6bade5020c3004e9c4a7786dbed3607e.ppt
- Количество слайдов: 28

Announcements 1. First lab report deadline extended by one week: X-linked cross lab report due 11/ 5, 6. 2. Bookstore is closed Sundays. Buy your bluebook early. 3. Get lab overview for PCR lab - quiz in lab next week. 4. If your transformation did not work, look at plates in lab refrigerator, both group “A” and “B”; make counts from these plates. Lab assignment due 10/29, 30 (extra week). 5. Group B presentations in lab 10/29, 30 - get sources approved. 6. Problems to look over Ch. 11: 4, 5, 8, 15. 7. Ch. 12: 3, Insights, 1.

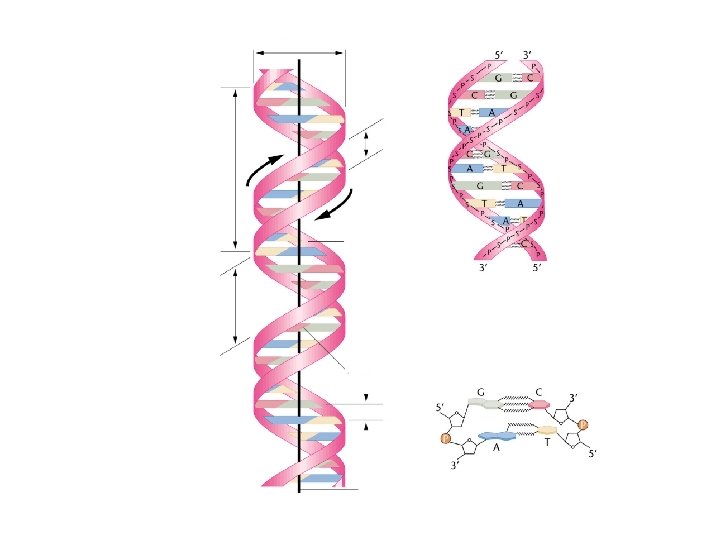
Review of Last Lecture 1. Structure of DNA 2. Analytical analysis of nucleic acids 3. DNA Replication is Semiconservative
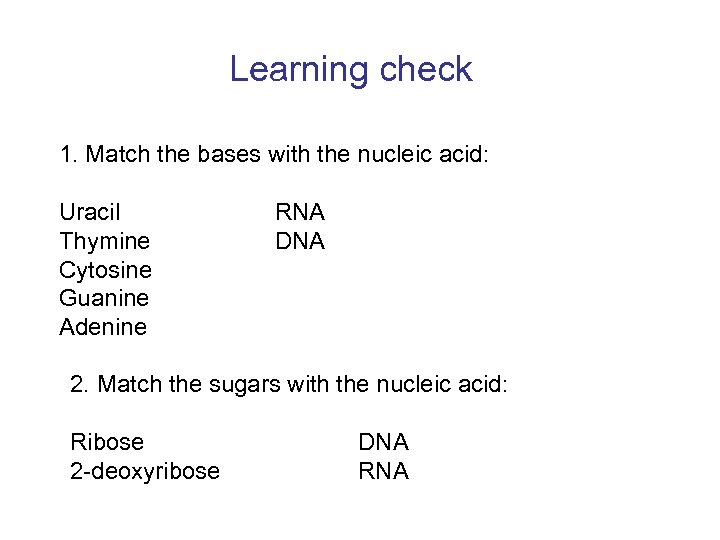
Learning check 1. Match the bases with the nucleic acid: Uracil Thymine Cytosine Guanine Adenine RNA DNA 2. Match the sugars with the nucleic acid: Ribose 2 -deoxyribose DNA RNA

Learning check, revisited The sequence of the dwarf gene in garden peas is as follows: 5’ - A G C T A C G T -3’ 3’ - T C G A T G C A -5’ Write the RNA sequence transcribed from the top strand of DNA, 5’- 3’.

