418213d99cc02eb37c0983a2f1a8493d.ppt
- Количество слайдов: 24
 APAN Workshop on Exploring e. Science Aug 26, 2005 Taipei, Taiwan Gfarm Grid File System for Distributed and Parallel Data Computing Osamu Tatebe o. tatebe@aist. go. jp Grid Technology Research Center, AIST National Institute of Advanced Industrial Science and Technology
APAN Workshop on Exploring e. Science Aug 26, 2005 Taipei, Taiwan Gfarm Grid File System for Distributed and Parallel Data Computing Osamu Tatebe o. tatebe@aist. go. jp Grid Technology Research Center, AIST National Institute of Advanced Industrial Science and Technology
![[Background] Petascale Data Intensive Computing High Energy Physics l CERN LHC, KEK-B Belle l [Background] Petascale Data Intensive Computing High Energy Physics l CERN LHC, KEK-B Belle l](https://present5.com/presentation/418213d99cc02eb37c0983a2f1a8493d/image-2.jpg) [Background] Petascale Data Intensive Computing High Energy Physics l CERN LHC, KEK-B Belle l ~MB/collision, 100 collisions/sec l ~PB/year l 2000 physicists, 35 countries Detector for LHCb experiment Detector for ALICE experiment Astronomical Data Analysis l data analysis of the whole data l TB~PB/year/telescope l Subaru telescope l 10 GB/night, 3 TB/year National Institute of Advanced Industrial Science and Technology
[Background] Petascale Data Intensive Computing High Energy Physics l CERN LHC, KEK-B Belle l ~MB/collision, 100 collisions/sec l ~PB/year l 2000 physicists, 35 countries Detector for LHCb experiment Detector for ALICE experiment Astronomical Data Analysis l data analysis of the whole data l TB~PB/year/telescope l Subaru telescope l 10 GB/night, 3 TB/year National Institute of Advanced Industrial Science and Technology
 Petascale Data-intensive Computing Requirements Peta/Exabyte scale files, millions of files Scalable computational power > 1 TFLOPS, hopefully > 10 TFLOPS Scalable parallel I/O throughput > 100 GB/s, hopefully > 1 TB/s within a system and between systems Efficiently global sharing with group-oriented authentication and access control Fault Tolerance / Dynamic re-configuration Resource Management and Scheduling System monitoring and administration Global Computing Environment National Institute of Advanced Industrial Science and Technology
Petascale Data-intensive Computing Requirements Peta/Exabyte scale files, millions of files Scalable computational power > 1 TFLOPS, hopefully > 10 TFLOPS Scalable parallel I/O throughput > 100 GB/s, hopefully > 1 TB/s within a system and between systems Efficiently global sharing with group-oriented authentication and access control Fault Tolerance / Dynamic re-configuration Resource Management and Scheduling System monitoring and administration Global Computing Environment National Institute of Advanced Industrial Science and Technology
 Goal and feature of Grid Datafarm Goal Dependable data sharing among multiple organizations High-speed data access, High-performance data computing Grid Datafarm Grid File System – Global dependable virtual file system Federates scratch disks in PCs Parallel & distributed data computing Associates Computational Grid with Data Grid Features Secured based on Grid Security Infrastructure Scalable depending on data size and usage scenarios Data location transparent data access Automatic and transparent replica selection for fault tolerance High-performance data access and computing by accessing multiple dispersed storages in parallel (file affinity scheduling) National Institute of Advanced Industrial Science and Technology
