26d61d2b0a47f1f0d473e7638e6cdc9b.ppt
- Количество слайдов: 44
 Analysis of Personal Genomes: Multi-scale Element Annotation & Variant Prioritization Lectures. Gerstein. Lab. org Mark Gerstein, Yale 1 - Slides freely downloadable from Lectures. Gerstein. Lab. org & “tweetable” (via @markgerstein). See last slide for more info.
Analysis of Personal Genomes: Multi-scale Element Annotation & Variant Prioritization Lectures. Gerstein. Lab. org Mark Gerstein, Yale 1 - Slides freely downloadable from Lectures. Gerstein. Lab. org & “tweetable” (via @markgerstein). See last slide for more info.
 Where is Waldo? 2 - Lectures. Gerstein. Lab. org (Finding the key mutations in ~3 M Germline variants & ~5 K Somatic Variants in a Tumor Sample)
Where is Waldo? 2 - Lectures. Gerstein. Lab. org (Finding the key mutations in ~3 M Germline variants & ~5 K Somatic Variants in a Tumor Sample)
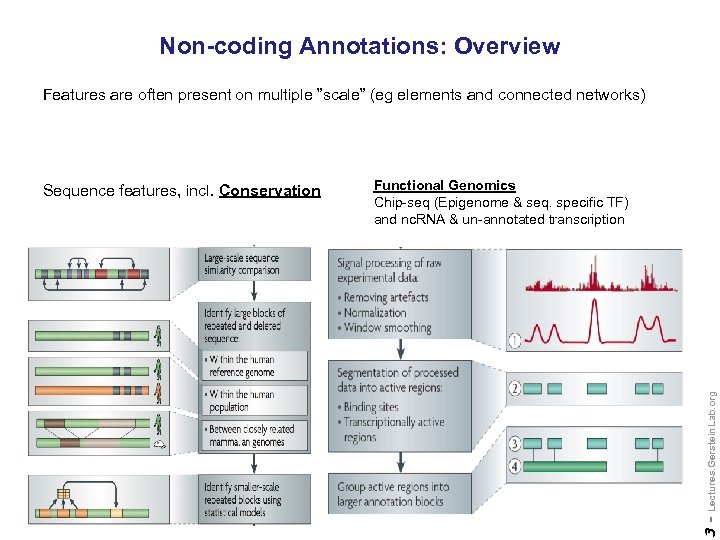 Non-coding Annotations: Overview Features are often present on multiple ”scale” (eg elements and connected networks) Lectures. Gerstein. Lab. org Functional Genomics Chip-seq (Epigenome & seq. specific TF) and nc. RNA & un-annotated transcription 3 - Sequence features, incl. Conservation
Non-coding Annotations: Overview Features are often present on multiple ”scale” (eg elements and connected networks) Lectures. Gerstein. Lab. org Functional Genomics Chip-seq (Epigenome & seq. specific TF) and nc. RNA & un-annotated transcription 3 - Sequence features, incl. Conservation
 Multi-scale Element Annotation & Variant Prioritization • Characterizing Regulatory • Features of Sites Multi-resolution TADs - Specific TFs & HMs associated at Multiple Scales - Multi-scale "site" calling (with Music) - Using high resolution conservation information to find sensitive sites • Characterizing TADs at Multiple Scales - Using modularity for identification - Developing an appropriate null expectation with TAD boundaries at different scales - Assoc. strong enough to build a predictor - HOT regions at boundaries • Fun. Seq Software Tool for Variant Prioritization - Systematically weighting all the features, for non-coding prioritization
Multi-scale Element Annotation & Variant Prioritization • Characterizing Regulatory • Features of Sites Multi-resolution TADs - Specific TFs & HMs associated at Multiple Scales - Multi-scale "site" calling (with Music) - Using high resolution conservation information to find sensitive sites • Characterizing TADs at Multiple Scales - Using modularity for identification - Developing an appropriate null expectation with TAD boundaries at different scales - Assoc. strong enough to build a predictor - HOT regions at boundaries • Fun. Seq Software Tool for Variant Prioritization - Systematically weighting all the features, for non-coding prioritization
 Multi-scale Element Annotation & Variant Prioritization • Characterizing Regulatory • Features of Sites Multi-resolution TADs - Specific TFs & HMs associated at Multiple Scales - Multi-scale "site" calling (with Music) - Using high resolution conservation information to find sensitive sites • Characterizing TADs at Multiple Scales - Using modularity for identification - Developing an appropriate null expectation with TAD boundaries at different scales - Assoc. strong enough to build a predictor - HOT regions at boundaries • Fun. Seq Software Tool for Variant Prioritization - Systematically weighting all the features, for non-coding prioritization
