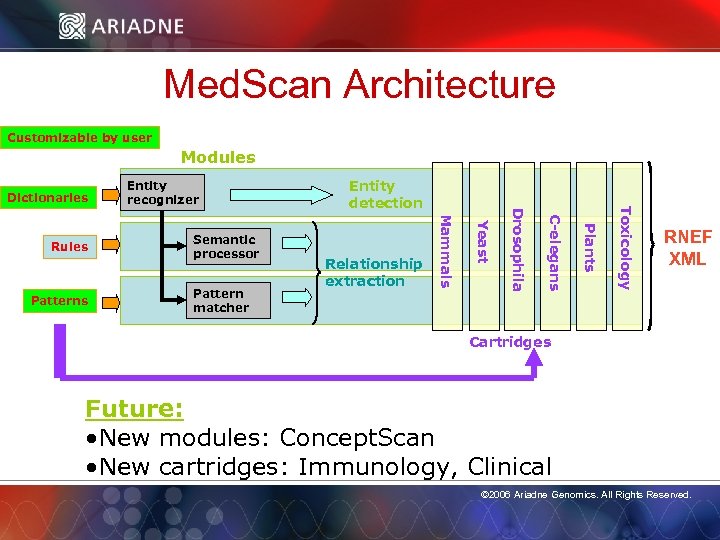 Med. Scan Architecture Customizable by user Modules Dictionaries Toxicology Plants C-elegans Drosophila Pattern matcher Relationship extraction Yeast Patterns Semantic processor Entity detection Mammals Rules Entity recognizer RNEF XML Cartridges Future: • New modules: Concept. Scan © 2006 Ariadne Genomics. All Rights Reserved. • New cartridges: Immunology, Clinical © 2006 Ariadne Genomics. All Rights Reserved. 15
Med. Scan Architecture Customizable by user Modules Dictionaries Toxicology Plants C-elegans Drosophila Pattern matcher Relationship extraction Yeast Patterns Semantic processor Entity detection Mammals Rules Entity recognizer RNEF XML Cartridges Future: • New modules: Concept. Scan © 2006 Ariadne Genomics. All Rights Reserved. • New cartridges: Immunology, Clinical © 2006 Ariadne Genomics. All Rights Reserved. 15
 Describing Med. Scan • Manually curated: dictionaries and grammar rules • Fast: 14 mln Pub. Med abstracts in 2 days on modern PC • Comprehensive: facts recovery rate > 90% = 70% sentence recovery rate + 20% literature redundancy • Removes redundancy: 7, 647, 282 non-distinct relations =>1, 000 distinct relations • Accurate: false positive rate – 10% • Customizable: dictionaries and patterns © 2006 Ariadne Genomics. All Rights Reserved. 16
Describing Med. Scan • Manually curated: dictionaries and grammar rules • Fast: 14 mln Pub. Med abstracts in 2 days on modern PC • Comprehensive: facts recovery rate > 90% = 70% sentence recovery rate + 20% literature redundancy • Removes redundancy: 7, 647, 282 non-distinct relations =>1, 000 distinct relations • Accurate: false positive rate – 10% • Customizable: dictionaries and patterns © 2006 Ariadne Genomics. All Rights Reserved. 16
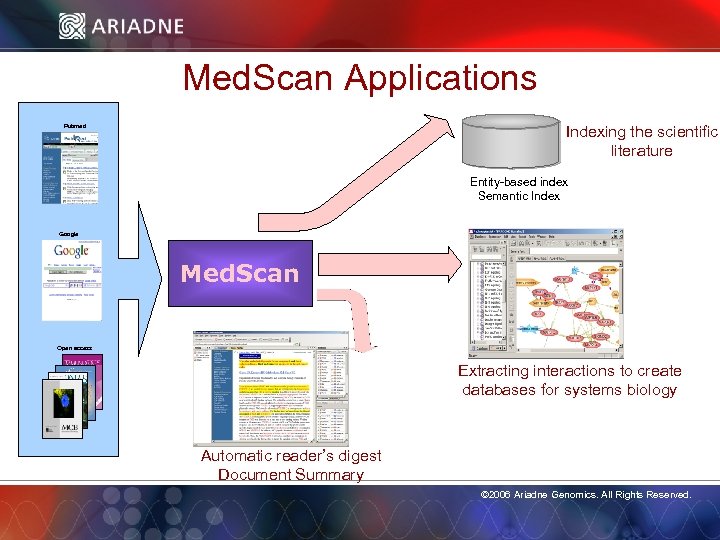 Med. Scan Applications Indexing the scientific literature Pubmed Entity-based index Semantic Index Google Med. Scan Open access Extracting interactions to create databases for systems biology Automatic reader’s digest Document Summary © 2006 Ariadne Genomics. All Rights Reserved. 17
Med. Scan Applications Indexing the scientific literature Pubmed Entity-based index Semantic Index Google Med. Scan Open access Extracting interactions to create databases for systems biology Automatic reader’s digest Document Summary © 2006 Ariadne Genomics. All Rights Reserved. 17
 Pathway Building in Pathway Studio • Manual • Automatic using Graph navigation tools • Using text-mining with Med. Scan © 2006 Ariadne Genomics. All Rights Reserved.
Pathway Building in Pathway Studio • Manual • Automatic using Graph navigation tools • Using text-mining with Med. Scan © 2006 Ariadne Genomics. All Rights Reserved.
 Viewing and editing pathways in Pathway Studio • • • Viewing entities in the List Pane Entity and relation tables Show all references Pathway Reference summary Export protein list Display styles: By type, By effect, By reference count • UI options: – magnifier – fit text to entities – simple and full graph view – fit to window – rotate – move – zoom by rectangle – advanced graph © 2006 Ariadne Genomics. All Rights Reserved. scaling • resizing nodes in pathway pane © 2006 Ariadne Genomics. All Rights Reserved. 19
Viewing and editing pathways in Pathway Studio • • • Viewing entities in the List Pane Entity and relation tables Show all references Pathway Reference summary Export protein list Display styles: By type, By effect, By reference count • UI options: – magnifier – fit text to entities – simple and full graph view – fit to window – rotate – move – zoom by rectangle – advanced graph © 2006 Ariadne Genomics. All Rights Reserved. scaling • resizing nodes in pathway pane © 2006 Ariadne Genomics. All Rights Reserved. 19
 Pathway Building by text-mining Non-melanoma skin cancer >1, 000 cases, (<2, 000 deaths), in USA © 2006 Ariadne Genomics. All Rights Reserved.
Pathway Building by text-mining Non-melanoma skin cancer >1, 000 cases, (<2, 000 deaths), in USA © 2006 Ariadne Genomics. All Rights Reserved.
