225524a85ad078b1b33ce44cbeca102c.ppt
- Количество слайдов: 21
 AQUEOUS COMPUTING - Writing on Molecules T. Head, M. Yamamura, and S. Gal Binghamton University 7/9/99 CEC'99
AQUEOUS COMPUTING - Writing on Molecules T. Head, M. Yamamura, and S. Gal Binghamton University 7/9/99 CEC'99
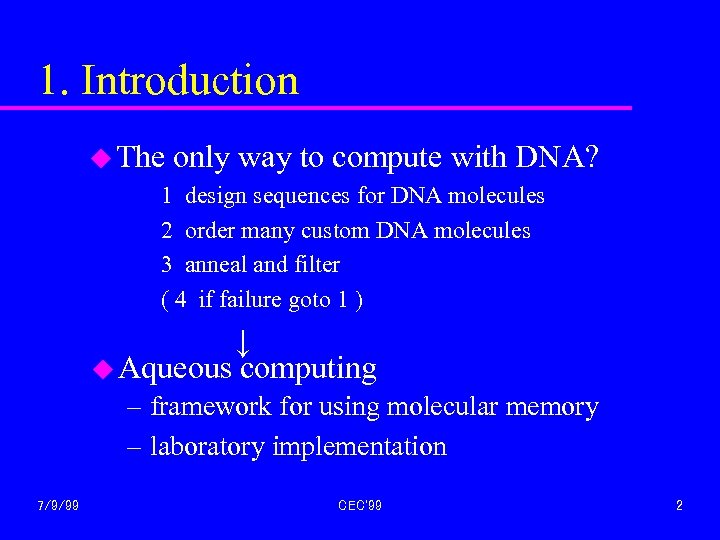 1. Introduction u The only way to compute with DNA? 1 design sequences for DNA molecules 2 order many custom DNA molecules 3 anneal and filter ( 4 if failure goto 1 ) ↓ u Aqueous computing – framework for using molecular memory – laboratory implementation 7/9/99 CEC'99 2
1. Introduction u The only way to compute with DNA? 1 design sequences for DNA molecules 2 order many custom DNA molecules 3 anneal and filter ( 4 if failure goto 1 ) ↓ u Aqueous computing – framework for using molecular memory – laboratory implementation 7/9/99 CEC'99 2
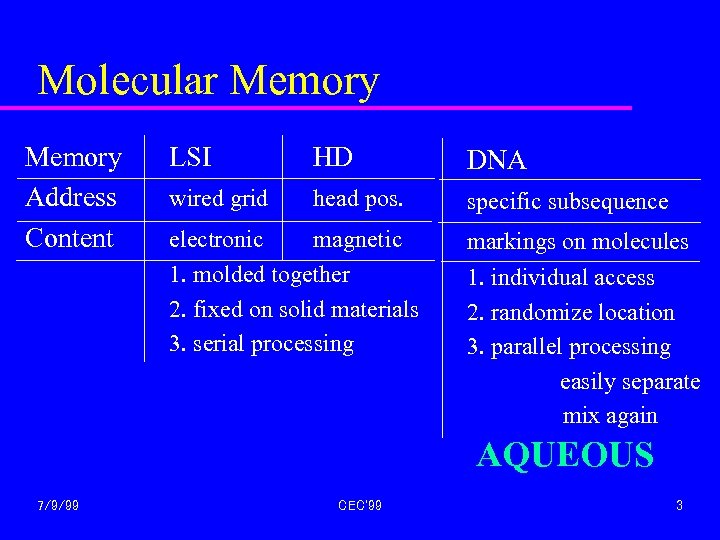 Molecular Memory Address Content LSI HD DNA wired grid head pos. specific subsequence electronic magnetic 1. molded together 2. fixed on solid materials 3. serial processing markings on molecules 1. individual access 2. randomize location 3. parallel processing easily separate mix again AQUEOUS 7/9/99 CEC'99 3
Molecular Memory Address Content LSI HD DNA wired grid head pos. specific subsequence electronic magnetic 1. molded together 2. fixed on solid materials 3. serial processing markings on molecules 1. individual access 2. randomize location 3. parallel processing easily separate mix again AQUEOUS 7/9/99 CEC'99 3
 2. Mathematical Basis u Common algorithmic problem (CAP) – a description of the pattern of the problem u Aqueous algorithm – a way to use molecular memory 7/9/99 CEC'99 4
2. Mathematical Basis u Common algorithmic problem (CAP) – a description of the pattern of the problem u Aqueous algorithm – a way to use molecular memory 7/9/99 CEC'99 4