Multi-scale Element Annotation & Variant Prioritization • Characterizing Regulatory • Features of Sites Multi-resolution TADs - Specific TFs & HMs associated at Multiple Scales - Multi-scale "site" calling (with Music) - Using high resolution conservation information to find sensitive sites • Characterizing TADs at Multiple Scales - Using modularity for identification - Developing an appropriate null expectation with TAD boundaries at different scales - Assoc. strong enough to build a predictor - HOT regions at boundaries • Fun. Seq Software Tool for Variant Prioritization - Systematically weighting all the features, for non-coding prioritization
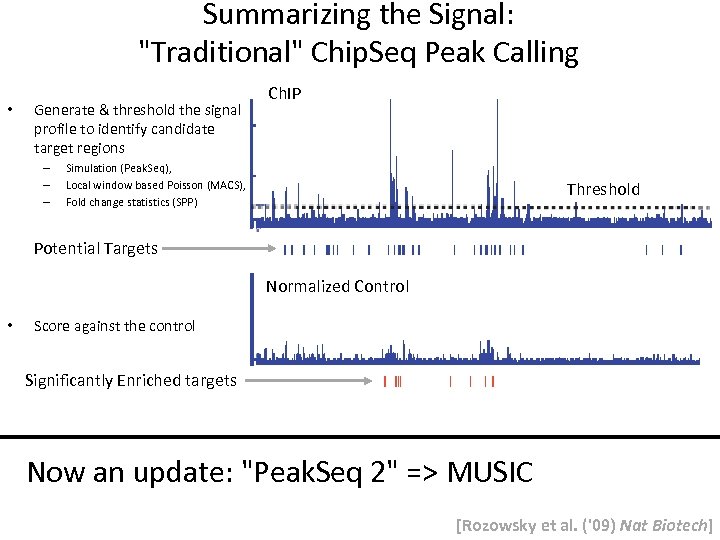 Summarizing the Signal: "Traditional" Chip. Seq Peak Calling • Generate & threshold the signal profile to identify candidate target regions – – – Ch. IP Simulation (Peak. Seq), Local window based Poisson (MACS), Fold change statistics (SPP) Threshold Potential Targets Normalized Control • Score against the control Significantly Enriched targets Now an update: "Peak. Seq 2" => MUSIC [Rozowsky et al. ('09) Nat Biotech]
Summarizing the Signal: "Traditional" Chip. Seq Peak Calling • Generate & threshold the signal profile to identify candidate target regions – – – Ch. IP Simulation (Peak. Seq), Local window based Poisson (MACS), Fold change statistics (SPP) Threshold Potential Targets Normalized Control • Score against the control Significantly Enriched targets Now an update: "Peak. Seq 2" => MUSIC [Rozowsky et al. ('09) Nat Biotech]
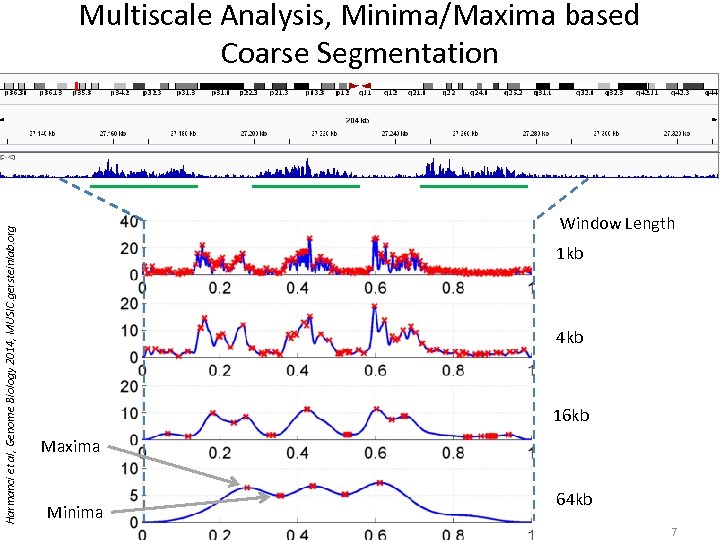 Multiscale Analysis, Minima/Maxima based Coarse Segmentation Harmanci et al, Genome Biology 2014, MUSIC. gersteinlab. org • Multiscale analysis is a natural way to analyze the Ch. IP-Seq data Window Length 1 kb 4 kb 16 kb Maxima Minima 64 kb 7
Multiscale Analysis, Minima/Maxima based Coarse Segmentation Harmanci et al, Genome Biology 2014, MUSIC. gersteinlab. org • Multiscale analysis is a natural way to analyze the Ch. IP-Seq data Window Length 1 kb 4 kb 16 kb Maxima Minima 64 kb 7
![Multiscale Decomposition 20 kb 0 b kb [Harmanci et al, Genome Biol. ('14)] 8 Multiscale Decomposition 20 kb 0 b kb [Harmanci et al, Genome Biol. ('14)] 8](https://present5.com/presentation/26d61d2b0a47f1f0d473e7638e6cdc9b/image-8.jpg) Multiscale Decomposition 20 kb 0 b kb [Harmanci et al, Genome Biol. ('14)] 8 - 0 10 Lectures. Gerstein. Lab. org Increasing Scale 10
Multiscale Decomposition 20 kb 0 b kb [Harmanci et al, Genome Biol. ('14)] 8 - 0 10 Lectures. Gerstein. Lab. org Increasing Scale 10
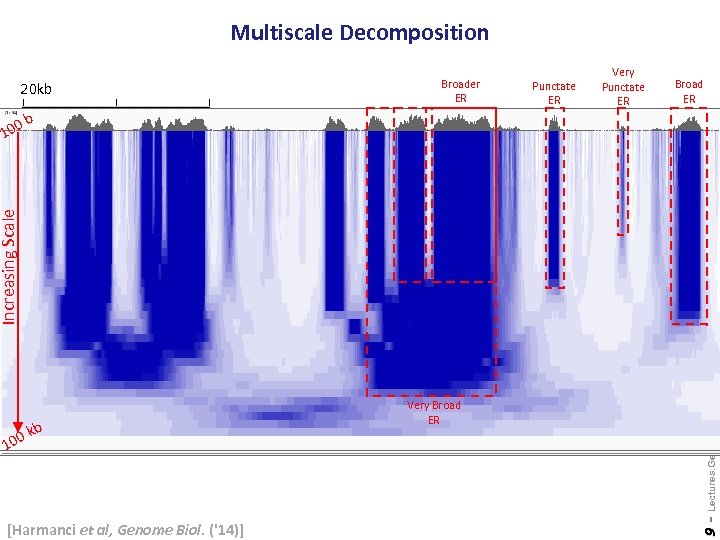 Multiscale Decomposition 20 kb Punctate ER Very Punctate ER Broad ER 0 b kb [Harmanci et al, Genome Biol. ('14)] 9 - 0 10 Very Broad ER Lectures. Gerstein. Lab. org Increasing Scale 10 Broader ER
Multiscale Decomposition 20 kb Punctate ER Very Punctate ER Broad ER 0 b kb [Harmanci et al, Genome Biol. ('14)] 9 - 0 10 Very Broad ER Lectures. Gerstein. Lab. org Increasing Scale 10 Broader ER
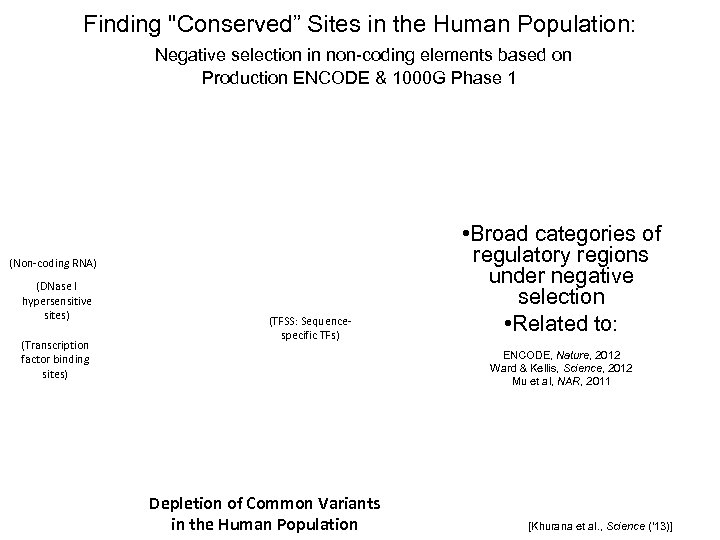 Finding "Conserved” Sites in the Human Population: Negative selection in non-coding elements based on Production ENCODE & 1000 G Phase 1 (Non-coding RNA) (DNase I hypersensitive sites) (Transcription factor binding sites) (TFSS: Sequencespecific TFs) • Broad categories of regulatory regions under negative selection • Related to: ENCODE, Nature, 2012 Ward & Kellis, Science, 2012 Mu et al, NAR, 2011 Depletion of Common Variants in the Human Population [Khurana et al. , Science (‘ 13)]