Outline of Lecture 21 1. I. Discussion: Watson and Crick paper 2. II. Bacterial DNA replication III. Many complex issues to resolve during DNA replication - DNA helix must be unwound, RNA primer needed - Synthesis is both continuous and discontinuous - Proofreading is critical IV. Eukaryotic DNA replication is similar, more complex V. The “end” problem and telomerase VI. DNA Recombination
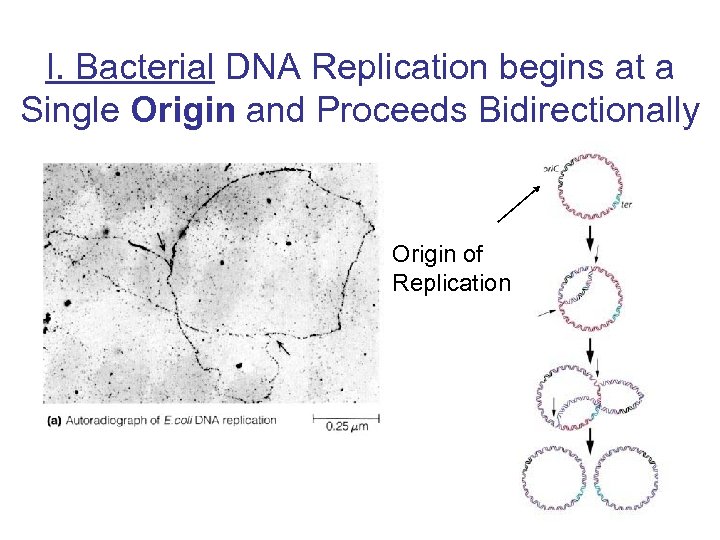
I. Bacterial DNA Replication begins at a Single Origin and Proceeds Bidirectionally Origin of Replication
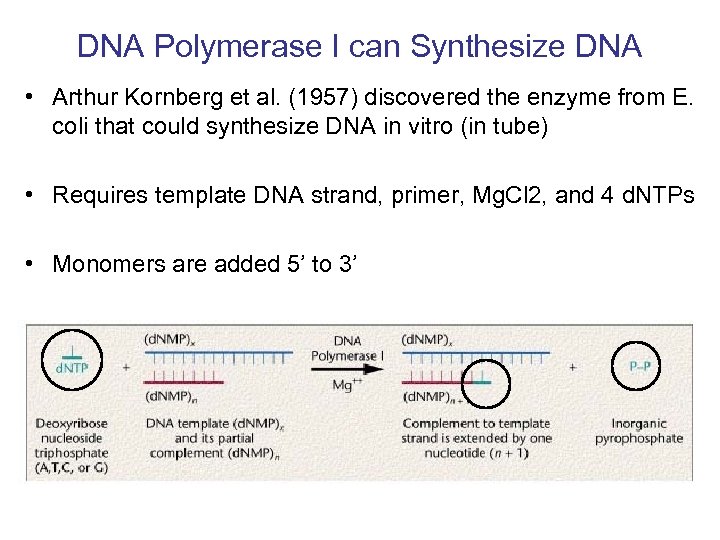
DNA Polymerase I can Synthesize DNA • Arthur Kornberg et al. (1957) discovered the enzyme from E. coli that could synthesize DNA in vitro (in tube) • Requires template DNA strand, primer, Mg. Cl 2, and 4 d. NTPs • Monomers are added 5’ to 3’
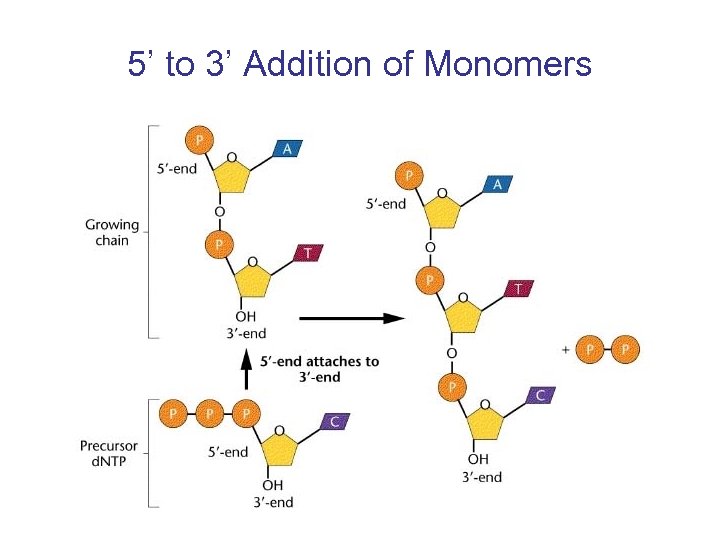
5’ to 3’ Addition of Monomers