 Common algorithmic problem u CAP given S: finite set F ⊂2 S (the forbidden subsets) find the largest cardinal number n for which there is a subset T of S for which: |T|=n, ∀U∈F U⊂T. – NP-complete problems having the CAP pattern » maximum independent set » minimum vertex cover » Hamiltonian cycles » Boolean satisfiability, etc. 7/9/99 CEC'99 5
Common algorithmic problem u CAP given S: finite set F ⊂2 S (the forbidden subsets) find the largest cardinal number n for which there is a subset T of S for which: |T|=n, ∀U∈F U⊂T. – NP-complete problems having the CAP pattern » maximum independent set » minimum vertex cover » Hamiltonian cycles » Boolean satisfiability, etc. 7/9/99 CEC'99 5
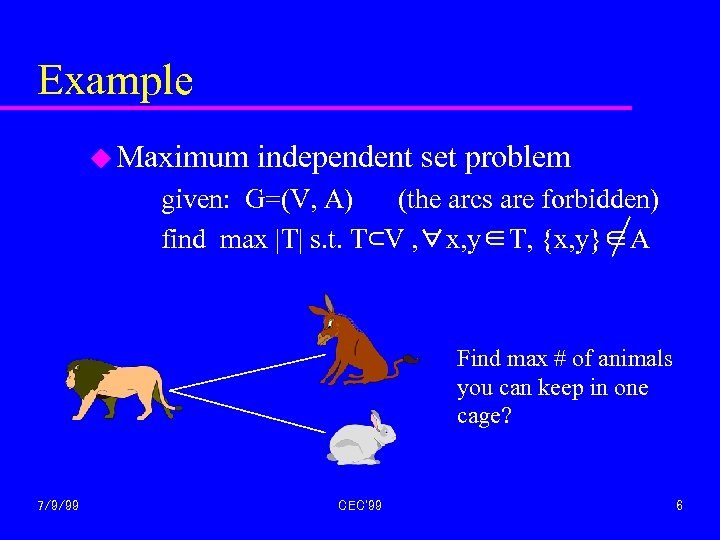 Example u Maximum independent set problem given: G=(V, A) (the arcs are forbidden) find max |T| s. t. T⊂V , ∀x, y∈T, {x, y}∈A Find max # of animals you can keep in one cage? 7/9/99 CEC'99 6
Example u Maximum independent set problem given: G=(V, A) (the arcs are forbidden) find max |T| s. t. T⊂V , ∀x, y∈T, {x, y}∈A Find max # of animals you can keep in one cage? 7/9/99 CEC'99 6
 Aqueous Algorithm Initialize; For each {s 1, s 2, . . . , sk} in F Do Pour (k) 1: Set. To. Zero( s 1 ) 2: Set. To. Zero( s 2 ). . . k: Set. To. Zero( sk ) Unite End. For; Max. Count. Of. Ones 7/9/99 CEC'99 7
Aqueous Algorithm Initialize; For each {s 1, s 2, . . . , sk} in F Do Pour (k) 1: Set. To. Zero( s 1 ) 2: Set. To. Zero( s 2 ). . . k: Set. To. Zero( sk ) Unite End. For; Max. Count. Of. Ones 7/9/99 CEC'99 7
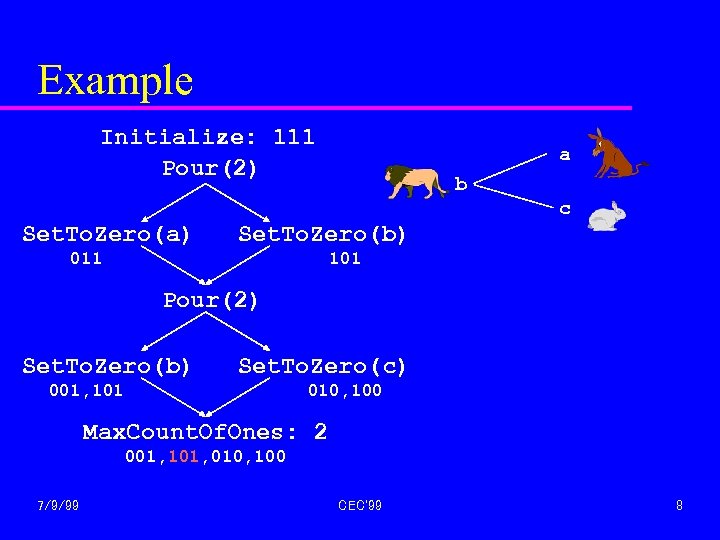 Example Initialize: 111 Pour(2) a b c Set. To. Zero(a) Set. To. Zero(b) 011 101 Pour(2) Set. To. Zero(b) Set. To. Zero(c) 001, 101 010, 100 Max. Count. Of. Ones: 2 001, 101, 010, 100 7/9/99 CEC'99 8