Finding "Conserved” Sites in the Human Population: Negative selection in non-coding elements based on Production ENCODE & 1000 G Phase 1 (Non-coding RNA) (DNase I hypersensitive sites) (Transcription factor binding sites) (TFSS: Sequencespecific TFs) • Broad categories of regulatory regions under negative selection • Related to: ENCODE, Nature, 2012 Ward & Kellis, Science, 2012 Mu et al, NAR, 2011 Depletion of Common Variants in the Human Population [Khurana et al. , Science (‘ 13)]
 Differential selective constraints among specific sub-categories Sub-categorization possible because of better statistics from 1000 G phase 1 v pilot [Khurana et al. , Science (‘ 13)]
Differential selective constraints among specific sub-categories Sub-categorization possible because of better statistics from 1000 G phase 1 v pilot [Khurana et al. , Science (‘ 13)]
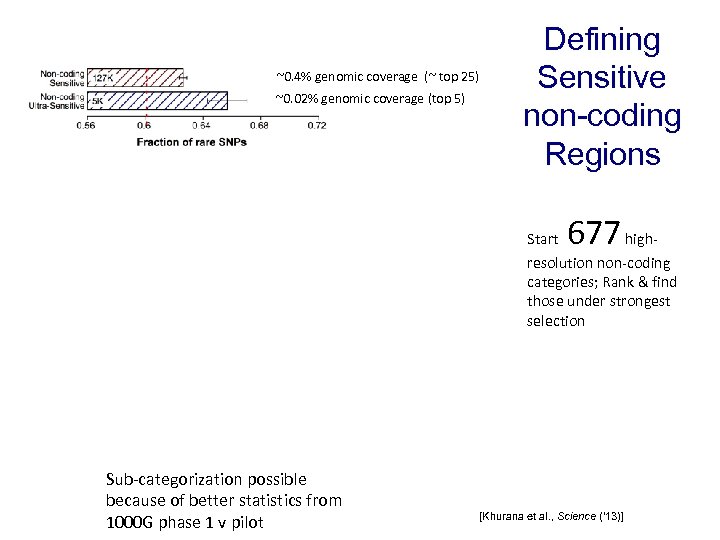 ~0. 4% genomic coverage (~ top 25) ~0. 02% genomic coverage (top 5) Defining Sensitive non-coding Regions Start 677 high- resolution non-coding categories; Rank & find those under strongest selection Sub-categorization possible because of better statistics from 1000 G phase 1 v pilot [Khurana et al. , Science (‘ 13)]
~0. 4% genomic coverage (~ top 25) ~0. 02% genomic coverage (top 5) Defining Sensitive non-coding Regions Start 677 high- resolution non-coding categories; Rank & find those under strongest selection Sub-categorization possible because of better statistics from 1000 G phase 1 v pilot [Khurana et al. , Science (‘ 13)]
 Multi-scale Element Annotation & Variant Prioritization • Characterizing Regulatory • Features of Sites Multi-resolution TADs - Specific TFs & HMs associated at Multiple Scales - Multi-scale "site" calling (with Music) - Using high resolution conservation information to find sensitive sites • Characterizing TADs at Multiple Scales - Using modularity for identification - Developing an appropriate null expectation with TAD boundaries at different scales - Assoc. strong enough to build a predictor - HOT regions at boundaries • Fun. Seq Software Tool for Variant Prioritization - Systematically weighting all the features, for non-coding prioritization
Multi-scale Element Annotation & Variant Prioritization • Characterizing Regulatory • Features of Sites Multi-resolution TADs - Specific TFs & HMs associated at Multiple Scales - Multi-scale "site" calling (with Music) - Using high resolution conservation information to find sensitive sites • Characterizing TADs at Multiple Scales - Using modularity for identification - Developing an appropriate null expectation with TAD boundaries at different scales - Assoc. strong enough to build a predictor - HOT regions at boundaries • Fun. Seq Software Tool for Variant Prioritization - Systematically weighting all the features, for non-coding prioritization
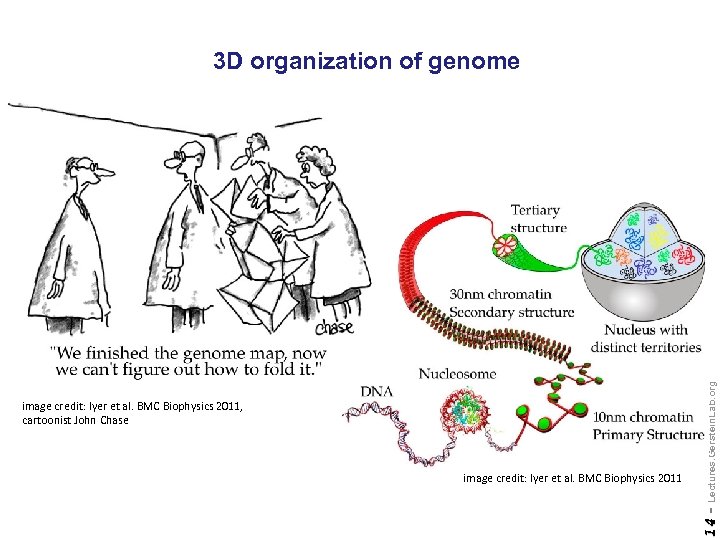 image credit: Iyer et al. BMC Biophysics 2011 14 - image credit: Iyer et al. BMC Biophysics 2011, cartoonist John Chase Lectures. Gerstein. Lab. org 3 D organization of genome
image credit: Iyer et al. BMC Biophysics 2011 14 - image credit: Iyer et al. BMC Biophysics 2011, cartoonist John Chase Lectures. Gerstein. Lab. org 3 D organization of genome