Skepticism regarding pol I as the enzyme that replicates DNA in vivo 1. Pol I slower in vitro than rate of replication observed in vivo 2. Pol I could degrade DNA 3. Pol I replicated ss DNA better than ds DNA 4. A mutant E. coli strain was isolated that had defective pol I activity, but still duplicated DNA and reproduced. 5. - mutant was sensitive to UV light and radiation A second enzyme must exist to replicate DNA Pol I may perform a different function in vivo
Bacterial DNA polymerases I, II and III • pol I – most abundant (400/cell) and very stable – RNA primer removal, polymerase, exonuclease • pol II – unknown abundance – Polymerase, exonuclese, DNA repair? • pol III – low abundance (15/cell) – Polymerase, exonuclease, DNA replication Pol III is very complex, many subunits

II. Problems of DNA Synthesis • • Unwinding double-stranded helix Tension must be relieved Priming Antiparallel strands RNA primer removal Backbone joining Proofreading

Steps of DNA Synthesis -how “problems” are solved (1) Denaturation and Unwinding (2) Priming and Initiation (3) Continuous and Discontinuous Synthesis - Including Proofreading and Error Correction (4) Removal of Primer (5) Ligation of nicks in backbone
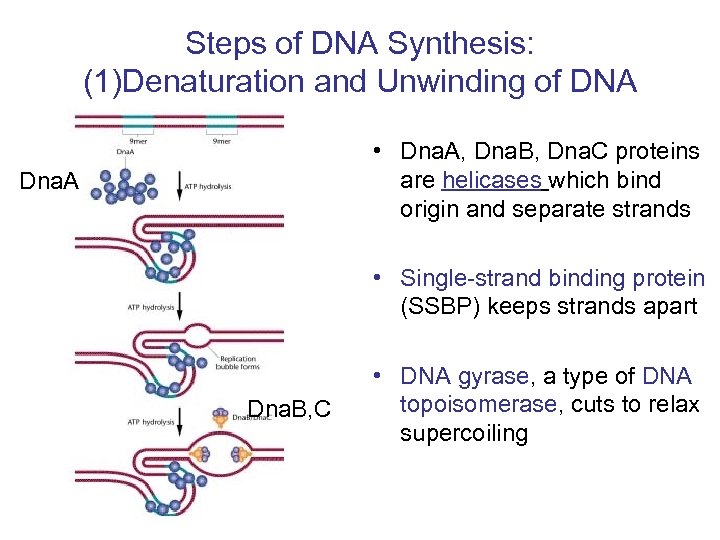
Steps of DNA Synthesis: (1)Denaturation and Unwinding of DNA • Dna. A, Dna. B, Dna. C proteins are helicases which bind origin and separate strands Dna. A • Single-strand binding protein (SSBP) keeps strands apart Dna. B, C • DNA gyrase, a type of DNA topoisomerase, cuts to relax supercoiling
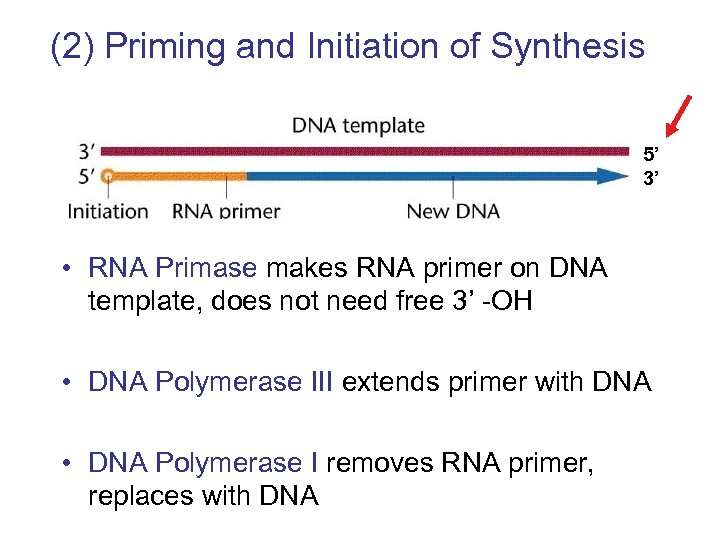
(2) Priming and Initiation of Synthesis 5’ 3’ • RNA Primase makes RNA primer on DNA template, does not need free 3’ -OH • DNA Polymerase III extends primer with DNA • DNA Polymerase I removes RNA primer, replaces with DNA
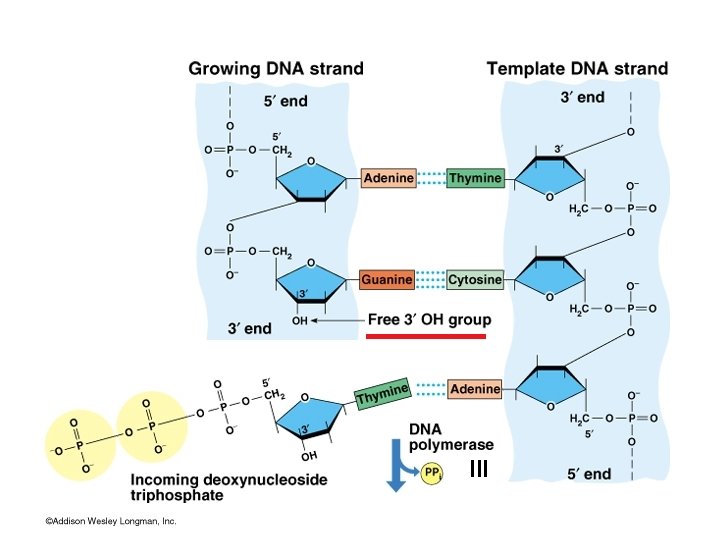
Directionality of DNA synthesis III
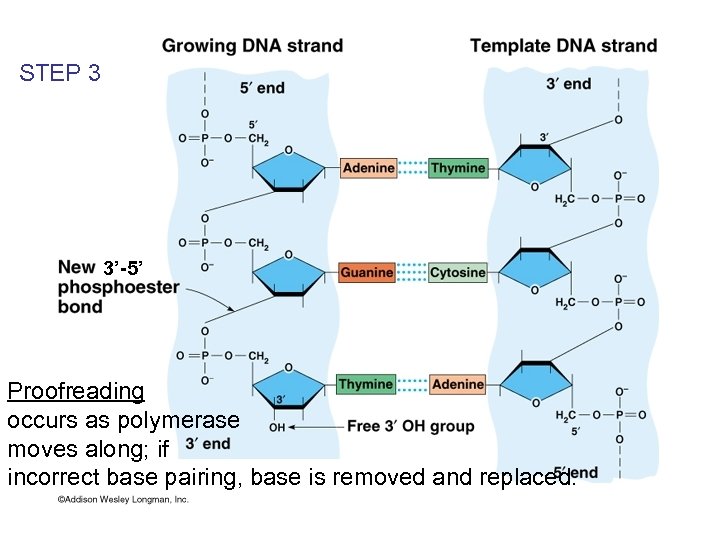
STEP 3 3’-5’ Proofreading occurs as polymerase moves along; if incorrect base pairing, base is removed and replaced.