Example Initialize: 111 Pour(2) a b c Set. To. Zero(a) Set. To. Zero(b) 011 101 Pour(2) Set. To. Zero(b) Set. To. Zero(c) 001, 101 010, 100 Max. Count. Of. Ones: 2 001, 101, 010, 100 7/9/99 CEC'99 8
 3. Biomolecular Implementation u DNA modification enzymes – how to write on molecules u DNA plasmid – use of bacteria and blue/white selection 7/9/99 CEC'99 9
3. Biomolecular Implementation u DNA modification enzymes – how to write on molecules u DNA plasmid – use of bacteria and blue/white selection 7/9/99 CEC'99 9
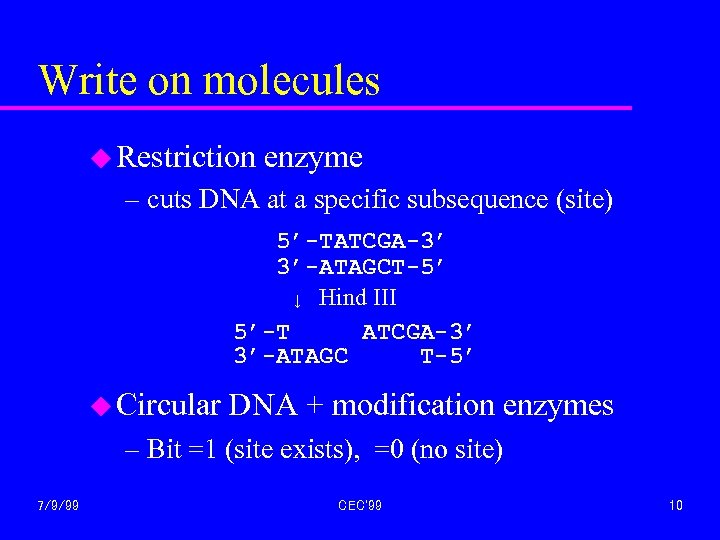 Write on molecules u Restriction enzyme – cuts DNA at a specific subsequence (site) 5’-TATCGA-3’ 3’-ATAGCT-5’ ↓ Hind III 5’-T ATCGA-3’ 3’-ATAGC T-5’ u Circular DNA + modification enzymes – Bit =1 (site exists), =0 (no site) 7/9/99 CEC'99 10
Write on molecules u Restriction enzyme – cuts DNA at a specific subsequence (site) 5’-TATCGA-3’ 3’-ATAGCT-5’ ↓ Hind III 5’-T ATCGA-3’ 3’-ATAGC T-5’ u Circular DNA + modification enzymes – Bit =1 (site exists), =0 (no site) 7/9/99 CEC'99 10
 Cut/fill/paste 5’-TATCGA-3’ Bit=1, circular 3’-ATAGCT-5’ cut ↓ restriction enzyme 5’-T ATCGA-3’ linear 3’-ATAGC T-5’ fill ↓ DNA polymerase 5’-TATCGA-3’ 3’-ATAGCT-5’ paste ↓ DNA ligase 5’-TATCGA-3’ 3’-ATAGCT-5’ Bit=0, circular 7/9/99 CEC'99 11
Cut/fill/paste 5’-TATCGA-3’ Bit=1, circular 3’-ATAGCT-5’ cut ↓ restriction enzyme 5’-T ATCGA-3’ linear 3’-ATAGC T-5’ fill ↓ DNA polymerase 5’-TATCGA-3’ 3’-ATAGCT-5’ paste ↓ DNA ligase 5’-TATCGA-3’ 3’-ATAGCT-5’ Bit=0, circular 7/9/99 CEC'99 11
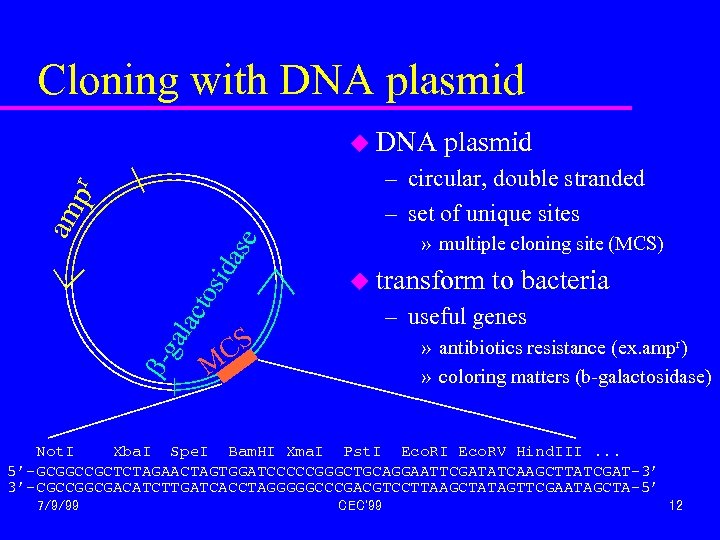 Cloning with DNA plasmid u DNA plasmid » multiple cloning site (MCS) b- ga lac tos ida se am pr – circular, double stranded – set of unique sites u transform to bacteria – useful genes CS » antibiotics resistance (ex. ampr) » coloring matters (b-galactosidase) M Not. I Xba. I Spe. I Bam. HI Xma. I Pst. I Eco. RV Hind. III. . . 5’-GCGGCCGCTCTAGAACTAGTGGATCCCCCGGGCTGCAGGAATTCGATATCAAGCTTATCGAT-3’ 3’-CGCCGGCGACATCTTGATCACCTAGGGGGCCCGACGTCCTTAAGCTATAGTTCGAATAGCTA-5’ 7/9/99 CEC'99 12