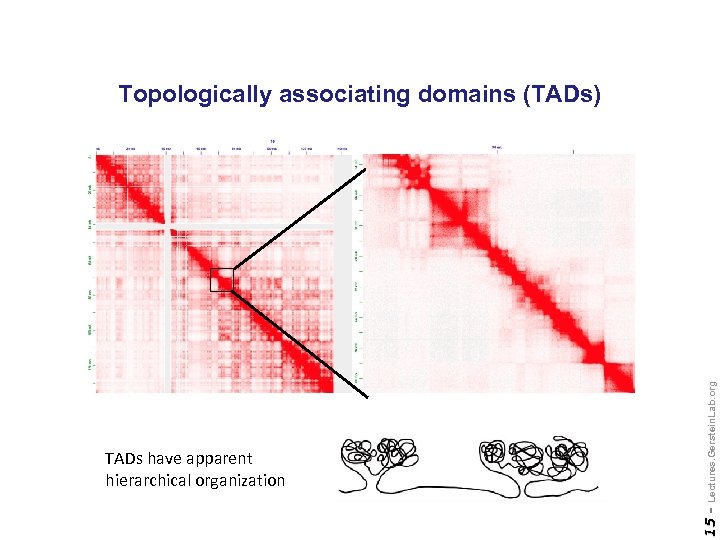 15 - TADs have apparent hierarchical organization Lectures. Gerstein. Lab. org Topologically associating domains (TADs)
15 - TADs have apparent hierarchical organization Lectures. Gerstein. Lab. org Topologically associating domains (TADs)
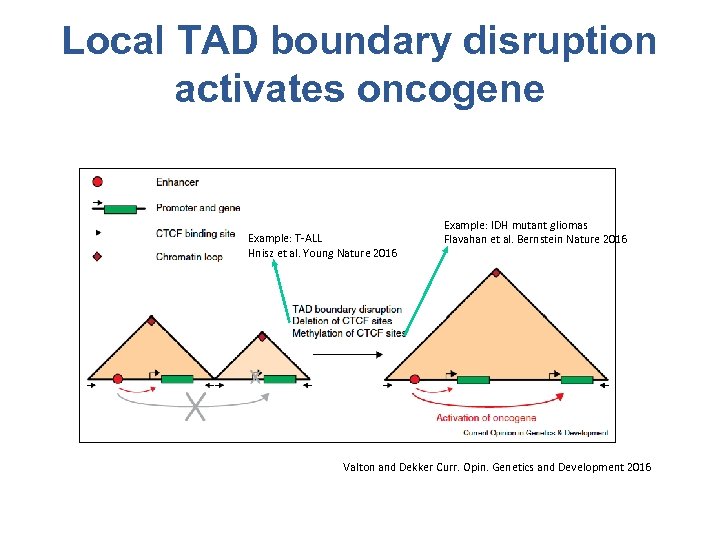 Local TAD boundary disruption activates oncogene Example: T-ALL Hnisz et al. Young Nature 2016 Example: IDH mutant gliomas Flavahan et al. Bernstein Nature 2016 Valton and Dekker Curr. Opin. Genetics and Development 2016
Local TAD boundary disruption activates oncogene Example: T-ALL Hnisz et al. Young Nature 2016 Example: IDH mutant gliomas Flavahan et al. Bernstein Nature 2016 Valton and Dekker Curr. Opin. Genetics and Development 2016
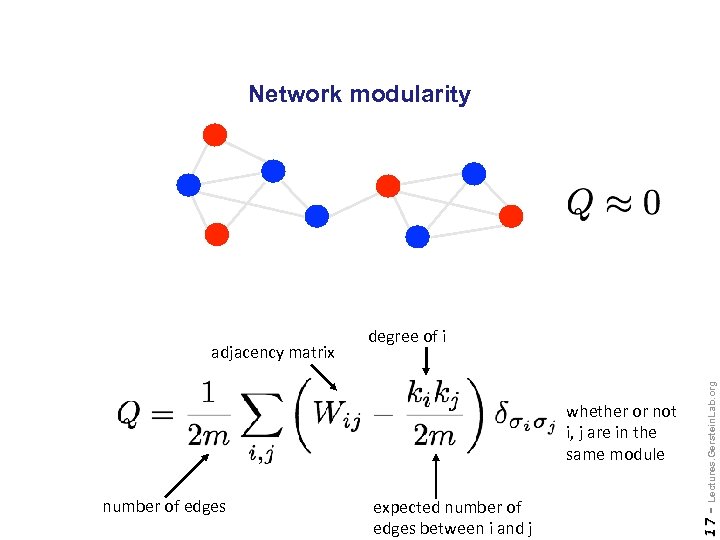 Network modularity number of edges expected number of edges between i and j Lectures. Gerstein. Lab. org whether or not i, j are in the same module 17 - adjacency matrix degree of i
Network modularity number of edges expected number of edges between i and j Lectures. Gerstein. Lab. org whether or not i, j are in the same module 17 - adjacency matrix degree of i
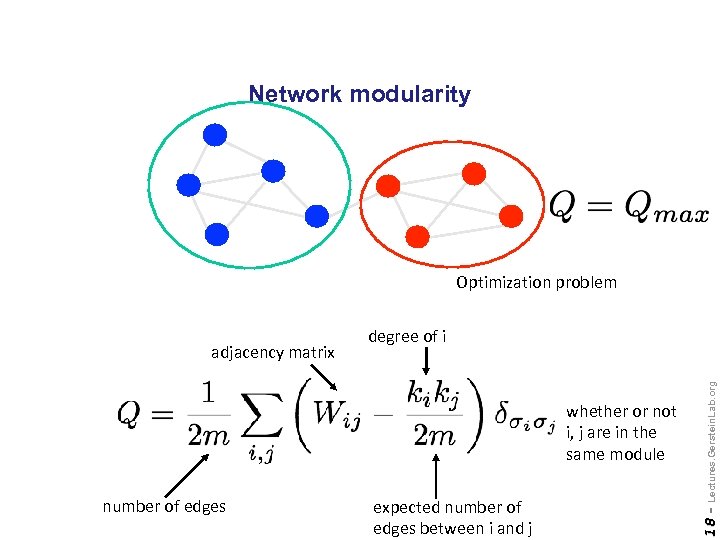 Network modularity Optimization problem number of edges expected number of edges between i and j Lectures. Gerstein. Lab. org whether or not i, j are in the same module 18 - adjacency matrix degree of i
Network modularity Optimization problem number of edges expected number of edges between i and j Lectures. Gerstein. Lab. org whether or not i, j are in the same module 18 - adjacency matrix degree of i
![[Yan et al. , PLOS Comp. Bio. (in revision, ‘ 17); bio. Rxiv 097345] [Yan et al. , PLOS Comp. Bio. (in revision, ‘ 17); bio. Rxiv 097345]](https://present5.com/presentation/26d61d2b0a47f1f0d473e7638e6cdc9b/image-19.jpg) [Yan et al. , PLOS Comp. Bio. (in revision, ‘ 17); bio. Rxiv 097345] 19 - Lectures. Gerstein. Lab. org Identifying TADs in multiple resolutions
[Yan et al. , PLOS Comp. Bio. (in revision, ‘ 17); bio. Rxiv 097345] 19 - Lectures. Gerstein. Lab. org Identifying TADs in multiple resolutions
![[Yan et al. , PLOS Comp. Bio. (in revision, ‘ 17); bio. Rxiv 097345] [Yan et al. , PLOS Comp. Bio. (in revision, ‘ 17); bio. Rxiv 097345]](https://present5.com/presentation/26d61d2b0a47f1f0d473e7638e6cdc9b/image-20.jpg) [Yan et al. , PLOS Comp. Bio. (in revision, ‘ 17); bio. Rxiv 097345] 20 - Lectures. Gerstein. Lab. org Identifying TADs in multiple resolutions
[Yan et al. , PLOS Comp. Bio. (in revision, ‘ 17); bio. Rxiv 097345] 20 - Lectures. Gerstein. Lab. org Identifying TADs in multiple resolutions
![[Yan et al. , PLOS Comp. Bio. (in revision, ‘ 17); bio. Rxiv 097345] [Yan et al. , PLOS Comp. Bio. (in revision, ‘ 17); bio. Rxiv 097345]](https://present5.com/presentation/26d61d2b0a47f1f0d473e7638e6cdc9b/image-21.jpg) [Yan et al. , PLOS Comp. Bio. (in revision, ‘ 17); bio. Rxiv 097345] Identifying TADs in multiple resolutions Lectures. Gerstein. Lab. org in equations 21 - Numerically solve for
[Yan et al. , PLOS Comp. Bio. (in revision, ‘ 17); bio. Rxiv 097345] Identifying TADs in multiple resolutions Lectures. Gerstein. Lab. org in equations 21 - Numerically solve for