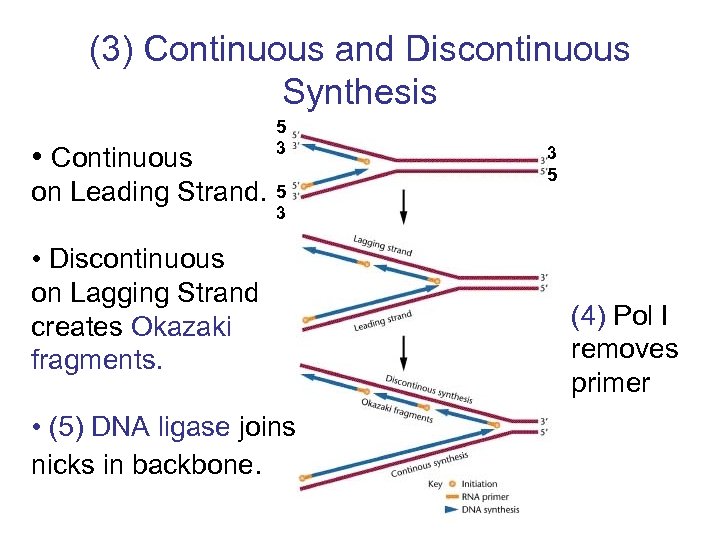
(3) Continuous and Discontinuous Synthesis • Continuous 5 3 on Leading Strand. 5 3 • Discontinuous on Lagging Strand creates Okazaki fragments. • (5) DNA ligase joins nicks in backbone. (4) Pol I removes primer
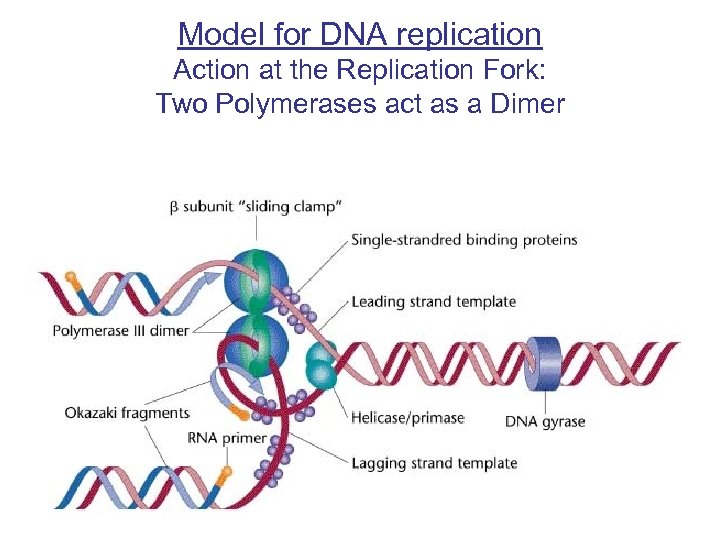
Model for DNA replication Action at the Replication Fork: Two Polymerases act as a Dimer
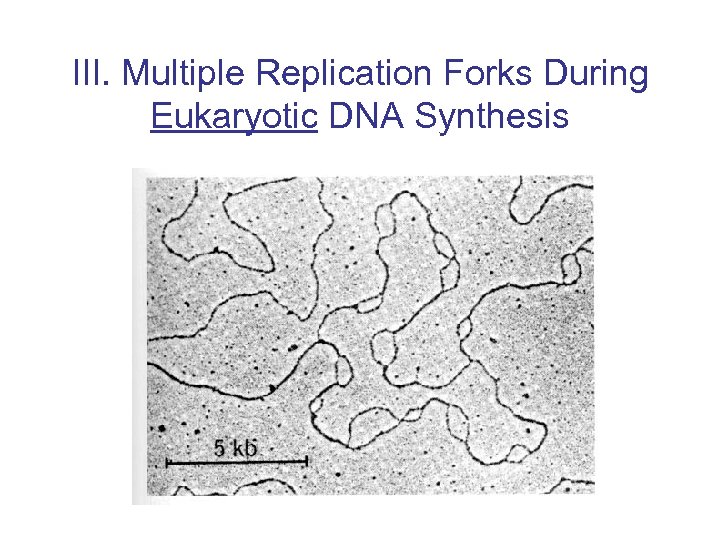
III. Multiple Replication Forks During Eukaryotic DNA Synthesis
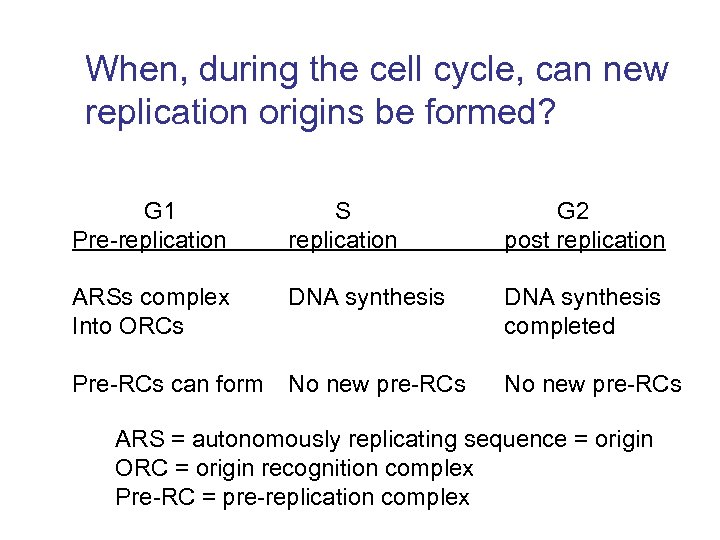
When, during the cell cycle, can new replication origins be formed? G 1 Pre-replication S replication G 2 post replication ARSs complex Into ORCs DNA synthesis completed Pre-RCs can form No new pre-RCs ARS = autonomously replicating sequence = origin ORC = origin recognition complex Pre-RC = pre-replication complex