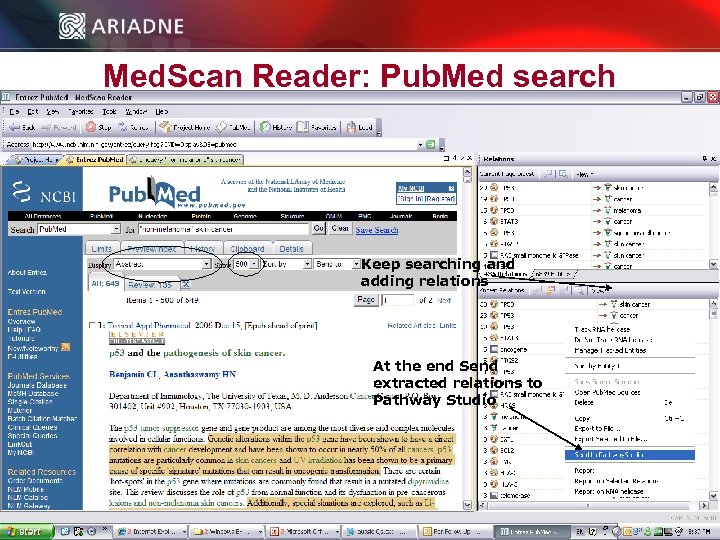 Med. Scan Reader: Pub. Med search Keep searching and adding relations At the end Send extracted relations to Pathway Studio © 2006 Ariadne Genomics. All Rights Reserved. 21
Med. Scan Reader: Pub. Med search Keep searching and adding relations At the end Send extracted relations to Pathway Studio © 2006 Ariadne Genomics. All Rights Reserved. 21
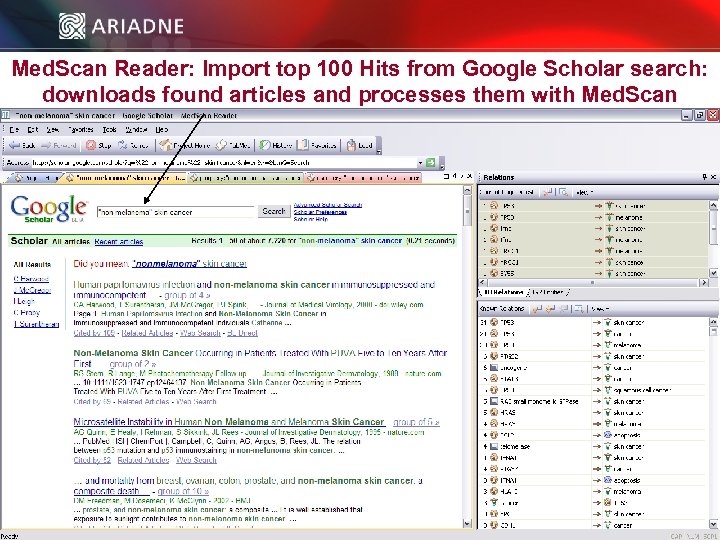 Med. Scan Reader: Import top 100 Hits from Google Scholar search: downloads found articles and processes them with Med. Scan © 2006 Ariadne Genomics. All Rights Reserved. 22
Med. Scan Reader: Import top 100 Hits from Google Scholar search: downloads found articles and processes them with Med. Scan © 2006 Ariadne Genomics. All Rights Reserved. 22
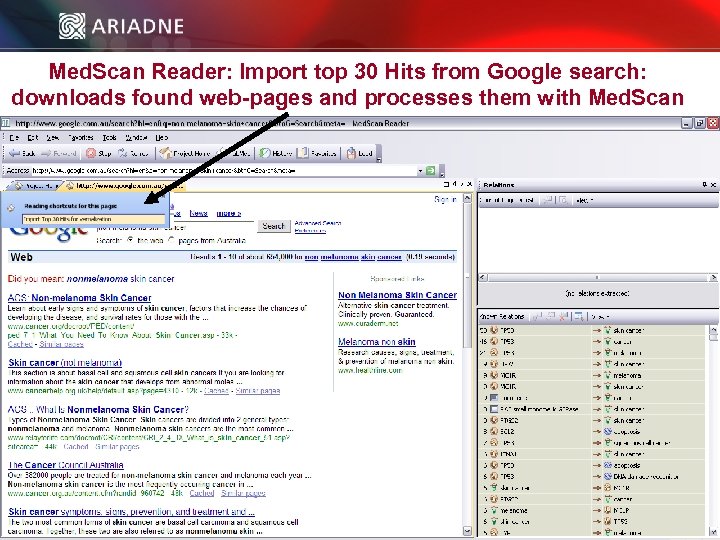 Med. Scan Reader: Import top 30 Hits from Google search: downloads found web-pages and processes them with Med. Scan © 2006 Ariadne Genomics. All Rights Reserved. 23
Med. Scan Reader: Import top 30 Hits from Google search: downloads found web-pages and processes them with Med. Scan © 2006 Ariadne Genomics. All Rights Reserved. 23
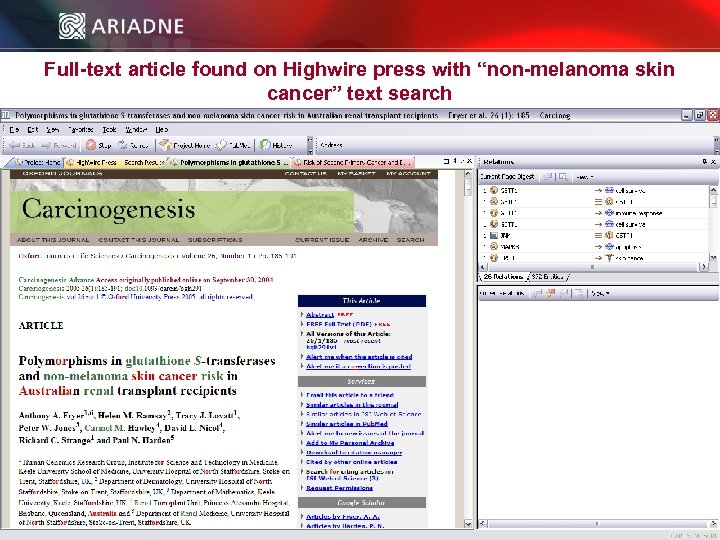 Full-text article found on Highwire press with “non-melanoma skin cancer” text search © 2006 Ariadne Genomics. All Rights Reserved. 24
Full-text article found on Highwire press with “non-melanoma skin cancer” text search © 2006 Ariadne Genomics. All Rights Reserved. 24
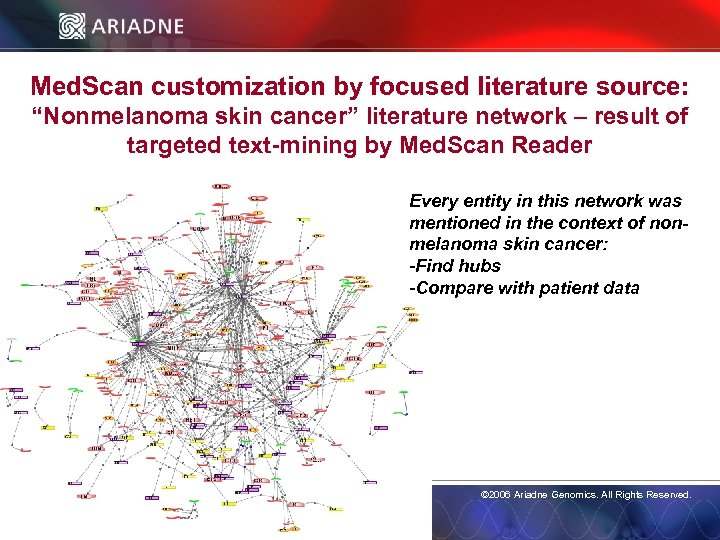 Med. Scan customization by focused literature source: “Nonmelanoma skin cancer” literature network – result of targeted text-mining by Med. Scan Reader Every entity in this network was mentioned in the context of nonmelanoma skin cancer: -Find hubs -Compare with patient data © 2006 Ariadne Genomics. All Rights Reserved. 25
Med. Scan customization by focused literature source: “Nonmelanoma skin cancer” literature network – result of targeted text-mining by Med. Scan Reader Every entity in this network was mentioned in the context of nonmelanoma skin cancer: -Find hubs -Compare with patient data © 2006 Ariadne Genomics. All Rights Reserved. 25
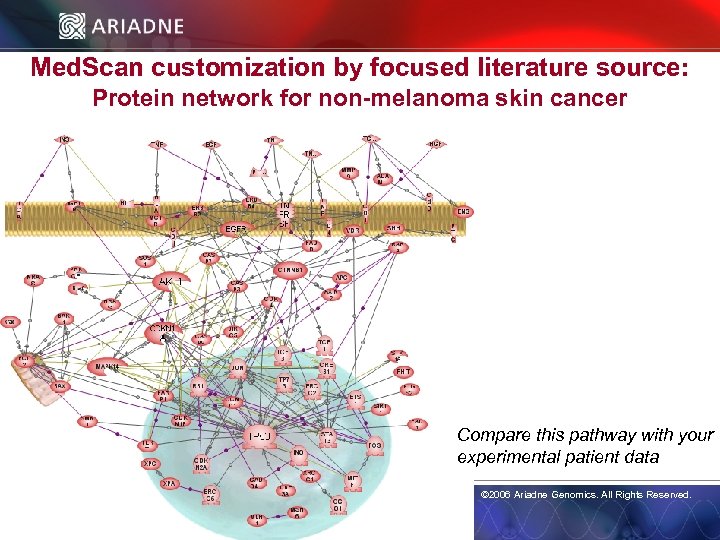 Med. Scan customization by focused literature source: Protein network for non-melanoma skin cancer Compare this pathway with your © 2006 Ariadne Genomics. All Rights Reserved. experimental patient data © 2006 Ariadne Genomics. All Rights Reserved. 26
Med. Scan customization by focused literature source: Protein network for non-melanoma skin cancer Compare this pathway with your © 2006 Ariadne Genomics. All Rights Reserved. experimental patient data © 2006 Ariadne Genomics. All Rights Reserved. 26
 Automatic Pathway Building using Graph navigation Build pathway tool © 2006 Ariadne Genomics. All Rights Reserved.