![[Yan et al. , PLOS Comp. Bio. (in revision, ‘ 17); bio. Rxiv 097345] [Yan et al. , PLOS Comp. Bio. (in revision, ‘ 17); bio. Rxiv 097345]](https://present5.com/presentation/26d61d2b0a47f1f0d473e7638e6cdc9b/image-22.jpg) [Yan et al. , PLOS Comp. Bio. (in revision, ‘ 17); bio. Rxiv 097345] 22 - Lectures. Gerstein. Lab. org Identifying TADs in multiple resolutions
[Yan et al. , PLOS Comp. Bio. (in revision, ‘ 17); bio. Rxiv 097345] 22 - Lectures. Gerstein. Lab. org Identifying TADs in multiple resolutions
![[Yan et al. , PLOS Comp. Bio. (in revision, ‘ 17); bio. Rxiv 097345] [Yan et al. , PLOS Comp. Bio. (in revision, ‘ 17); bio. Rxiv 097345]](https://present5.com/presentation/26d61d2b0a47f1f0d473e7638e6cdc9b/image-23.jpg) [Yan et al. , PLOS Comp. Bio. (in revision, ‘ 17); bio. Rxiv 097345] Identifying TADs in multiple resolutions 23 - Lectures. Gerstein. Lab. org a modified Louvain algorithm
[Yan et al. , PLOS Comp. Bio. (in revision, ‘ 17); bio. Rxiv 097345] Identifying TADs in multiple resolutions 23 - Lectures. Gerstein. Lab. org a modified Louvain algorithm
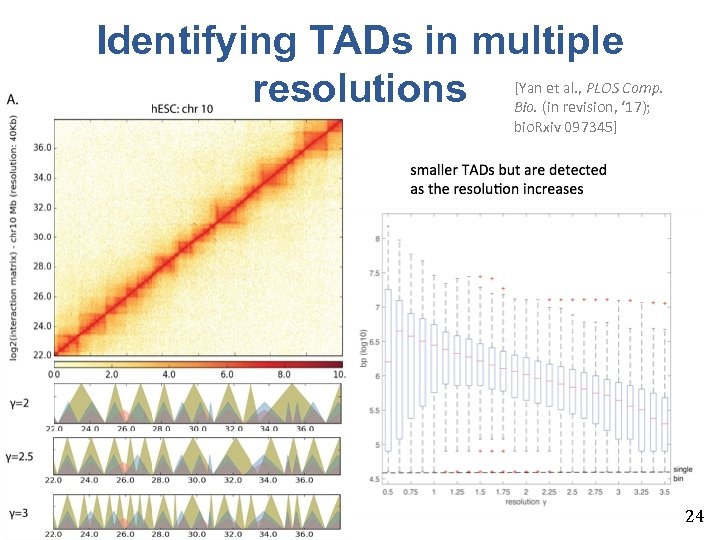 Identifying TADs in multiple resolutions [Yan et al. , PLOS Comp. Bio. (in revision, ‘ 17); bio. Rxiv 097345] 24
Identifying TADs in multiple resolutions [Yan et al. , PLOS Comp. Bio. (in revision, ‘ 17); bio. Rxiv 097345] 24
 Multi-scale Element Annotation & Variant Prioritization • Characterizing Regulatory • Features of Sites Multi-resolution TADs - Specific TFs & HMs associated at Multiple Scales - Multi-scale "site" calling (with Music) - Using high resolution conservation information to find sensitive sites • Characterizing TADs at Multiple Scales - Using modularity for identification - Developing an appropriate null expectation with TAD boundaries at different scales - Assoc. strong enough to build a predictor - HOT regions at boundaries • Fun. Seq Software Tool for Variant Prioritization - Systematically weighting all the features, for non-coding prioritization
Multi-scale Element Annotation & Variant Prioritization • Characterizing Regulatory • Features of Sites Multi-resolution TADs - Specific TFs & HMs associated at Multiple Scales - Multi-scale "site" calling (with Music) - Using high resolution conservation information to find sensitive sites • Characterizing TADs at Multiple Scales - Using modularity for identification - Developing an appropriate null expectation with TAD boundaries at different scales - Assoc. strong enough to build a predictor - HOT regions at boundaries • Fun. Seq Software Tool for Variant Prioritization - Systematically weighting all the features, for non-coding prioritization
 Enrichment of histone features at different resolution [Yan et al. , PLOS Comp. Bio. (in revision, ‘ 17); bio. Rxiv 097345] 26
Enrichment of histone features at different resolution [Yan et al. , PLOS Comp. Bio. (in revision, ‘ 17); bio. Rxiv 097345] 26
![[Yan et al. , PLOS Comp. Bio. (in revision, ‘ 17); bio. Rxiv 097345] [Yan et al. , PLOS Comp. Bio. (in revision, ‘ 17); bio. Rxiv 097345]](https://present5.com/presentation/26d61d2b0a47f1f0d473e7638e6cdc9b/image-27.jpg) [Yan et al. , PLOS Comp. Bio. (in revision, ‘ 17); bio. Rxiv 097345] Enrichment of histone features at different resolution characteristic length scale 27
[Yan et al. , PLOS Comp. Bio. (in revision, ‘ 17); bio. Rxiv 097345] Enrichment of histone features at different resolution characteristic length scale 27