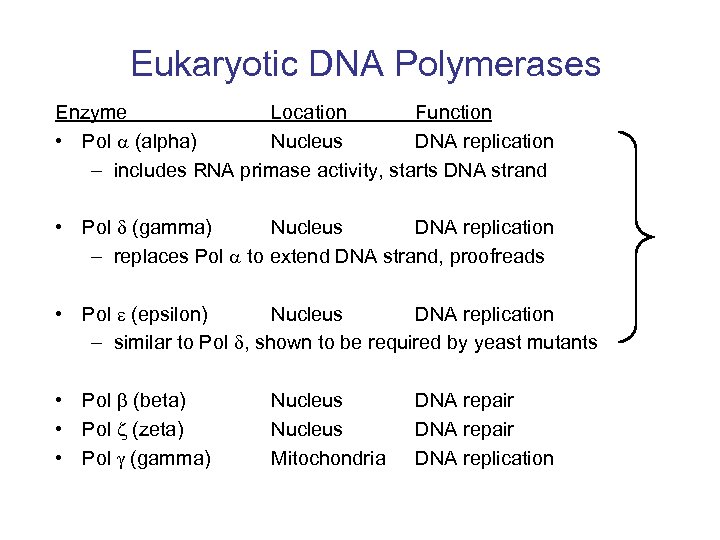
Eukaryotic DNA Polymerases Enzyme Location Function • Pol (alpha) Nucleus DNA replication – includes RNA primase activity, starts DNA strand • Pol (gamma) Nucleus DNA replication – replaces Pol to extend DNA strand, proofreads • Pol (epsilon) Nucleus DNA replication – similar to Pol , shown to be required by yeast mutants • Pol (beta) • Pol (zeta) • Pol (gamma) Nucleus Mitochondria DNA repair DNA replication
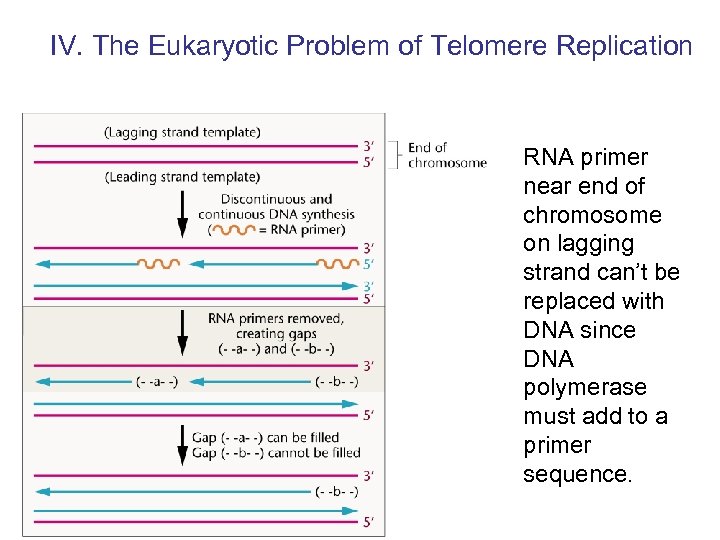
IV. The Eukaryotic Problem of Telomere Replication RNA primer near end of chromosome on lagging strand can’t be replaced with DNA since DNA polymerase must add to a primer sequence.