Goal and feature of Grid Datafarm Goal Dependable data sharing among multiple organizations High-speed data access, High-performance data computing Grid Datafarm Grid File System – Global dependable virtual file system Federates scratch disks in PCs Parallel & distributed data computing Associates Computational Grid with Data Grid Features Secured based on Grid Security Infrastructure Scalable depending on data size and usage scenarios Data location transparent data access Automatic and transparent replica selection for fault tolerance High-performance data access and computing by accessing multiple dispersed storages in parallel (file affinity scheduling) National Institute of Advanced Industrial Science and Technology
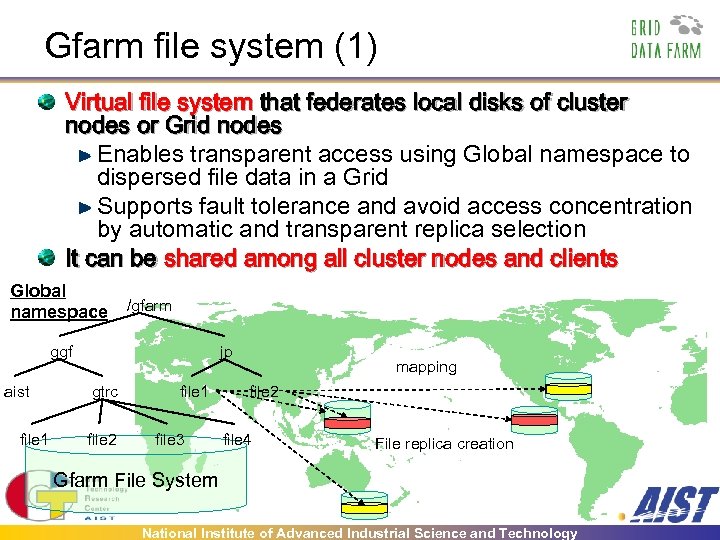 Gfarm file system (1) Virtual file system that federates local disks of cluster nodes or Grid nodes Enables transparent access using Global namespace to dispersed file data in a Grid Supports fault tolerance and avoid access concentration by automatic and transparent replica selection It can be shared among all cluster nodes and clients Global namespace /gfarm ggf aist file 1 jp gtrc file 2 file 1 file 3 mapping file 2 file 4 File replica creation Gfarm File System National Institute of Advanced Industrial Science and Technology
Gfarm file system (1) Virtual file system that federates local disks of cluster nodes or Grid nodes Enables transparent access using Global namespace to dispersed file data in a Grid Supports fault tolerance and avoid access concentration by automatic and transparent replica selection It can be shared among all cluster nodes and clients Global namespace /gfarm ggf aist file 1 jp gtrc file 2 file 1 file 3 mapping file 2 file 4 File replica creation Gfarm File System National Institute of Advanced Industrial Science and Technology
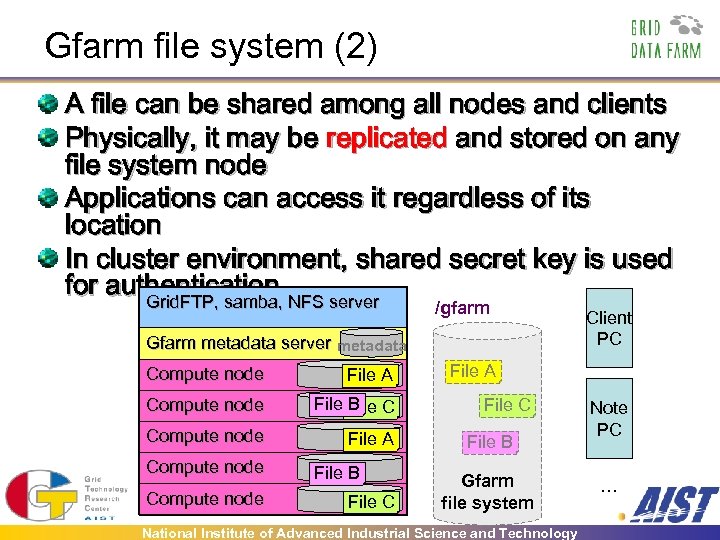 Gfarm file system (2) A file can be shared among all nodes and clients Physically, it may be replicated and stored on any file system node Applications can access it regardless of its location In cluster environment, shared secret key is used for authentication NFS server Grid. FTP, samba, /gfarm Gfarm metadata server metadata Compute node File A Compute node File C B Compute node File A Compute node File B File C Client PC File A File C File B Gfarm file system National Institute of Advanced Industrial Science and Technology Note PC …
Gfarm file system (2) A file can be shared among all nodes and clients Physically, it may be replicated and stored on any file system node Applications can access it regardless of its location In cluster environment, shared secret key is used for authentication NFS server Grid. FTP, samba, /gfarm Gfarm metadata server metadata Compute node File A Compute node File C B Compute node File A Compute node File B File C Client PC File A File C File B Gfarm file system National Institute of Advanced Industrial Science and Technology Note PC …
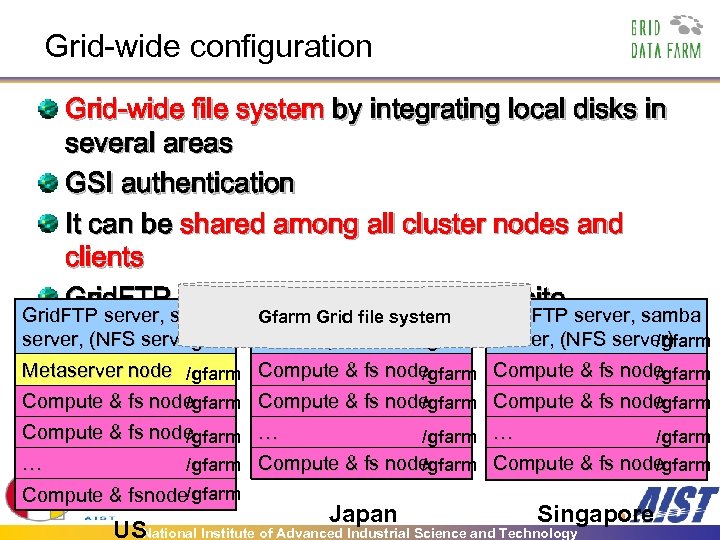 Grid-wide configuration Grid-wide file system by integrating local disks in several areas GSI authentication It can be shared among all cluster nodes and clients Grid. FTP and samba servers in each site Grid. FTP server, samba Gfarm Grid file system Grid. FTP server, samba server, (NFS server) /gfarm Metaserver node /gfarm Compute & fs node /gfarm Compute & fs node /gfarm … … /gfarm Compute & fs node /gfarm Compute & fsnode/gfarm Japan Singapore USNational Institute of Advanced Industrial Science and Technology