Automatic Pathway Building using Graph navigation Build pathway tool © 2006 Ariadne Genomics. All Rights Reserved.
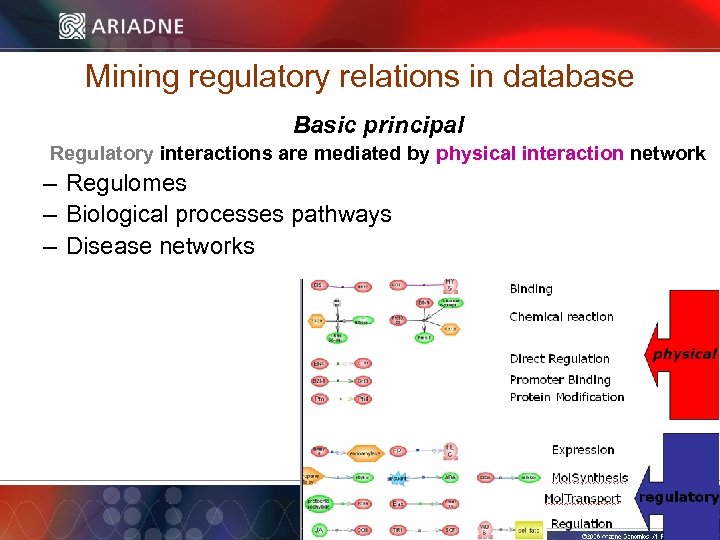 Mining regulatory relations in database Basic principal Regulatory interactions are mediated by physical interaction network – Regulomes – Biological processes pathways – Disease networks © 2006 Ariadne Genomics. All Rights Reserved. 28
Mining regulatory relations in database Basic principal Regulatory interactions are mediated by physical interaction network – Regulomes – Biological processes pathways – Disease networks © 2006 Ariadne Genomics. All Rights Reserved. 28
 Regulome pathways: algorithm input © 2006 Ariadne Genomics. All Rights Reserved. 29
Regulome pathways: algorithm input © 2006 Ariadne Genomics. All Rights Reserved. 29
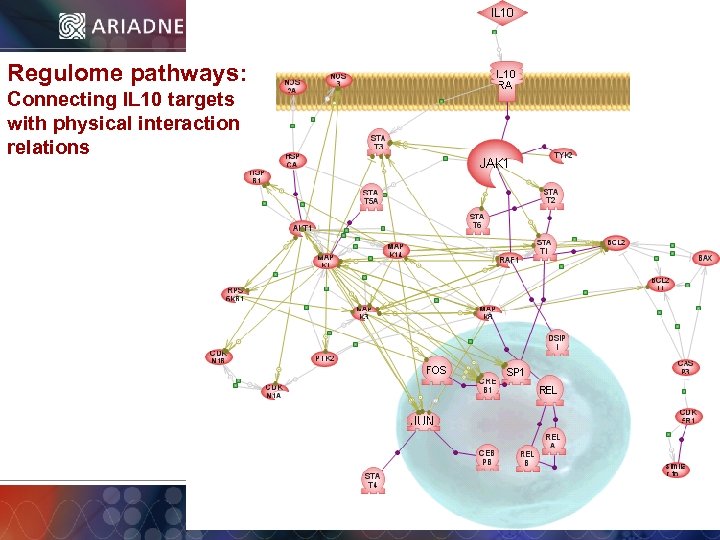 Regulome pathways: Connecting IL 10 targets with physical interaction relations © 2006 Ariadne Genomics. All Rights Reserved. 30
Regulome pathways: Connecting IL 10 targets with physical interaction relations © 2006 Ariadne Genomics. All Rights Reserved. 30
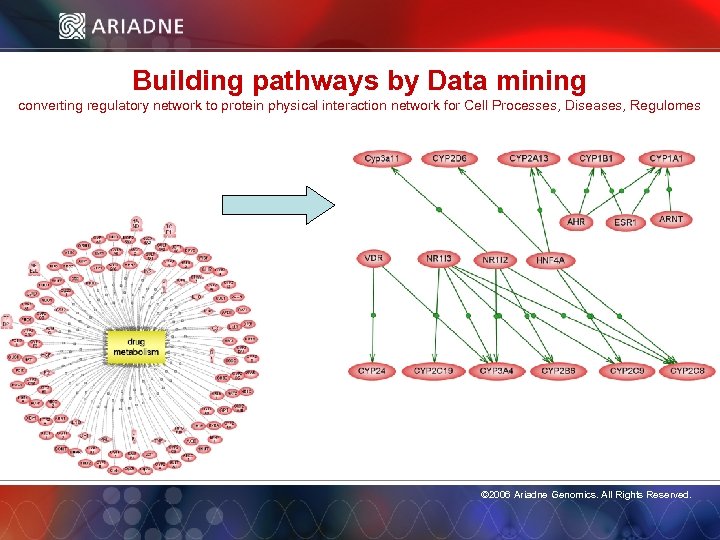 Building pathways by Data mining converting regulatory network to protein physical interaction network for Cell Processes, Diseases, Regulomes © 2006 Ariadne Genomics. All Rights Reserved. 31
Building pathways by Data mining converting regulatory network to protein physical interaction network for Cell Processes, Diseases, Regulomes © 2006 Ariadne Genomics. All Rights Reserved. 31
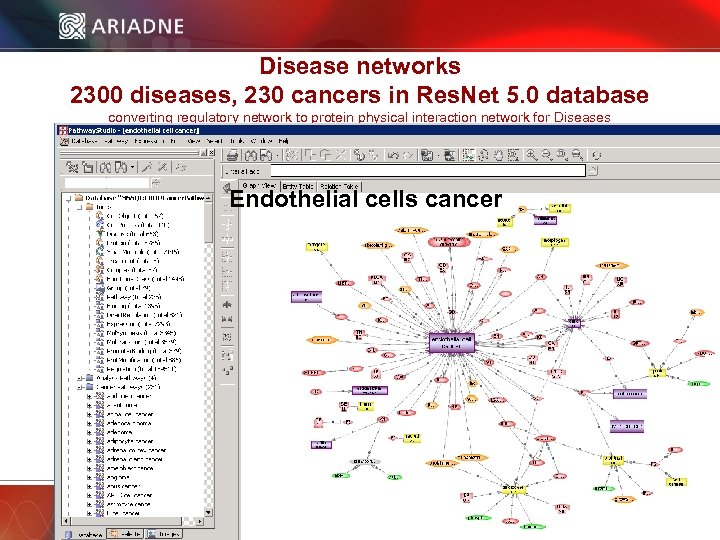 Disease networks 2300 diseases, 230 cancers in Res. Net 5. 0 database converting regulatory network to protein physical interaction network for Diseases Endothelial cells cancer © 2006 Ariadne Genomics. All Rights Reserved. 32
Disease networks 2300 diseases, 230 cancers in Res. Net 5. 0 database converting regulatory network to protein physical interaction network for Diseases Endothelial cells cancer © 2006 Ariadne Genomics. All Rights Reserved. 32
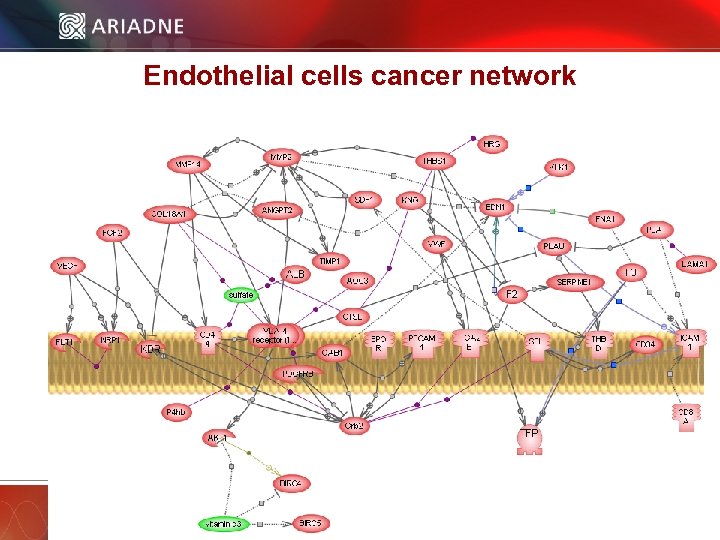 Endothelial cells cancer network © 2006 Ariadne Genomics. All Rights Reserved. 33
Endothelial cells cancer network © 2006 Ariadne Genomics. All Rights Reserved. 33
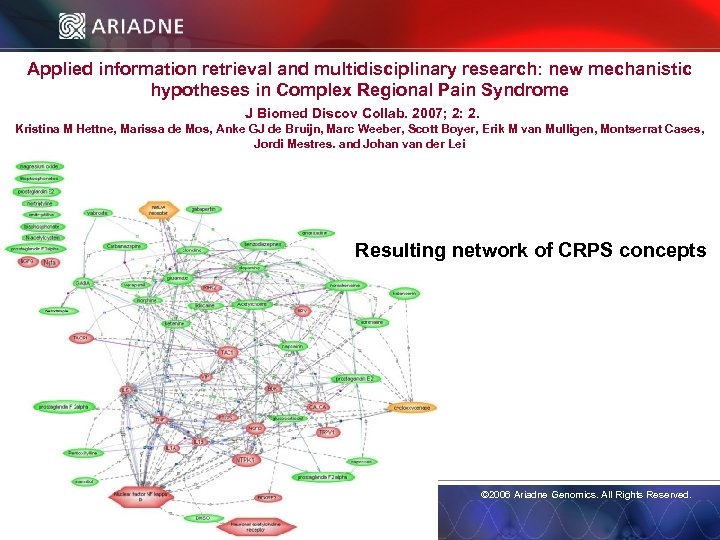 Applied information retrieval and multidisciplinary research: new mechanistic hypotheses in Complex Regional Pain Syndrome J Biomed Discov Collab. 2007; 2: 2. Kristina M Hettne, Marissa de Mos, Anke GJ de Bruijn, Marc Weeber, Scott Boyer, Erik M van Mulligen, Montserrat Cases, Jordi Mestres, and Johan van der Lei Resulting network of CRPS concepts © 2006 Ariadne Genomics. All Rights Reserved. 34
Applied information retrieval and multidisciplinary research: new mechanistic hypotheses in Complex Regional Pain Syndrome J Biomed Discov Collab. 2007; 2: 2. Kristina M Hettne, Marissa de Mos, Anke GJ de Bruijn, Marc Weeber, Scott Boyer, Erik M van Mulligen, Montserrat Cases, Jordi Mestres, and Johan van der Lei Resulting network of CRPS concepts © 2006 Ariadne Genomics. All Rights Reserved. 34