Cloning with DNA plasmid u DNA plasmid » multiple cloning site (MCS) b- ga lac tos ida se am pr – circular, double stranded – set of unique sites u transform to bacteria – useful genes CS » antibiotics resistance (ex. ampr) » coloring matters (b-galactosidase) M Not. I Xba. I Spe. I Bam. HI Xma. I Pst. I Eco. RV Hind. III. . . 5’-GCGGCCGCTCTAGAACTAGTGGATCCCCCGGGCTGCAGGAATTCGATATCAAGCTTATCGAT-3’ 3’-CGCCGGCGACATCTTGATCACCTAGGGGGCCCGACGTCCTTAAGCTATAGTTCGAATAGCTA-5’ 7/9/99 CEC'99 12
 Genetic code translation u Genetic code – translated into a series of amino acids by groups of 3 base pairs (codon) u Reading frame – 3 different meanings ex) 7/9/99 5’-GCTCTAGAACTAGTGGATCCCCCGGGCTGCAGGAATTCGATATC A L E L V D P P G C R N S I. . . . (under construction) CEC'99 13
Genetic code translation u Genetic code – translated into a series of amino acids by groups of 3 base pairs (codon) u Reading frame – 3 different meanings ex) 7/9/99 5’-GCTCTAGAACTAGTGGATCCCCCGGGCTGCAGGAATTCGATATC A L E L V D P P G C R N S I. . . . (under construction) CEC'99 13
 Blue / white selection u initial DNA plasmid express b-galactosidase gene → blue ↓ u 1 st cut/fill/paste +4 bp ⇒ reading frame shift → white ↓ u 2 nd cut/fill/paste +8 bp ⇒ reading frame still shift → white ↓ u 3 rd cut/fill/paste +12 bp ⇒ readinf frame restored → blue » useful as a debugging tool 7/9/99 CEC'99 14
Blue / white selection u initial DNA plasmid express b-galactosidase gene → blue ↓ u 1 st cut/fill/paste +4 bp ⇒ reading frame shift → white ↓ u 2 nd cut/fill/paste +8 bp ⇒ reading frame still shift → white ↓ u 3 rd cut/fill/paste +12 bp ⇒ readinf frame restored → blue » useful as a debugging tool 7/9/99 CEC'99 14