Grid-wide configuration Grid-wide file system by integrating local disks in several areas GSI authentication It can be shared among all cluster nodes and clients Grid. FTP and samba servers in each site Grid. FTP server, samba Gfarm Grid file system Grid. FTP server, samba server, (NFS server) /gfarm Metaserver node /gfarm Compute & fs node /gfarm Compute & fs node /gfarm … … /gfarm Compute & fs node /gfarm Compute & fsnode/gfarm Japan Singapore USNational Institute of Advanced Industrial Science and Technology
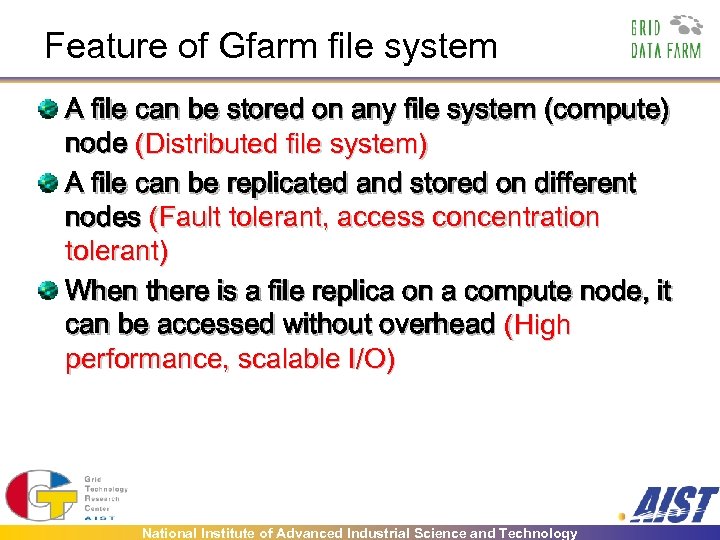 Feature of Gfarm file system A file can be stored on any file system (compute) node (Distributed file system) A file can be replicated and stored on different nodes (Fault tolerant, access concentration tolerant) When there is a file replica on a compute node, it can be accessed without overhead (High performance, scalable I/O) National Institute of Advanced Industrial Science and Technology
Feature of Gfarm file system A file can be stored on any file system (compute) node (Distributed file system) A file can be replicated and stored on different nodes (Fault tolerant, access concentration tolerant) When there is a file replica on a compute node, it can be accessed without overhead (High performance, scalable I/O) National Institute of Advanced Industrial Science and Technology
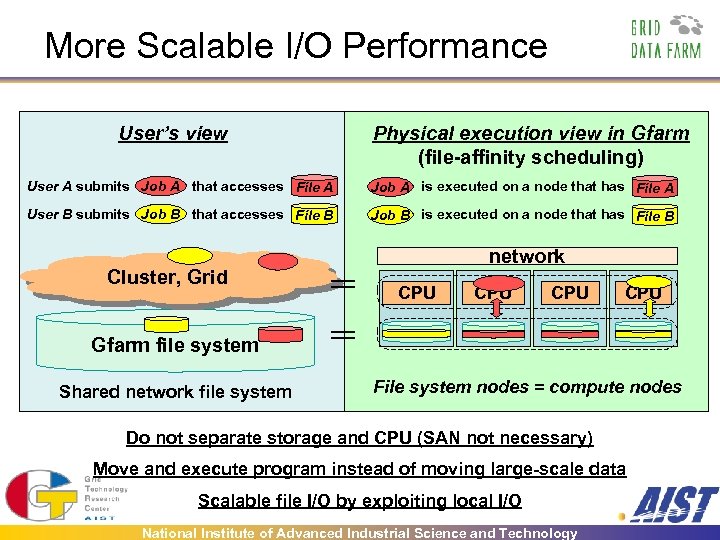 More Scalable I/O Performance User’s view Physical execution view in Gfarm (file-affinity scheduling) User A submits Job A that accesses File A Job A is executed on a node that has File A User B submits Job B that accesses File B Job B is executed on a node that has File B Cluster, Grid network CPU CPU Gfarm file system Shared network file system File system nodes = compute nodes Do not separate storage and CPU (SAN not necessary) Move and execute program instead of moving large-scale data Scalable file I/O by exploiting local I/O National Institute of Advanced Industrial Science and Technology
More Scalable I/O Performance User’s view Physical execution view in Gfarm (file-affinity scheduling) User A submits Job A that accesses File A Job A is executed on a node that has File A User B submits Job B that accesses File B Job B is executed on a node that has File B Cluster, Grid network CPU CPU Gfarm file system Shared network file system File system nodes = compute nodes Do not separate storage and CPU (SAN not necessary) Move and execute program instead of moving large-scale data Scalable file I/O by exploiting local I/O National Institute of Advanced Industrial Science and Technology
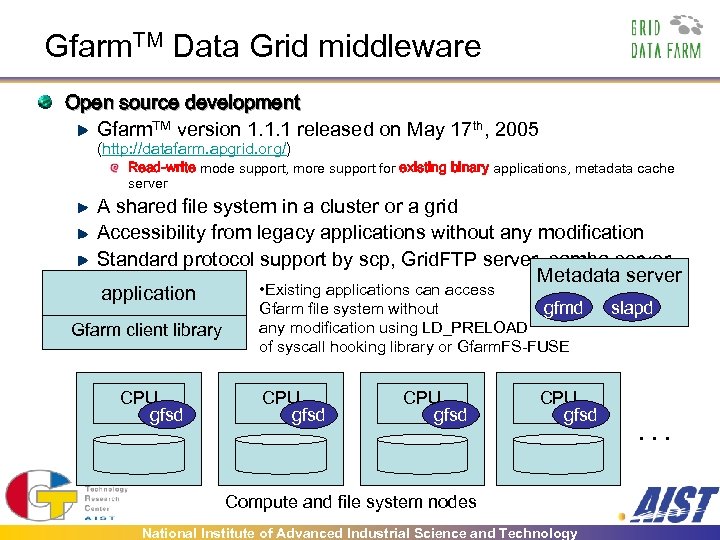 Gfarm. TM Data Grid middleware Open source development Gfarm. TM version 1. 1. 1 released on May 17 th, 2005 (http: //datafarm. apgrid. org/) Read-write mode support, more support for existing binary applications, metadata cache server A shared file system in a cluster or a grid Accessibility from legacy applications without any modification Standard protocol support by scp, Grid. FTP server, samba server, . Metadata server. . • Existing applications can access application Gfarm client library CPU gfsd gfmd Gfarm file system without any modification using LD_PRELOAD of syscall hooking library or Gfarm. FS-FUSE CPU gfsd Compute and file system nodes National Institute of Advanced Industrial Science and Technology slapd . . .