 High-throughput data analysis in Pathway Studio • Identification of responsive genes • Functional ontology analysis • Network analysis © 2006 Ariadne Genomics. All Rights Reserved.
High-throughput data analysis in Pathway Studio • Identification of responsive genes • Functional ontology analysis • Network analysis © 2006 Ariadne Genomics. All Rights Reserved.
 Supports analysis of all types of experiment data • • • Gene expression Metabolomics Proteomics SNP and CNV analysis Methylation arrays Phosphorylation arrays Support for all microarray platforms: • Affymetrix • Agilent • Illumina • Nimblegen • Superarray © 2006 Ariadne Genomics. All Rights Reserved. • Custom design chips © 2006 Ariadne Genomics. All Rights Reserved. 36
Supports analysis of all types of experiment data • • • Gene expression Metabolomics Proteomics SNP and CNV analysis Methylation arrays Phosphorylation arrays Support for all microarray platforms: • Affymetrix • Agilent • Illumina • Nimblegen • Superarray © 2006 Ariadne Genomics. All Rights Reserved. • Custom design chips © 2006 Ariadne Genomics. All Rights Reserved. 36
 Analysis of gene expression microarray data: STEP 1: Identification of responsive genes • Expression data import (tab, xls, cel) • Selection of responsive genes – Find differentially expressed genes (significance analysis via t-test) – Gene clustering via correlation networks – Find responsive genes in the third party software for statistical analysis of microarray data and import it as a protein list (Tools->Import protein list) © 2006 Ariadne Genomics. All Rights Reserved. 37
Analysis of gene expression microarray data: STEP 1: Identification of responsive genes • Expression data import (tab, xls, cel) • Selection of responsive genes – Find differentially expressed genes (significance analysis via t-test) – Gene clustering via correlation networks – Find responsive genes in the third party software for statistical analysis of microarray data and import it as a protein list (Tools->Import protein list) © 2006 Ariadne Genomics. All Rights Reserved. 37
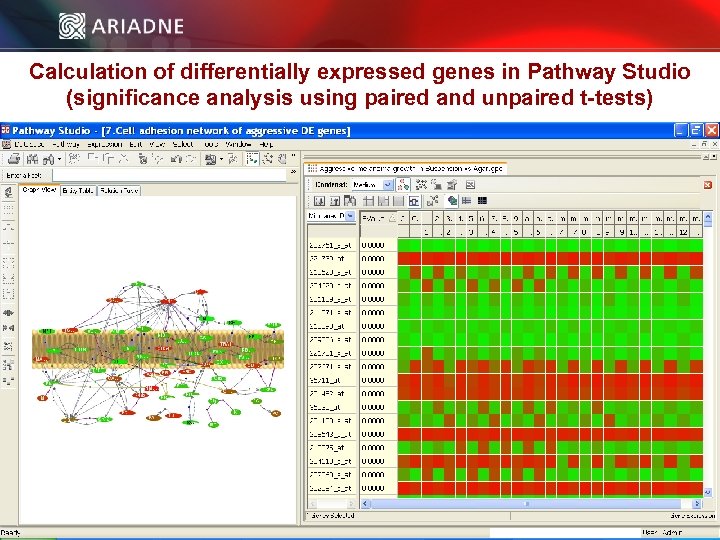 Calculation of differentially expressed genes in Pathway Studio (significance analysis using paired and unpaired t-tests) © 2006 Ariadne Genomics. All Rights Reserved. 38
Calculation of differentially expressed genes in Pathway Studio (significance analysis using paired and unpaired t-tests) © 2006 Ariadne Genomics. All Rights Reserved. 38
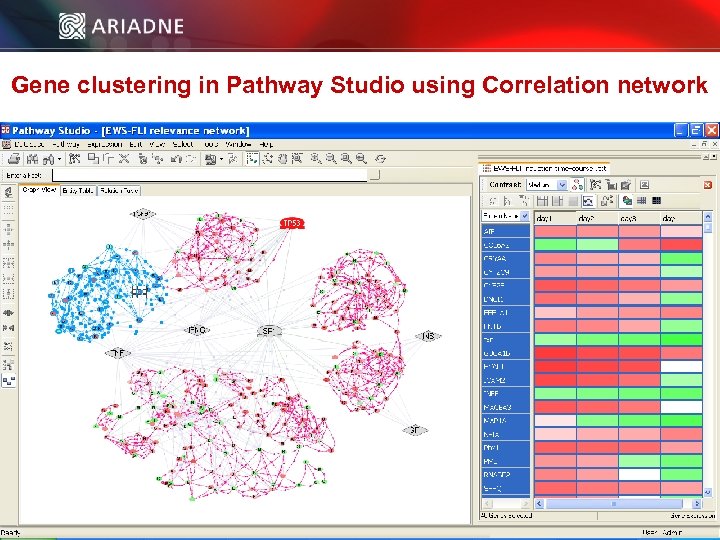 Gene clustering in Pathway Studio using Correlation network © 2006 Ariadne Genomics. All Rights Reserved. 39
Gene clustering in Pathway Studio using Correlation network © 2006 Ariadne Genomics. All Rights Reserved. 39