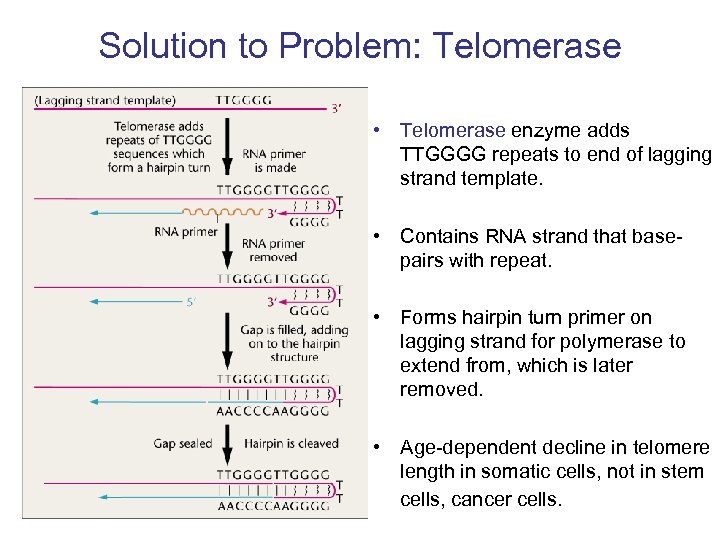
Solution to Problem: Telomerase • Telomerase enzyme adds TTGGGG repeats to end of lagging strand template. • Contains RNA strand that basepairs with repeat. • Forms hairpin turn primer on lagging strand for polymerase to extend from, which is later removed. • Age-dependent decline in telomere length in somatic cells, not in stem cells, cancer cells.
V. Recombination at the Molecular Level • Breakage and joining also directed by enzymes. • Homologous recombination occurs during synapsis in meiosis I, general recombination in bacteria, and viral genetic exchange. • Molecular mechanism proposed by Holliday and Whitehouse (1964). • Depends on complementary base pairing.