Gfarm. TM Data Grid middleware Open source development Gfarm. TM version 1. 1. 1 released on May 17 th, 2005 (http: //datafarm. apgrid. org/) Read-write mode support, more support for existing binary applications, metadata cache server A shared file system in a cluster or a grid Accessibility from legacy applications without any modification Standard protocol support by scp, Grid. FTP server, samba server, . Metadata server. . • Existing applications can access application Gfarm client library CPU gfsd gfmd Gfarm file system without any modification using LD_PRELOAD of syscall hooking library or Gfarm. FS-FUSE CPU gfsd Compute and file system nodes National Institute of Advanced Industrial Science and Technology slapd . . .
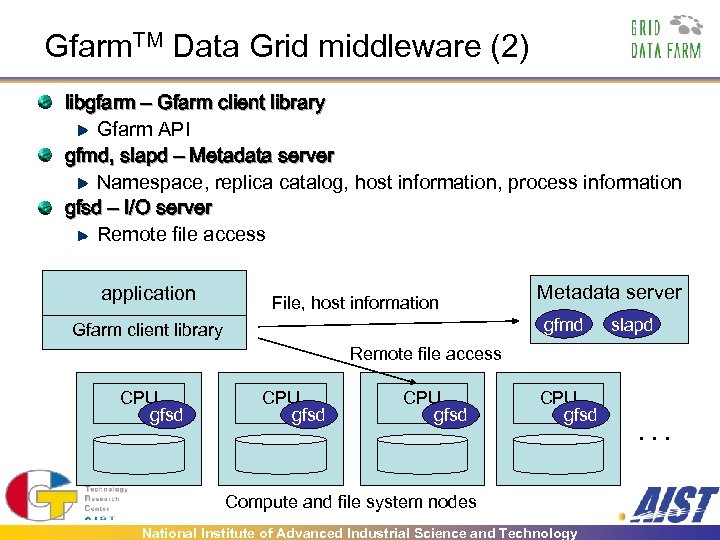 Gfarm. TM Data Grid middleware (2) libgfarm – Gfarm client library Gfarm API gfmd, slapd – Metadata server Namespace, replica catalog, host information, process information gfsd – I/O server Remote file access application File, host information Metadata server gfmd Gfarm client library slapd Remote file access CPU gfsd Compute and file system nodes National Institute of Advanced Industrial Science and Technology . . .
Gfarm. TM Data Grid middleware (2) libgfarm – Gfarm client library Gfarm API gfmd, slapd – Metadata server Namespace, replica catalog, host information, process information gfsd – I/O server Remote file access application File, host information Metadata server gfmd Gfarm client library slapd Remote file access CPU gfsd Compute and file system nodes National Institute of Advanced Industrial Science and Technology . . .