 Analysis of gene expression microarray data: STEP 2: Pathway Analysis of responsive genes • Network analysis – Identification of DE expressed protein complexes and physical networks – Identification of major regulators and targets in expression network • Via network querying (Build pathway tool) • Via Network enrichment analysis (in PS Enterprise only) • Functional analysis – Comparison of responsive genes with ontologies and pathway collection • • • Via Fisher exact test Via Gene Set Enrichment analysis (GSEA in PS Enterprise only) Gene ontology analysis (via Fisher’s test or GSEA) Comparative gene ontology analysis Via network querying (Build pathway tool) © 2006 Ariadne Genomics. All Rights Reserved. 40
Analysis of gene expression microarray data: STEP 2: Pathway Analysis of responsive genes • Network analysis – Identification of DE expressed protein complexes and physical networks – Identification of major regulators and targets in expression network • Via network querying (Build pathway tool) • Via Network enrichment analysis (in PS Enterprise only) • Functional analysis – Comparison of responsive genes with ontologies and pathway collection • • • Via Fisher exact test Via Gene Set Enrichment analysis (GSEA in PS Enterprise only) Gene ontology analysis (via Fisher’s test or GSEA) Comparative gene ontology analysis Via network querying (Build pathway tool) © 2006 Ariadne Genomics. All Rights Reserved. 40
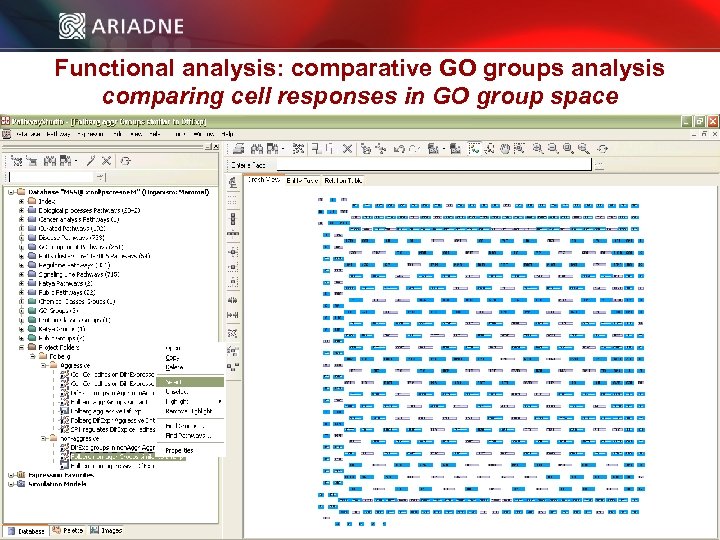 Functional analysis: comparative GO groups analysis comparing cell responses in GO group space © 2006 Ariadne Genomics. All Rights Reserved. 41
Functional analysis: comparative GO groups analysis comparing cell responses in GO group space © 2006 Ariadne Genomics. All Rights Reserved. 41
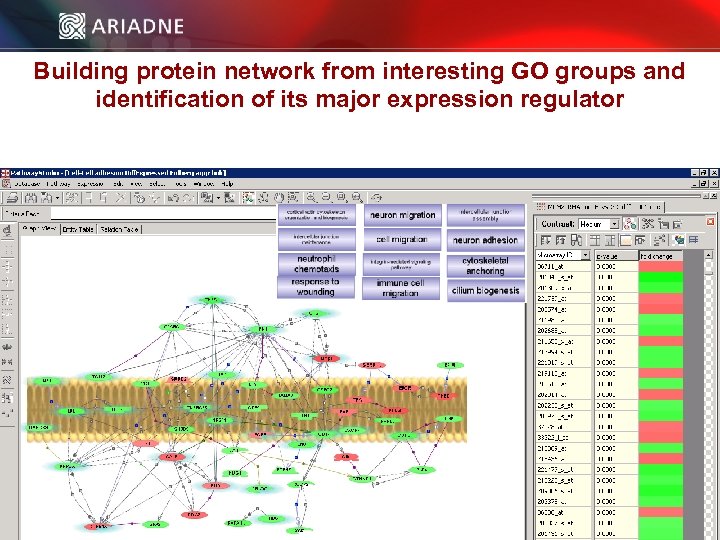 Building protein network from interesting GO groups and identification of its major expression regulator © 2006 Ariadne Genomics. All Rights Reserved. 42
Building protein network from interesting GO groups and identification of its major expression regulator © 2006 Ariadne Genomics. All Rights Reserved. 42
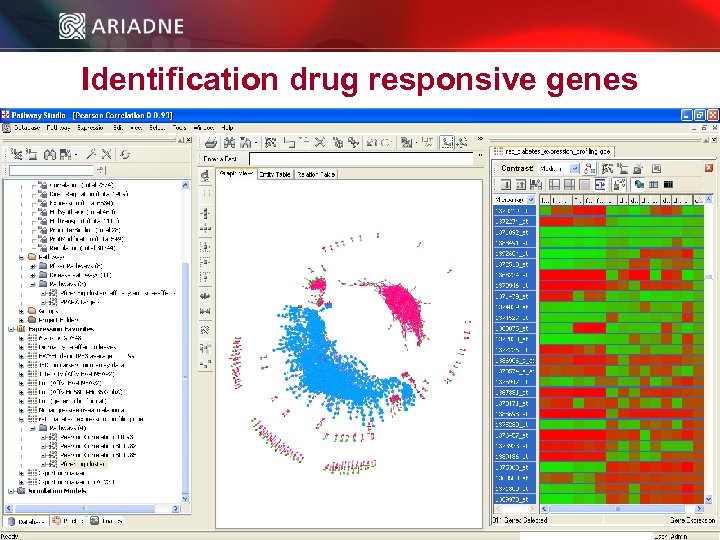 Identification drug responsive genes © 2006 Ariadne Genomics. All Rights Reserved. 43
Identification drug responsive genes © 2006 Ariadne Genomics. All Rights Reserved. 43
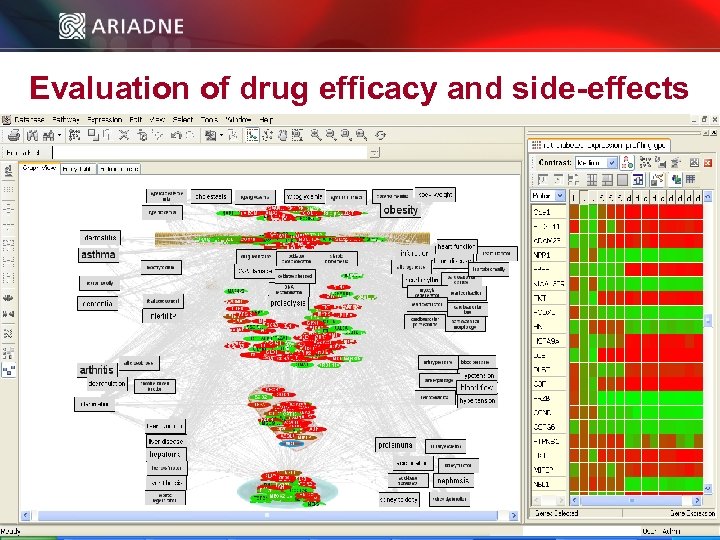 Evaluation of drug efficacy and side-effects © 2006 Ariadne Genomics. All Rights Reserved. 44
Evaluation of drug efficacy and side-effects © 2006 Ariadne Genomics. All Rights Reserved. 44
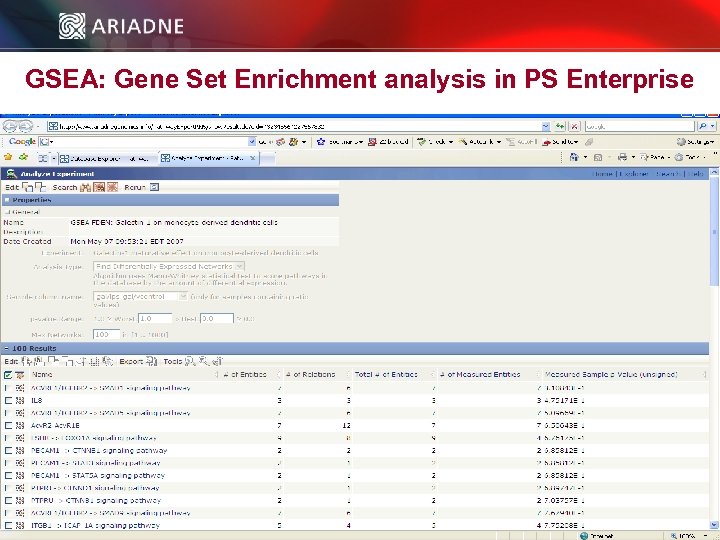 GSEA: Gene Set Enrichment analysis in PS Enterprise © 2006 Ariadne Genomics. All Rights Reserved. 45
GSEA: Gene Set Enrichment analysis in PS Enterprise © 2006 Ariadne Genomics. All Rights Reserved. 45
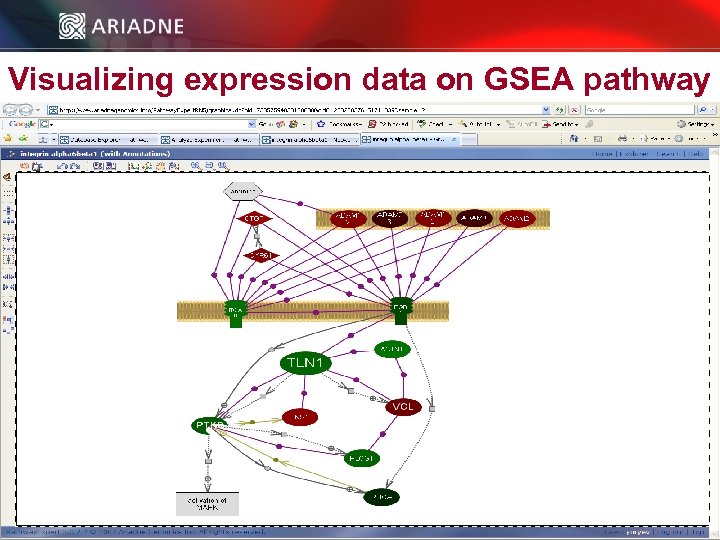 Visualizing expression data on GSEA pathway © 2006 Ariadne Genomics. All Rights Reserved. 46
Visualizing expression data on GSEA pathway © 2006 Ariadne Genomics. All Rights Reserved. 46
 High-throughput data analysis in Pathway Studio • Functional ontology analysis • Network analysis © 2006 Ariadne Genomics. All Rights Reserved.