![[Yan et al. , PLOS Comp. Bio. (in revision, ‘ 17); bio. Rxiv 097345] [Yan et al. , PLOS Comp. Bio. (in revision, ‘ 17); bio. Rxiv 097345]](https://present5.com/presentation/26d61d2b0a47f1f0d473e7638e6cdc9b/image-28.jpg) [Yan et al. , PLOS Comp. Bio. (in revision, ‘ 17); bio. Rxiv 097345] House-keeping vs tissue-specific genes 28
[Yan et al. , PLOS Comp. Bio. (in revision, ‘ 17); bio. Rxiv 097345] House-keeping vs tissue-specific genes 28
![[Yan et al. , PLOS Comp. Bio. (in revision, ‘ 17); bio. Rxiv 097345] [Yan et al. , PLOS Comp. Bio. (in revision, ‘ 17); bio. Rxiv 097345]](https://present5.com/presentation/26d61d2b0a47f1f0d473e7638e6cdc9b/image-29.jpg) [Yan et al. , PLOS Comp. Bio. (in revision, ‘ 17); bio. Rxiv 097345] Enrichment of TF binding sites near boundaries Question: Causes or Consequences? 29
[Yan et al. , PLOS Comp. Bio. (in revision, ‘ 17); bio. Rxiv 097345] Enrichment of TF binding sites near boundaries Question: Causes or Consequences? 29
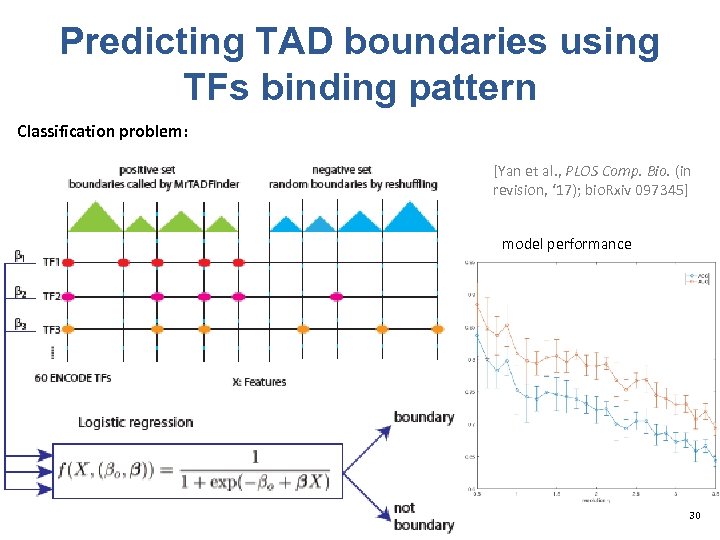 Predicting TAD boundaries using TFs binding pattern Classification problem: [Yan et al. , PLOS Comp. Bio. (in revision, ‘ 17); bio. Rxiv 097345] model performance 30
Predicting TAD boundaries using TFs binding pattern Classification problem: [Yan et al. , PLOS Comp. Bio. (in revision, ‘ 17); bio. Rxiv 097345] model performance 30
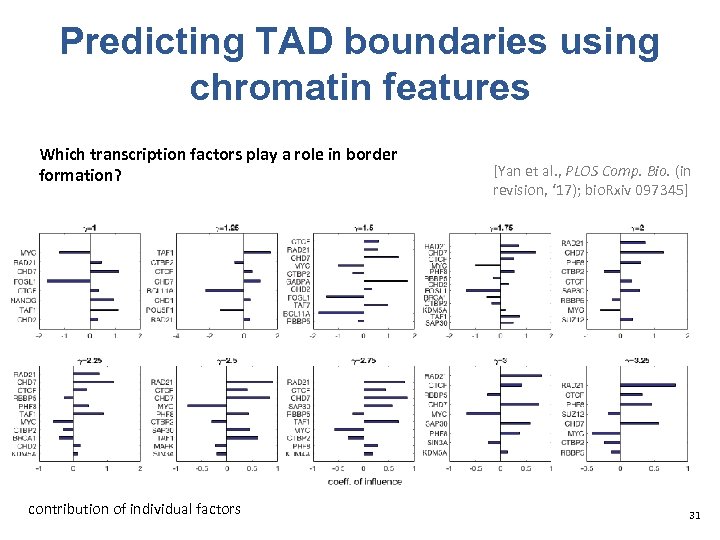 Predicting TAD boundaries using chromatin features Which transcription factors play a role in border formation? contribution of individual factors [Yan et al. , PLOS Comp. Bio. (in revision, ‘ 17); bio. Rxiv 097345] 31
Predicting TAD boundaries using chromatin features Which transcription factors play a role in border formation? contribution of individual factors [Yan et al. , PLOS Comp. Bio. (in revision, ‘ 17); bio. Rxiv 097345] 31
 Multi-scale Element Annotation & Variant Prioritization • Characterizing Regulatory • Features of Sites Multi-resolution TADs - Specific TFs & HMs associated at Multiple Scales - Multi-scale "site" calling (with Music) - Using high resolution conservation information to find sensitive sites • Characterizing TADs at Multiple Scales - Using modularity for identification - Developing an appropriate null expectation with TAD boundaries at different scales - Assoc. strong enough to build a predictor - HOT regions at boundaries • Fun. Seq Software Tool for Variant Prioritization - Systematically weighting all the features, for non-coding prioritization
Multi-scale Element Annotation & Variant Prioritization • Characterizing Regulatory • Features of Sites Multi-resolution TADs - Specific TFs & HMs associated at Multiple Scales - Multi-scale "site" calling (with Music) - Using high resolution conservation information to find sensitive sites • Characterizing TADs at Multiple Scales - Using modularity for identification - Developing an appropriate null expectation with TAD boundaries at different scales - Assoc. strong enough to build a predictor - HOT regions at boundaries • Fun. Seq Software Tool for Variant Prioritization - Systematically weighting all the features, for non-coding prioritization
![33 [Khurana et al. , Science (‘ 13)] 33 - Lectures. Gerstein. Lab. org 33 [Khurana et al. , Science (‘ 13)] 33 - Lectures. Gerstein. Lab. org](https://present5.com/presentation/26d61d2b0a47f1f0d473e7638e6cdc9b/image-33.jpg) 33 [Khurana et al. , Science (‘ 13)] 33 - Lectures. Gerstein. Lab. org Identification of non-coding candidate drivers amongst somatic variants: Scheme