 Access from legacy applications libgfs_hook. so – system call hooking library It emulates to mount Gfarm file system at /gfarm hooking open(2), read(2), write(2), … When it accesses under /gfarm, call appropriate Gfarm API Otherwise, call ordinal system call Re-link not necessary by specifying LD_PRELOAD Linux, Free. BSD, Net. BSD, … Higher portability than developing kernel module Mounting Gfarm file system Gfarm. FS-FUSE enables to mount Gfarm file system using FUSE mechanism in Linux released on Jul 12, 2005 Need to develop a kernel module for other OSs Need volunteers National Institute of Advanced Industrial Science and Technology
Access from legacy applications libgfs_hook. so – system call hooking library It emulates to mount Gfarm file system at /gfarm hooking open(2), read(2), write(2), … When it accesses under /gfarm, call appropriate Gfarm API Otherwise, call ordinal system call Re-link not necessary by specifying LD_PRELOAD Linux, Free. BSD, Net. BSD, … Higher portability than developing kernel module Mounting Gfarm file system Gfarm. FS-FUSE enables to mount Gfarm file system using FUSE mechanism in Linux released on Jul 12, 2005 Need to develop a kernel module for other OSs Need volunteers National Institute of Advanced Industrial Science and Technology
 Gfarm – Application and performance result http: //datafarm. apgrid. org/ National Institute of Advanced Industrial Science and Technology
Gfarm – Application and performance result http: //datafarm. apgrid. org/ National Institute of Advanced Industrial Science and Technology
 Scientific Application (1) ATLAS Data Production Distribution kit (binary) Atlfast – fast simulation Input data stored in Gfarm file system not NFS G 4 sim – full simulation (Collaboration with ICEPP, KEK) Belle Monte-Carlo/Data Production Online data processing Distributed data processing Realtime histgram display 10 M events generated in a few days using a 50 -node PC cluster (Collaboration with KEK, U-Tokyo) National Institute of Advanced Industrial Science and Technology
Scientific Application (1) ATLAS Data Production Distribution kit (binary) Atlfast – fast simulation Input data stored in Gfarm file system not NFS G 4 sim – full simulation (Collaboration with ICEPP, KEK) Belle Monte-Carlo/Data Production Online data processing Distributed data processing Realtime histgram display 10 M events generated in a few days using a 50 -node PC cluster (Collaboration with KEK, U-Tokyo) National Institute of Advanced Industrial Science and Technology
 Scientific Application (2) Astronomical Object Survey Data analysis on the whole archive 652 GBytes data observed by SUBARU telescope Large configuration data from Lattice QCD Three sets of hundreds of gluon field configurations on a 24^3*48 4 -D space-time lattice (3 sets x 364. 5 MB x 800 = 854. 3 GB) Generated by the CP-PACS parallel computer at Center for Computational Physics, Univ. of Tsukuba (300 Gflops x years of CPU time) National Institute of Advanced Industrial Science and Technology