High-throughput data analysis in Pathway Studio • Functional ontology analysis • Network analysis © 2006 Ariadne Genomics. All Rights Reserved.
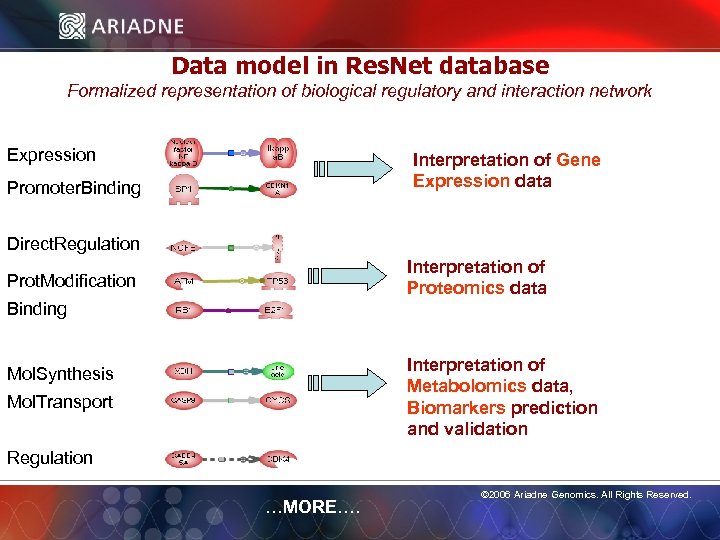 Data model in Res. Net database Formalized representation of biological regulatory and interaction network Expression Interpretation of Gene Expression data Promoter. Binding Direct. Regulation Interpretation of Proteomics data Prot. Modification Binding Interpretation of Metabolomics data, Biomarkers prediction and validation Mol. Synthesis Mol. Transport © 2006 Ariadne Genomics. All Rights Reserved. Regulation …MORE…. © 2006 Ariadne Genomics. All Rights Reserved. 48
Data model in Res. Net database Formalized representation of biological regulatory and interaction network Expression Interpretation of Gene Expression data Promoter. Binding Direct. Regulation Interpretation of Proteomics data Prot. Modification Binding Interpretation of Metabolomics data, Biomarkers prediction and validation Mol. Synthesis Mol. Transport © 2006 Ariadne Genomics. All Rights Reserved. Regulation …MORE…. © 2006 Ariadne Genomics. All Rights Reserved. 48
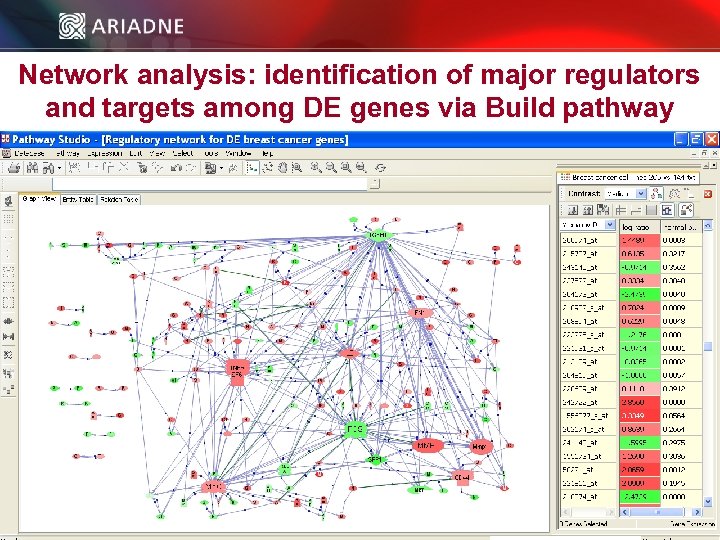 Network analysis: identification of major regulators and targets among DE genes via Build pathway © 2006 Ariadne Genomics. All Rights Reserved. 49
Network analysis: identification of major regulators and targets among DE genes via Build pathway © 2006 Ariadne Genomics. All Rights Reserved. 49
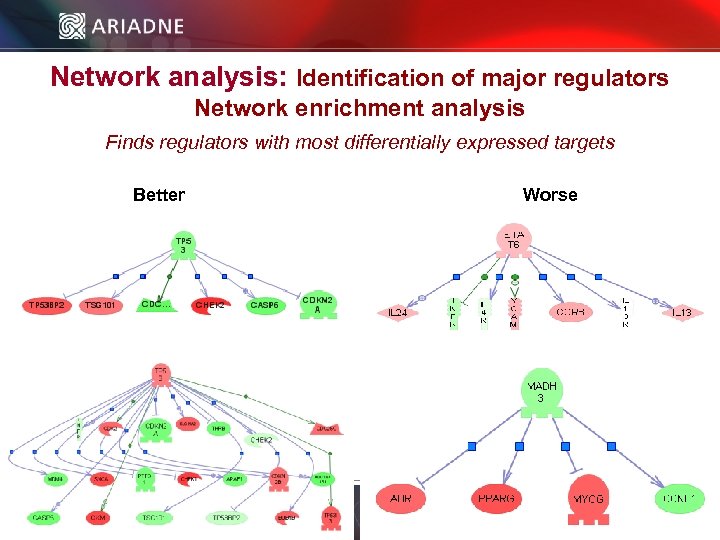 Network analysis: Identification of major regulators Network enrichment analysis Finds regulators with most differentially expressed targets Better Worse © 2006 Ariadne Genomics. All Rights Reserved. 50
Network analysis: Identification of major regulators Network enrichment analysis Finds regulators with most differentially expressed targets Better Worse © 2006 Ariadne Genomics. All Rights Reserved. 50
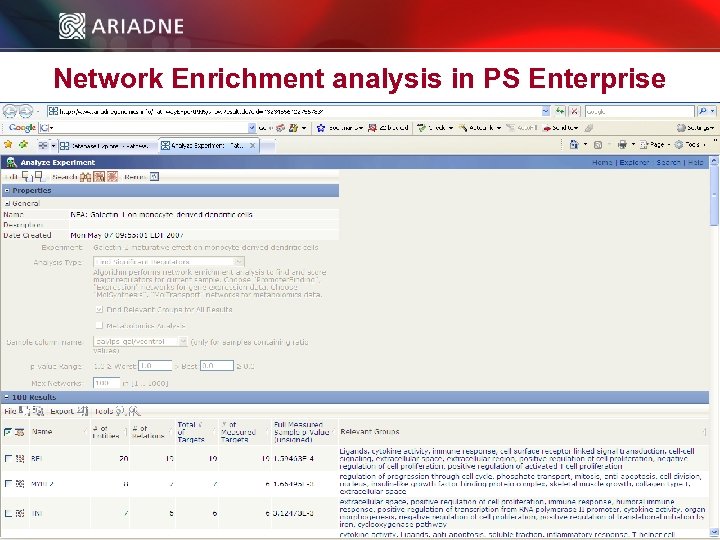 Network Enrichment analysis in PS Enterprise © 2006 Ariadne Genomics. All Rights Reserved. 51
Network Enrichment analysis in PS Enterprise © 2006 Ariadne Genomics. All Rights Reserved. 51
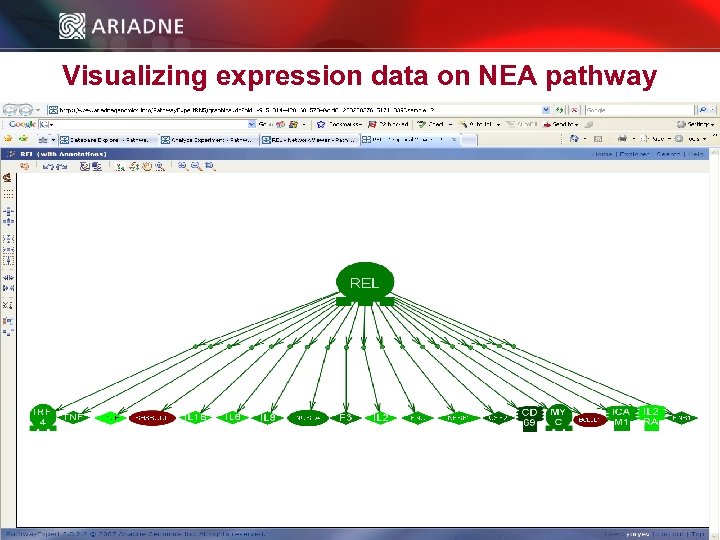 Visualizing expression data on NEA pathway © 2006 Ariadne Genomics. All Rights Reserved. 52
Visualizing expression data on NEA pathway © 2006 Ariadne Genomics. All Rights Reserved. 52
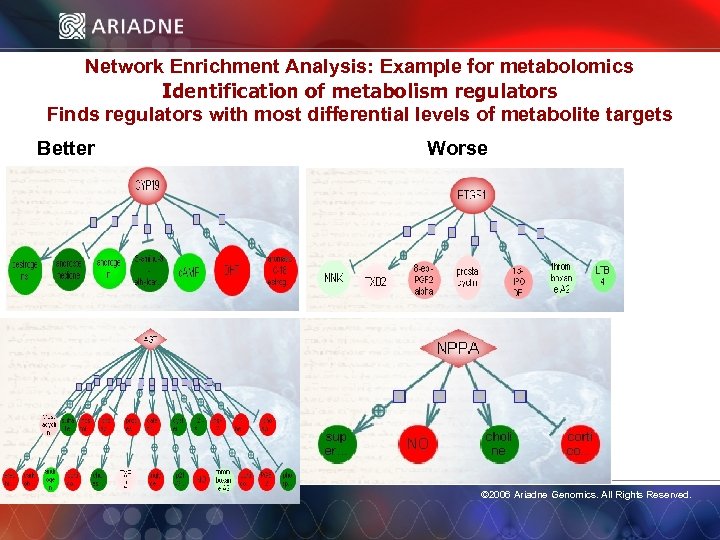 Network Enrichment Analysis: Example for metabolomics Identification of metabolism regulators Finds regulators with most differential levels of metabolite targets Better Worse © 2006 Ariadne Genomics. All Rights Reserved. 53