33 [Khurana et al. , Science (‘ 13)] 33 - Lectures. Gerstein. Lab. org Identification of non-coding candidate drivers amongst somatic variants: Scheme
![34 - Lectures. Gerstein. Lab. org [Khurana et al. , Science (‘ 13)] Flowchart 34 - Lectures. Gerstein. Lab. org [Khurana et al. , Science (‘ 13)] Flowchart](https://present5.com/presentation/26d61d2b0a47f1f0d473e7638e6cdc9b/image-34.jpg) 34 - Lectures. Gerstein. Lab. org [Khurana et al. , Science (‘ 13)] Flowchart for 1 Prostate Cancer Genome (from Berger et al. '11)
34 - Lectures. Gerstein. Lab. org [Khurana et al. , Science (‘ 13)] Flowchart for 1 Prostate Cancer Genome (from Berger et al. '11)
![[Fu et al. , Genome. Biology ('14)] 35 - Fun. Seq. gersteinlab. org User [Fu et al. , Genome. Biology ('14)] 35 - Fun. Seq. gersteinlab. org User](https://present5.com/presentation/26d61d2b0a47f1f0d473e7638e6cdc9b/image-35.jpg) [Fu et al. , Genome. Biology ('14)] 35 - Fun. Seq. gersteinlab. org User Variants Lectures. Gerstein. Lab. org Site integrates user variants with large-scale context
[Fu et al. , Genome. Biology ('14)] 35 - Fun. Seq. gersteinlab. org User Variants Lectures. Gerstein. Lab. org Site integrates user variants with large-scale context
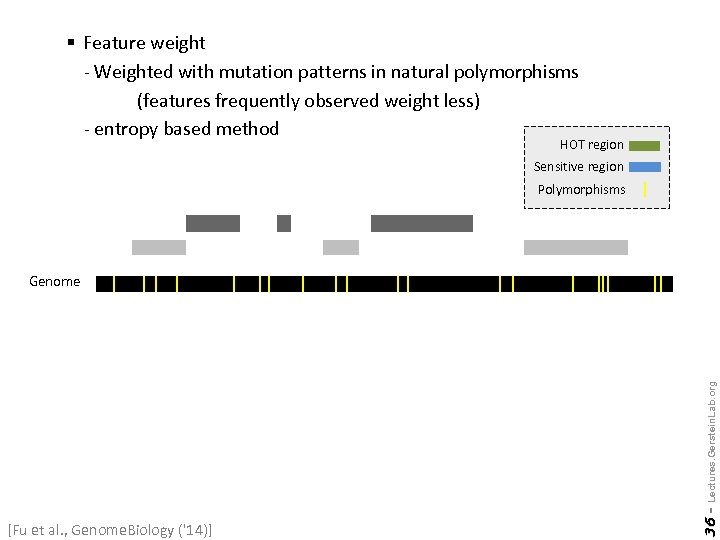 § Feature weight - Weighted with mutation patterns in natural polymorphisms (features frequently observed weight less) - entropy based method HOT region Sensitive region Polymorphisms [Fu et al. , Genome. Biology ('14)] 36 - Lectures. Gerstein. Lab. org Genome
§ Feature weight - Weighted with mutation patterns in natural polymorphisms (features frequently observed weight less) - entropy based method HOT region Sensitive region Polymorphisms [Fu et al. , Genome. Biology ('14)] 36 - Lectures. Gerstein. Lab. org Genome
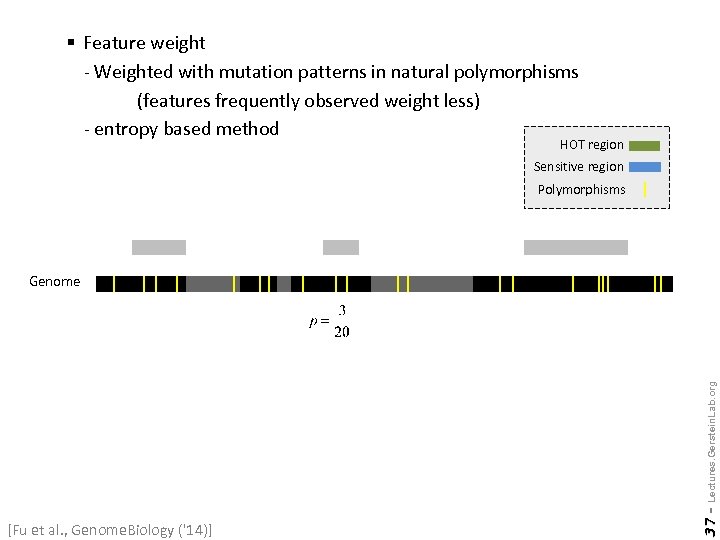 § Feature weight - Weighted with mutation patterns in natural polymorphisms (features frequently observed weight less) - entropy based method HOT region Sensitive region Polymorphisms [Fu et al. , Genome. Biology ('14)] 37 - Lectures. Gerstein. Lab. org Genome
§ Feature weight - Weighted with mutation patterns in natural polymorphisms (features frequently observed weight less) - entropy based method HOT region Sensitive region Polymorphisms [Fu et al. , Genome. Biology ('14)] 37 - Lectures. Gerstein. Lab. org Genome
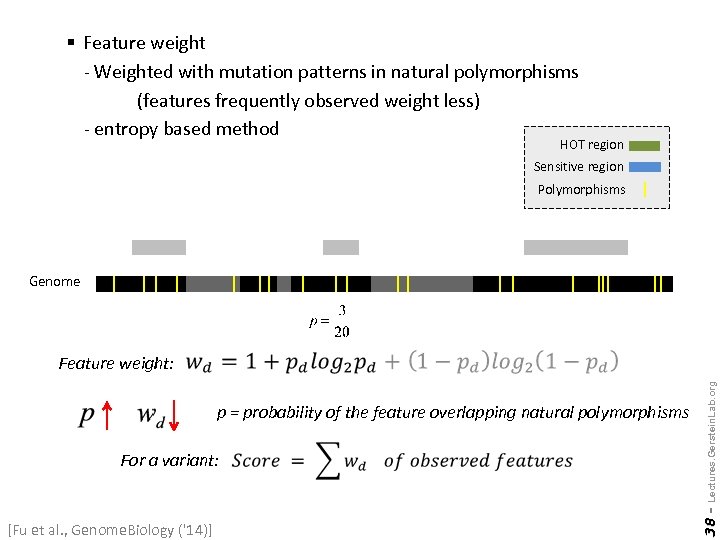 § Feature weight - Weighted with mutation patterns in natural polymorphisms (features frequently observed weight less) - entropy based method HOT region Sensitive region Polymorphisms Genome For a variant: [Fu et al. , Genome. Biology ('14)] 38 - p = probability of the feature overlapping natural polymorphisms Lectures. Gerstein. Lab. org Feature weight:
§ Feature weight - Weighted with mutation patterns in natural polymorphisms (features frequently observed weight less) - entropy based method HOT region Sensitive region Polymorphisms Genome For a variant: [Fu et al. , Genome. Biology ('14)] 38 - p = probability of the feature overlapping natural polymorphisms Lectures. Gerstein. Lab. org Feature weight:
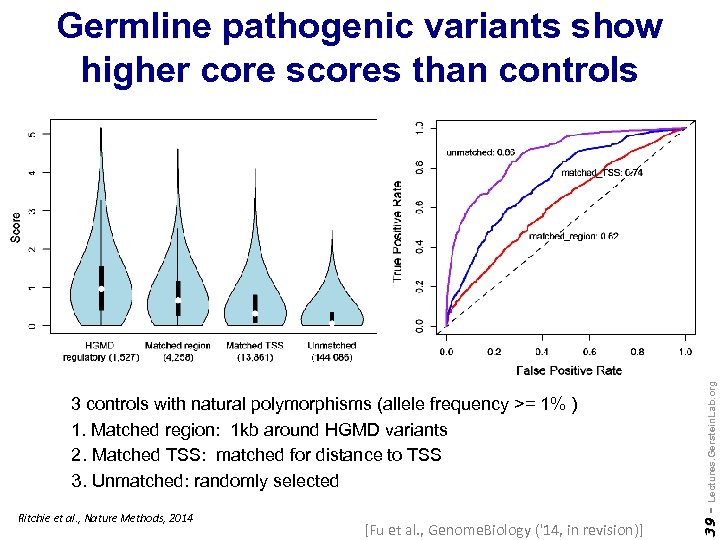 Ritchie et al. , Nature Methods, 2014 [Fu et al. , Genome. Biology ('14, in revision)] 39 - 3 controls with natural polymorphisms (allele frequency >= 1% ) 1. Matched region: 1 kb around HGMD variants 2. Matched TSS: matched for distance to TSS 3. Unmatched: randomly selected Lectures. Gerstein. Lab. org Germline pathogenic variants show higher core scores than controls