Scientific Application (2) Astronomical Object Survey Data analysis on the whole archive 652 GBytes data observed by SUBARU telescope Large configuration data from Lattice QCD Three sets of hundreds of gluon field configurations on a 24^3*48 4 -D space-time lattice (3 sets x 364. 5 MB x 800 = 854. 3 GB) Generated by the CP-PACS parallel computer at Center for Computational Physics, Univ. of Tsukuba (300 Gflops x years of CPU time) National Institute of Advanced Industrial Science and Technology
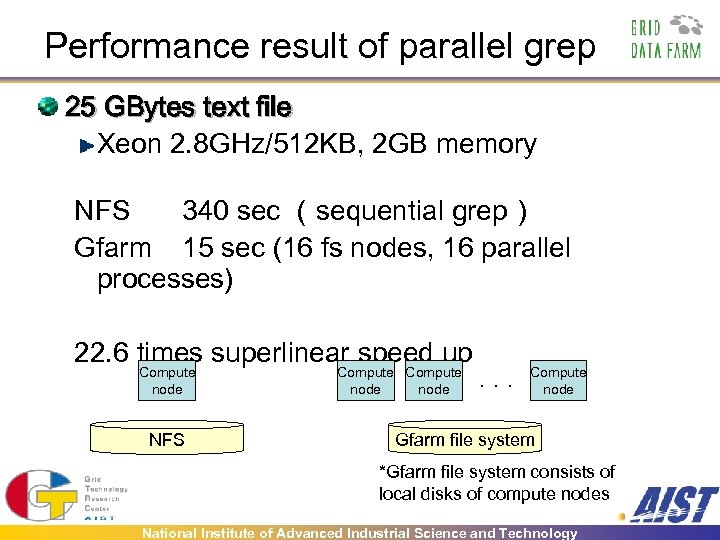 Performance result of parallel grep 25 GBytes text file Xeon 2. 8 GHz/512 KB, 2 GB memory NFS 340 sec (sequential grep) Gfarm 15 sec (16 fs nodes, 16 parallel processes) 22. 6 times superlinear speed up Compute node NFS Compute node . . . Compute node Gfarm file system *Gfarm file system consists of local disks of compute nodes National Institute of Advanced Industrial Science and Technology
Performance result of parallel grep 25 GBytes text file Xeon 2. 8 GHz/512 KB, 2 GB memory NFS 340 sec (sequential grep) Gfarm 15 sec (16 fs nodes, 16 parallel processes) 22. 6 times superlinear speed up Compute node NFS Compute node . . . Compute node Gfarm file system *Gfarm file system consists of local disks of compute nodes National Institute of Advanced Industrial Science and Technology
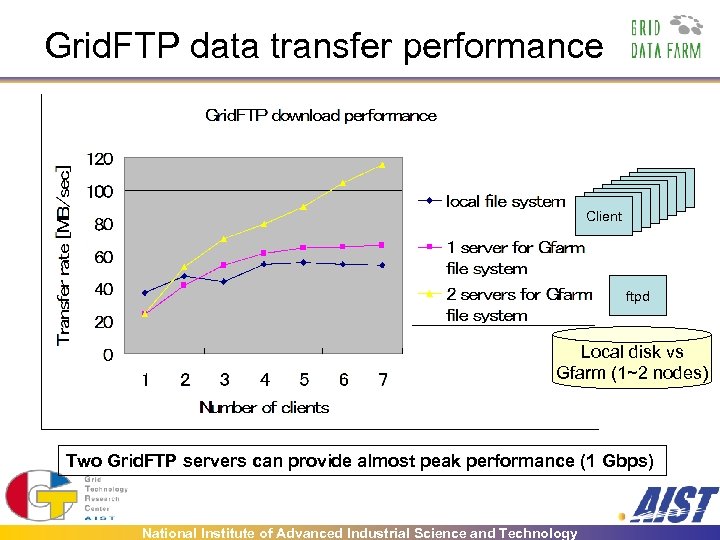 Grid. FTP data transfer performance Client Client ftpd Local disk vs Gfarm (1~2 nodes) Two Grid. FTP servers can provide almost peak performance (1 Gbps) National Institute of Advanced Industrial Science and Technology
Grid. FTP data transfer performance Client Client ftpd Local disk vs Gfarm (1~2 nodes) Two Grid. FTP servers can provide almost peak performance (1 Gbps) National Institute of Advanced Industrial Science and Technology
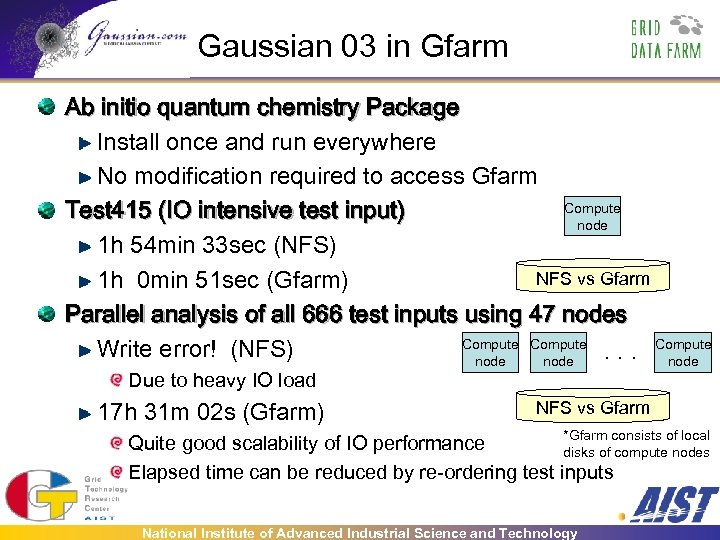 Gaussian 03 in Gfarm Ab initio quantum chemistry Package Install once and run everywhere No modification required to access Gfarm Compute Test 415 (IO intensive test input) node 1 h 54 min 33 sec (NFS) NFS vs Gfarm 1 h 0 min 51 sec (Gfarm) Parallel analysis of all 666 test inputs using 47 nodes Compute Write error! (NFS). . . node Due to heavy IO load 17 h 31 m 02 s (Gfarm) NFS vs Gfarm *Gfarm consists of local Quite good scalability of IO performance disks of compute nodes Elapsed time can be reduced by re-ordering test inputs National Institute of Advanced Industrial Science and Technology
Gaussian 03 in Gfarm Ab initio quantum chemistry Package Install once and run everywhere No modification required to access Gfarm Compute Test 415 (IO intensive test input) node 1 h 54 min 33 sec (NFS) NFS vs Gfarm 1 h 0 min 51 sec (Gfarm) Parallel analysis of all 666 test inputs using 47 nodes Compute Write error! (NFS). . . node Due to heavy IO load 17 h 31 m 02 s (Gfarm) NFS vs Gfarm *Gfarm consists of local Quite good scalability of IO performance disks of compute nodes Elapsed time can be reduced by re-ordering test inputs National Institute of Advanced Industrial Science and Technology