Network Enrichment Analysis: Example for metabolomics Identification of metabolism regulators Finds regulators with most differential levels of metabolite targets Better Worse © 2006 Ariadne Genomics. All Rights Reserved. 53
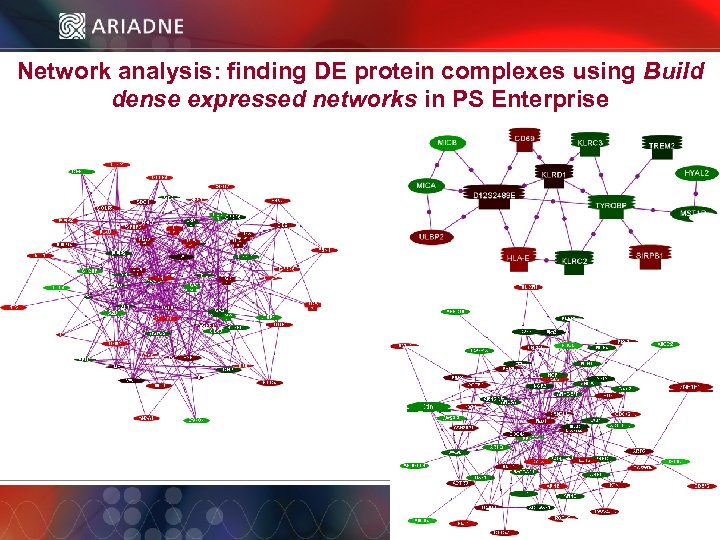 Network analysis: finding DE protein complexes using Build dense expressed networks in PS Enterprise © 2006 Ariadne Genomics. All Rights Reserved. 54
Network analysis: finding DE protein complexes using Build dense expressed networks in PS Enterprise © 2006 Ariadne Genomics. All Rights Reserved. 54
 >200 publications using AGI software and Res. Net database • • • Analysis of gene expression microarray data (139) Pathway Analysis (97) Disease mechanism (84) Publication by Ariadne Authors (18) Human genetics (7) Text processing (6) Reviews (7) Databases (3) Drug discovery (21) Toxicogenomics (4) © 2006 Ariadne Genomics. All Rights Reserved. 55
>200 publications using AGI software and Res. Net database • • • Analysis of gene expression microarray data (139) Pathway Analysis (97) Disease mechanism (84) Publication by Ariadne Authors (18) Human genetics (7) Text processing (6) Reviews (7) Databases (3) Drug discovery (21) Toxicogenomics (4) © 2006 Ariadne Genomics. All Rights Reserved. 55
 Most common workflow for microarray analysis in Pathway Studio for disease • Identify genes differentially expressed in disease (DE genes) • Identify genes known to associate to disease according to the literature using Pathway Studio • Identify DE genes that are linked to known diseases genes using Pathway Studio • Report novel disease genes © 2006 Ariadne Genomics. All Rights Reserved. 56
Most common workflow for microarray analysis in Pathway Studio for disease • Identify genes differentially expressed in disease (DE genes) • Identify genes known to associate to disease according to the literature using Pathway Studio • Identify DE genes that are linked to known diseases genes using Pathway Studio • Report novel disease genes © 2006 Ariadne Genomics. All Rights Reserved. 56
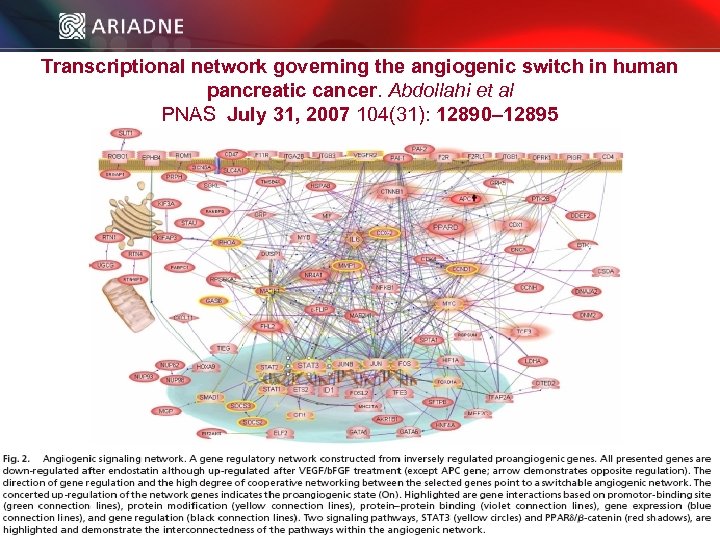 Transcriptional network governing the angiogenic switch in human pancreatic cancer. Abdollahi et al PNAS July 31, 2007 104(31): 12890– 12895 © 2006 Ariadne Genomics. All Rights Reserved. 57
Transcriptional network governing the angiogenic switch in human pancreatic cancer. Abdollahi et al PNAS July 31, 2007 104(31): 12890– 12895 © 2006 Ariadne Genomics. All Rights Reserved. 57
 High-throughput data analysis in Pathway Studio Extras © 2006 Ariadne Genomics. All Rights Reserved.