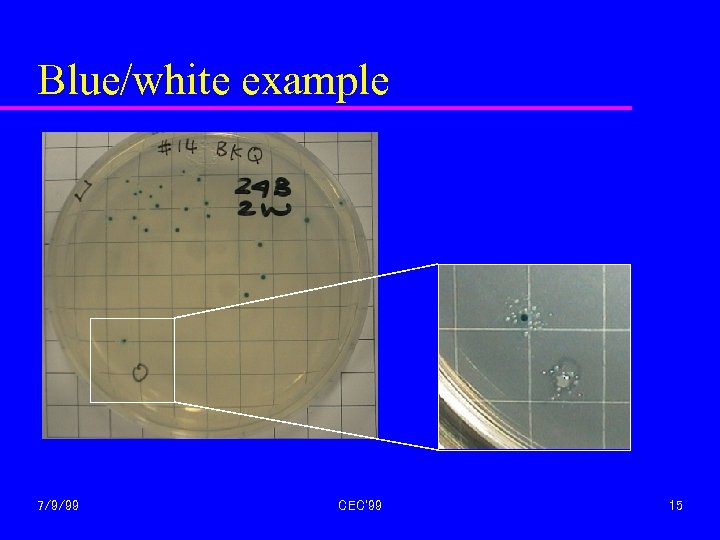 Blue/white example 7/9/99 CEC'99 15
Blue/white example 7/9/99 CEC'99 15
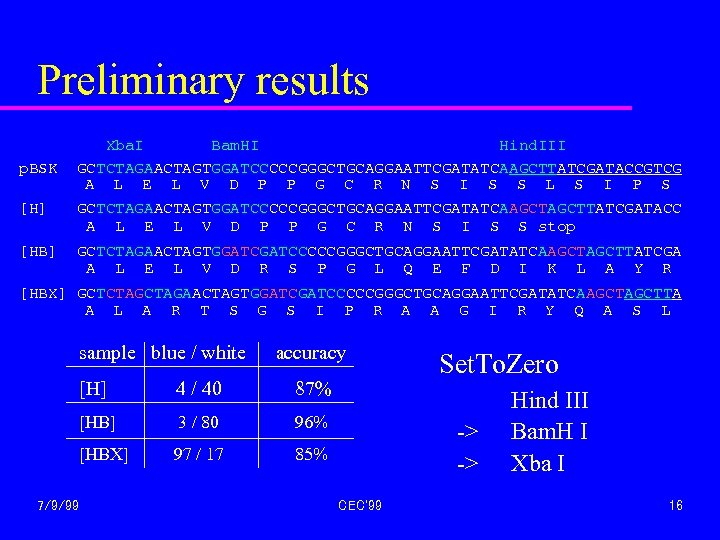 Preliminary results p. BSK Xba. I Bam. HI Hind. III GCTCTAGAACTAGTGGATCCCCCGGGCTGCAGGAATTCGATATCAAGCTTATCGATACCGTCG A L E L V D P P G C R N S I S S L S I P S [H] GCTCTAGAACTAGTGGATCCCCCGGGCTGCAGGAATTCGATATCAAGCTTATCGATACC A L E L V D P P G C R N S I S S stop [HB] GCTCTAGAACTAGTGGATCCCCCGGGCTGCAGGAATTCGATATCAAGCTTATCGA A L E L V D R S P G L Q E F D I K L A Y R [HBX] GCTCTAGAACTAGTGGATCCCCCGGGCTGCAGGAATTCGATATCAAGCTTA A L A R T S G S I P R A A G I R Y Q A S L sample blue / white accuracy [H] 4 / 40 87% [HB] 3 / 80 96% [HBX] 97 / 17 85% 7/9/99 Set. To. Zero -> -> CEC'99 Hind III Bam. H I Xba I 16
Preliminary results p. BSK Xba. I Bam. HI Hind. III GCTCTAGAACTAGTGGATCCCCCGGGCTGCAGGAATTCGATATCAAGCTTATCGATACCGTCG A L E L V D P P G C R N S I S S L S I P S [H] GCTCTAGAACTAGTGGATCCCCCGGGCTGCAGGAATTCGATATCAAGCTTATCGATACC A L E L V D P P G C R N S I S S stop [HB] GCTCTAGAACTAGTGGATCCCCCGGGCTGCAGGAATTCGATATCAAGCTTATCGA A L E L V D R S P G L Q E F D I K L A Y R [HBX] GCTCTAGAACTAGTGGATCCCCCGGGCTGCAGGAATTCGATATCAAGCTTA A L A R T S G S I P R A A G I R Y Q A S L sample blue / white accuracy [H] 4 / 40 87% [HB] 3 / 80 96% [HBX] 97 / 17 85% 7/9/99 Set. To. Zero -> -> CEC'99 Hind III Bam. H I Xba I 16
![Example a b c [HB] (+8, white) a=0 (Spe. I) b=0 (Xho. I) c=0 Example a b c [HB] (+8, white) a=0 (Spe. I) b=0 (Xho. I) c=0](https://present5.com/presentation/225524a85ad078b1b33ce44cbeca102c/image-17.jpg) Example a b c [HB] (+8, white) a=0 (Spe. I) b=0 (Xho. I) c=0 (Xba. I) 7/9/99 mix; +12 & +16 (solution = +12, white) 0 +4 +8 +12 under construction CEC'99 17