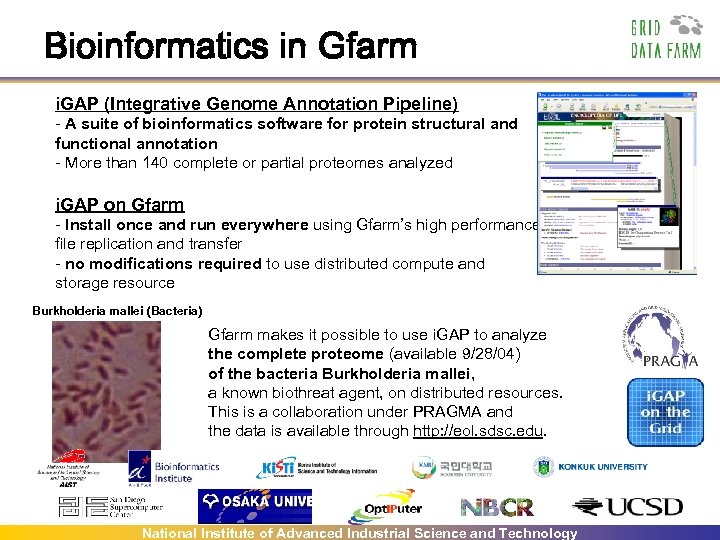 Bioinformatics in Gfarm i. GAP (Integrative Genome Annotation Pipeline) - A suite of bioinformatics software for protein structural and functional annotation - More than 140 complete or partial proteomes analyzed i. GAP on Gfarm - Install once and run everywhere using Gfarm’s high performance file replication and transfer - no modifications required to use distributed compute and storage resource Burkholderia mallei (Bacteria) Gfarm makes it possible to use i. GAP to analyze the complete proteome (available 9/28/04) of the bacteria Burkholderia mallei, a known biothreat agent, on distributed resources. This is a collaboration under PRAGMA and the data is available through http: //eol. sdsc. edu. Participating sites: SDSC/UCSD (US), BII (Singapore), Osaka Univ, AIST (Japan), Konkuk Univ, Kookmin Univ, KISTI (Korea) National Institute of Advanced Industrial Science and Technology
Bioinformatics in Gfarm i. GAP (Integrative Genome Annotation Pipeline) - A suite of bioinformatics software for protein structural and functional annotation - More than 140 complete or partial proteomes analyzed i. GAP on Gfarm - Install once and run everywhere using Gfarm’s high performance file replication and transfer - no modifications required to use distributed compute and storage resource Burkholderia mallei (Bacteria) Gfarm makes it possible to use i. GAP to analyze the complete proteome (available 9/28/04) of the bacteria Burkholderia mallei, a known biothreat agent, on distributed resources. This is a collaboration under PRAGMA and the data is available through http: //eol. sdsc. edu. Participating sites: SDSC/UCSD (US), BII (Singapore), Osaka Univ, AIST (Japan), Konkuk Univ, Kookmin Univ, KISTI (Korea) National Institute of Advanced Industrial Science and Technology
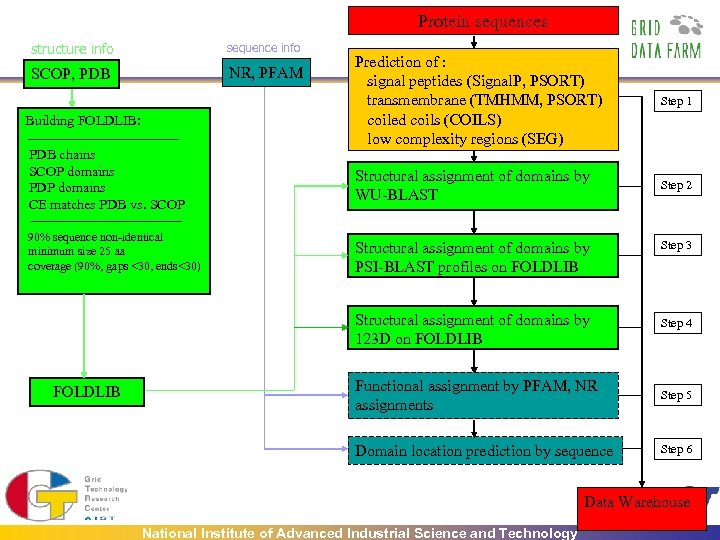 Protein sequences structure info sequence info SCOP, PDB NR, PFAM Building FOLDLIB: PDB chains SCOP domains PDP domains CE matches PDB vs. SCOP 90% sequence non-identical minimum size 25 aa coverage (90%, gaps <30, ends<30) Prediction of : signal peptides (Signal. P, PSORT) transmembrane (TMHMM, PSORT) coiled coils (COILS) low complexity regions (SEG) Step 1 Structural assignment of domains by WU-BLAST Step 2 Step 3 Structural assignment of domains by 123 D on FOLDLIB Structural assignment of domains by PSI-BLAST profiles on FOLDLIB Step 4 Functional assignment by PFAM, NR assignments Step 5 Domain location prediction by sequence Step 6 Data Warehouse National Institute of Advanced Industrial Science and Technology
Protein sequences structure info sequence info SCOP, PDB NR, PFAM Building FOLDLIB: PDB chains SCOP domains PDP domains CE matches PDB vs. SCOP 90% sequence non-identical minimum size 25 aa coverage (90%, gaps <30, ends<30) Prediction of : signal peptides (Signal. P, PSORT) transmembrane (TMHMM, PSORT) coiled coils (COILS) low complexity regions (SEG) Step 1 Structural assignment of domains by WU-BLAST Step 2 Step 3 Structural assignment of domains by 123 D on FOLDLIB Structural assignment of domains by PSI-BLAST profiles on FOLDLIB Step 4 Functional assignment by PFAM, NR assignments Step 5 Domain location prediction by sequence Step 6 Data Warehouse National Institute of Advanced Industrial Science and Technology