High-throughput data analysis in Pathway Studio Extras © 2006 Ariadne Genomics. All Rights Reserved.
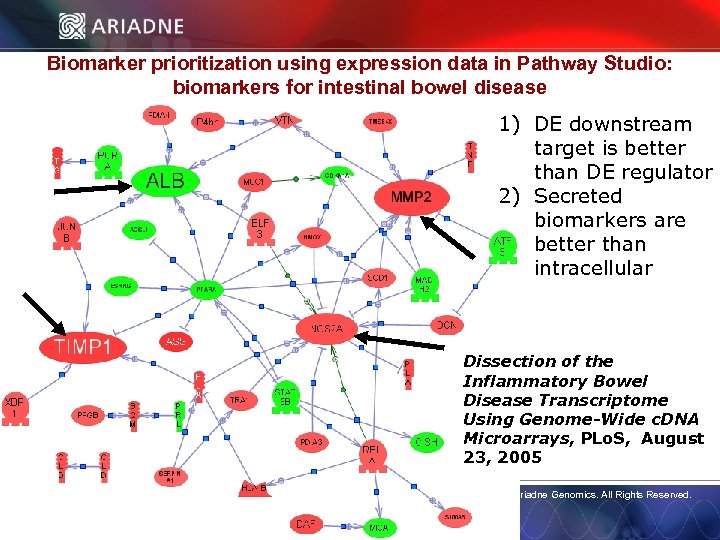 Biomarker prioritization using expression data in Pathway Studio: biomarkers for intestinal bowel disease 1) DE downstream target is better than DE regulator 2) Secreted biomarkers are better than intracellular Dissection of the Inflammatory Bowel Disease Transcriptome Using Genome-Wide c. DNA Microarrays, PLo. S, August © 2006 Ariadne Genomics. All Rights Reserved. 23, 2005 © 2006 Ariadne Genomics. All Rights Reserved. 59
Biomarker prioritization using expression data in Pathway Studio: biomarkers for intestinal bowel disease 1) DE downstream target is better than DE regulator 2) Secreted biomarkers are better than intracellular Dissection of the Inflammatory Bowel Disease Transcriptome Using Genome-Wide c. DNA Microarrays, PLo. S, August © 2006 Ariadne Genomics. All Rights Reserved. 23, 2005 © 2006 Ariadne Genomics. All Rights Reserved. 59
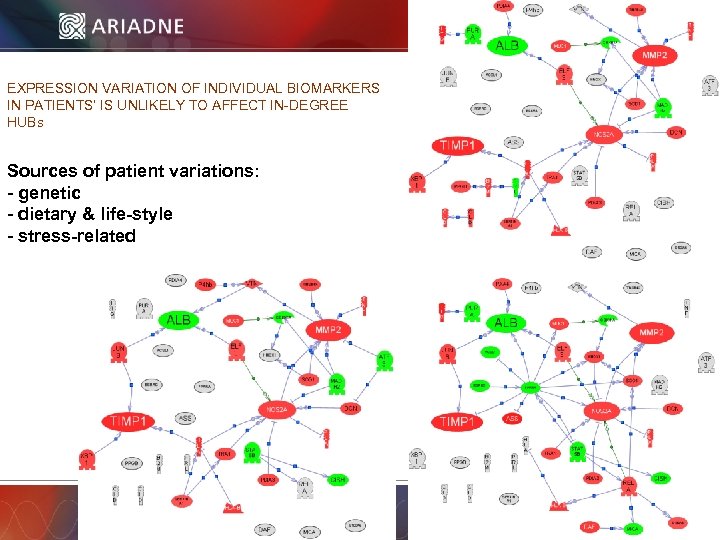 EXPRESSION VARIATION OF INDIVIDUAL BIOMARKERS IN PATIENTS’ IS UNLIKELY TO AFFECT IN-DEGREE HUBs Sources of patient variations: - genetic - dietary & life-style - stress-related © 2006 Ariadne Genomics. All Rights Reserved. 60
EXPRESSION VARIATION OF INDIVIDUAL BIOMARKERS IN PATIENTS’ IS UNLIKELY TO AFFECT IN-DEGREE HUBs Sources of patient variations: - genetic - dietary & life-style - stress-related © 2006 Ariadne Genomics. All Rights Reserved. 60
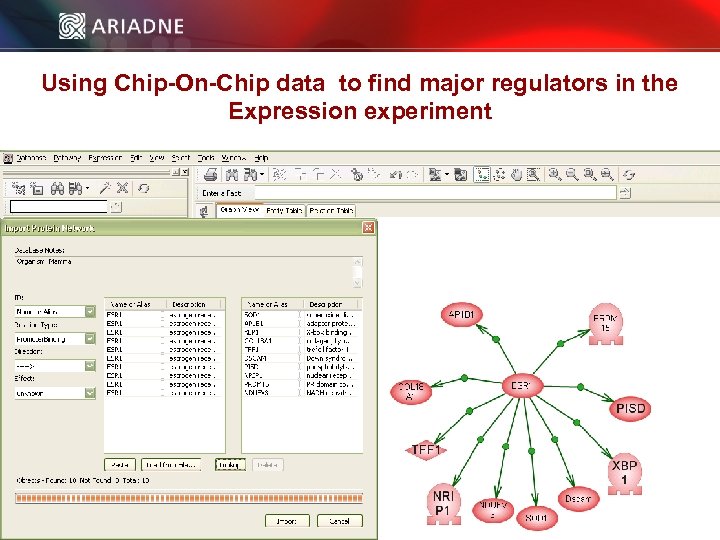 Using Chip-On-Chip data to find major regulators in the Expression experiment © 2006 Ariadne Genomics. All Rights Reserved. 61
Using Chip-On-Chip data to find major regulators in the Expression experiment © 2006 Ariadne Genomics. All Rights Reserved. 61
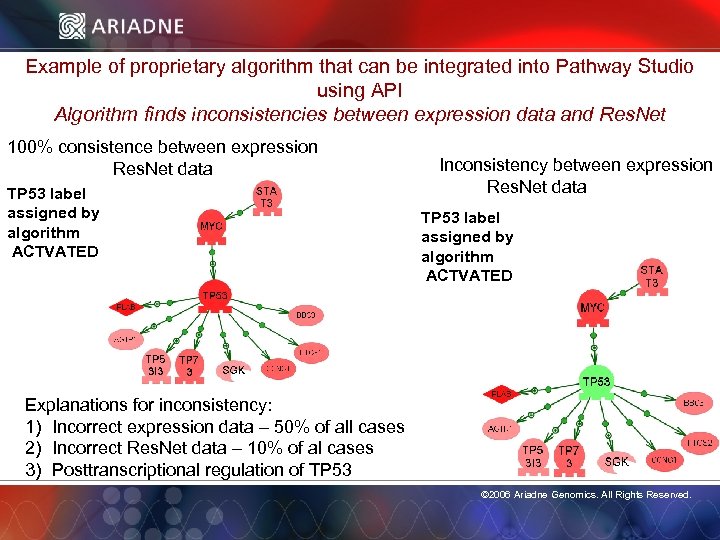 Example of proprietary algorithm that can be integrated into Pathway Studio using API Algorithm finds inconsistencies between expression data and Res. Net 100% consistence between expression Res. Net data TP 53 label assigned by algorithm ACTVATED Explanations for inconsistency: 1) Incorrect expression data – 50% of all cases 2) Incorrect Res. Net data – 10% of al cases 3) Posttranscriptional regulation of TP 53 Inconsistency between expression Res. Net data TP 53 label assigned by algorithm ACTVATED © 2006 Ariadne Genomics. All Rights Reserved. 62
Example of proprietary algorithm that can be integrated into Pathway Studio using API Algorithm finds inconsistencies between expression data and Res. Net 100% consistence between expression Res. Net data TP 53 label assigned by algorithm ACTVATED Explanations for inconsistency: 1) Incorrect expression data – 50% of all cases 2) Incorrect Res. Net data – 10% of al cases 3) Posttranscriptional regulation of TP 53 Inconsistency between expression Res. Net data TP 53 label assigned by algorithm ACTVATED © 2006 Ariadne Genomics. All Rights Reserved. 62