Example a b c [HB] (+8, white) a=0 (Spe. I) b=0 (Xho. I) c=0 (Xba. I) 7/9/99 mix; +12 & +16 (solution = +12, white) 0 +4 +8 +12 under construction CEC'99 17
 4. Discussion u Advantages as DNA computing – start with one DNA plasmid » no custom DNA for individual problem – amplify in bacteria » blue/white selection as debugging tool » preserving the distribution of DNA plasmids 7/9/99 CEC'99 18
4. Discussion u Advantages as DNA computing – start with one DNA plasmid » no custom DNA for individual problem – amplify in bacteria » blue/white selection as debugging tool » preserving the distribution of DNA plasmids 7/9/99 CEC'99 18
 5. Conclusion u Molecular Memory – Aqueous Algorithm » general framework to use molecular memory – Cut/fill/paste » laboratory implementation u Further issues – scale up & speed up – new algorithm fits bacteria 7/9/99 CEC'99 19
5. Conclusion u Molecular Memory – Aqueous Algorithm » general framework to use molecular memory – Cut/fill/paste » laboratory implementation u Further issues – scale up & speed up – new algorithm fits bacteria 7/9/99 CEC'99 19
 International Connection Binghamton University (USA) Aqueous Computing Leiden University (Netherlands) 7/9/99 Tokyo Institute of Technology (Japan) CEC'99 20
International Connection Binghamton University (USA) Aqueous Computing Leiden University (Netherlands) 7/9/99 Tokyo Institute of Technology (Japan) CEC'99 20
 Acknowledgement u Xia Chen & Shalini Aggarwal in S. Gal Laboratory at Binghamton University u NSF CCR-9509831 u DARPA/NSF CCR-9725021 u JSPS-RFTF 96100101 u LCNC at Leiden University 7/9/99 CEC'99 21
Acknowledgement u Xia Chen & Shalini Aggarwal in S. Gal Laboratory at Binghamton University u NSF CCR-9509831 u DARPA/NSF CCR-9725021 u JSPS-RFTF 96100101 u LCNC at Leiden University 7/9/99 CEC'99 21