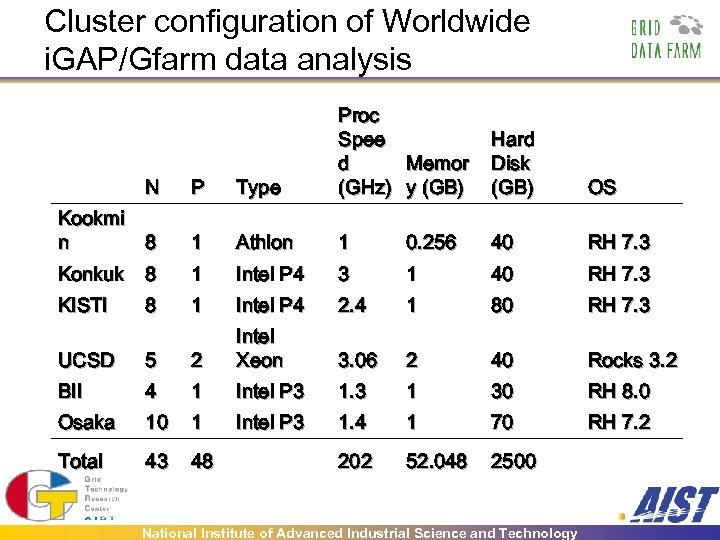 Cluster configuration of Worldwide i. GAP/Gfarm data analysis N P Type Proc Spee Memor d (GHz) y (GB) Kookmi n 8 1 Athlon 1 0. 256 40 RH 7. 3 Konkuk 8 1 Intel P 4 3 1 40 RH 7. 3 KISTI 8 1 Intel P 4 2. 4 1 80 RH 7. 3 3. 06 2 40 Rocks 3. 2 Hard Disk (GB) OS UCSD 5 2 Intel Xeon BII 4 1 Intel P 3 1 30 RH 8. 0 Osaka 10 1 Intel P 3 1. 4 1 70 RH 7. 2 Total 43 48 202 52. 048 2500 National Institute of Advanced Industrial Science and Technology
Cluster configuration of Worldwide i. GAP/Gfarm data analysis N P Type Proc Spee Memor d (GHz) y (GB) Kookmi n 8 1 Athlon 1 0. 256 40 RH 7. 3 Konkuk 8 1 Intel P 4 3 1 40 RH 7. 3 KISTI 8 1 Intel P 4 2. 4 1 80 RH 7. 3 3. 06 2 40 Rocks 3. 2 Hard Disk (GB) OS UCSD 5 2 Intel Xeon BII 4 1 Intel P 3 1 30 RH 8. 0 Osaka 10 1 Intel P 3 1. 4 1 70 RH 7. 2 Total 43 48 202 52. 048 2500 National Institute of Advanced Industrial Science and Technology
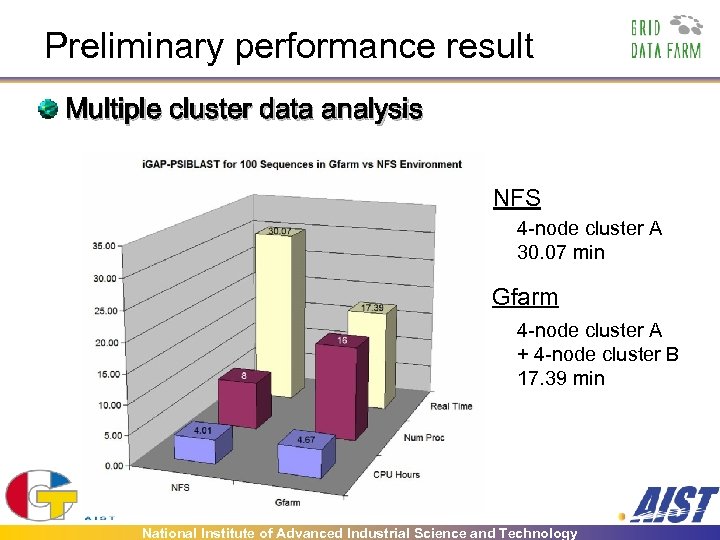 Preliminary performance result Multiple cluster data analysis NFS 4 -node cluster A 30. 07 min Gfarm 4 -node cluster A + 4 -node cluster B 17. 39 min National Institute of Advanced Industrial Science and Technology
Preliminary performance result Multiple cluster data analysis NFS 4 -node cluster A 30. 07 min Gfarm 4 -node cluster A + 4 -node cluster B 17. 39 min National Institute of Advanced Industrial Science and Technology
 National Institute of Advanced Industrial Science and Technology
National Institute of Advanced Industrial Science and Technology
 Development Status and Future Plan Gfarm – Grid file system Global virtual file system A dependable network shared file system in a cluster or a grid High performance data computing support Associates Computational Grid with Data Grid Gfarm Grid software Version 1. 1. 1 released on May 17, 2005 (http: //datafarm. apgrid. org/) Version 1. 2 available real soon now Existing programs can access Gfarm file system using syscall hooking library or Gfarm. FS-FUSE Distribute analysis shows scalable I/O performance i. GAP/Gfarm – bioinformatics package Gaussian 03 – Ab initio quantum chemistry package Standardization effort with GGF Grid File System WG (GFS-WG) https: //datafarm. apgrid. org/ National Institute of Advanced Industrial Science and Technology
Development Status and Future Plan Gfarm – Grid file system Global virtual file system A dependable network shared file system in a cluster or a grid High performance data computing support Associates Computational Grid with Data Grid Gfarm Grid software Version 1. 1. 1 released on May 17, 2005 (http: //datafarm. apgrid. org/) Version 1. 2 available real soon now Existing programs can access Gfarm file system using syscall hooking library or Gfarm. FS-FUSE Distribute analysis shows scalable I/O performance i. GAP/Gfarm – bioinformatics package Gaussian 03 – Ab initio quantum chemistry package Standardization effort with GGF Grid File System WG (GFS-WG) https: //datafarm. apgrid. org/ National Institute of Advanced Industrial Science and Technology