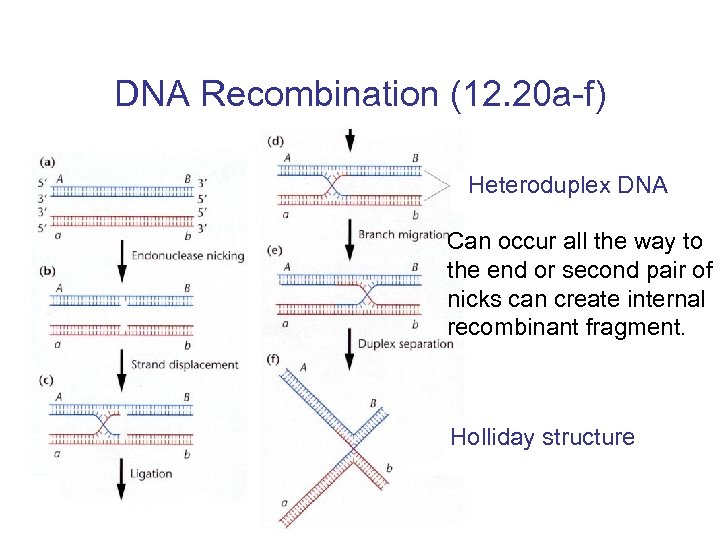
DNA Recombination (12. 20 a-f) Heteroduplex DNA Can occur all the way to the end or second pair of nicks can create internal recombinant fragment. Holliday structure
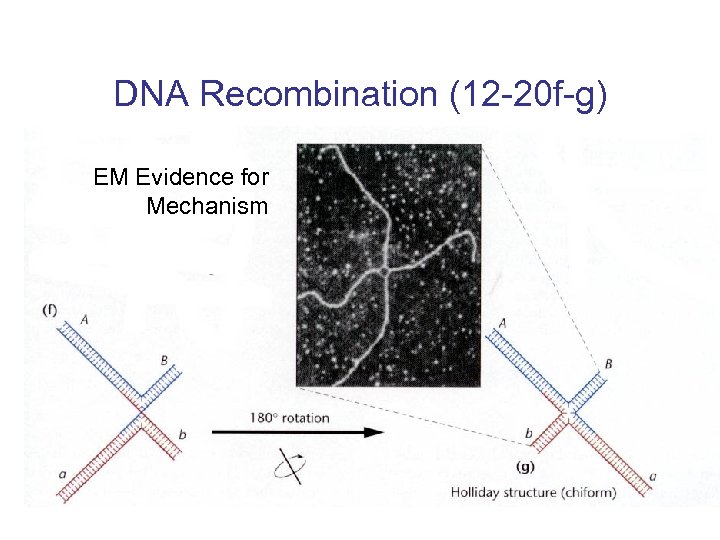
DNA Recombination (12 -20 f-g) EM Evidence for Mechanism
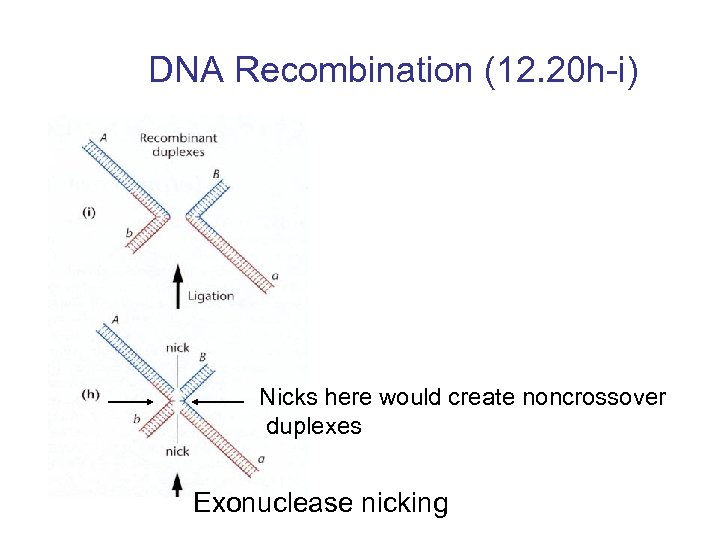
DNA Recombination (12. 20 h-i) Nicks here would create noncrossover duplexes Exonuclease nicking
6bade5020c3004e9c4a7786dbed3607e.ppt