Ritchie et al. , Nature Methods, 2014 [Fu et al. , Genome. Biology ('14, in revision)] 39 - 3 controls with natural polymorphisms (allele frequency >= 1% ) 1. Matched region: 1 kb around HGMD variants 2. Matched TSS: matched for distance to TSS 3. Unmatched: randomly selected Lectures. Gerstein. Lab. org Germline pathogenic variants show higher core scores than controls
 Multi-scale Element Annotation & Variant Prioritization • Characterizing Regulatory • Features of Sites Multi-resolution TADs - Specific TFs & HMs associated at Multiple Scales - Multi-scale "site" calling (with Music) - Using high resolution conservation information to find sensitive sites • Characterizing TADs at Multiple Scales - Using modularity for identification - Developing an appropriate null expectation with TAD boundaries at different scales - Assoc. strong enough to build a predictor - HOT regions at boundaries • Fun. Seq Software Tool for Variant Prioritization - Systematically weighting all the features, for non-coding prioritization
Multi-scale Element Annotation & Variant Prioritization • Characterizing Regulatory • Features of Sites Multi-resolution TADs - Specific TFs & HMs associated at Multiple Scales - Multi-scale "site" calling (with Music) - Using high resolution conservation information to find sensitive sites • Characterizing TADs at Multiple Scales - Using modularity for identification - Developing an appropriate null expectation with TAD boundaries at different scales - Assoc. strong enough to build a predictor - HOT regions at boundaries • Fun. Seq Software Tool for Variant Prioritization - Systematically weighting all the features, for non-coding prioritization
 Multi-scale Element Annotation & Variant Prioritization • Characterizing Regulatory • Features of Sites Multi-resolution TADs - Specific TFs & HMs associated at Multiple Scales - Multi-scale "site" calling (with Music) - Using high resolution conservation information to find sensitive sites • Characterizing TADs at Multiple Scales - Using modularity for identification - Developing an appropriate null expectation with TAD boundaries at different scales - Assoc. strong enough to build a predictor - HOT regions at boundaries • Fun. Seq Software Tool for Variant Prioritization - Systematically weighting all the features, for non-coding prioritization
Multi-scale Element Annotation & Variant Prioritization • Characterizing Regulatory • Features of Sites Multi-resolution TADs - Specific TFs & HMs associated at Multiple Scales - Multi-scale "site" calling (with Music) - Using high resolution conservation information to find sensitive sites • Characterizing TADs at Multiple Scales - Using modularity for identification - Developing an appropriate null expectation with TAD boundaries at different scales - Assoc. strong enough to build a predictor - HOT regions at boundaries • Fun. Seq Software Tool for Variant Prioritization - Systematically weighting all the features, for non-coding prioritization
 MUSIC. gersteinlab. org A Harmanci, J Rozowsky github. com/gersteinlab/Mr. TADfinder K Yan, S Lou Fun. Seq. gersteinlab. org Y Fu, E Khurana, XJ Mu, Z Liu, S Lou, J Bedford, Jobs. gersteinlab. org Acknowledgments 42 - Hiring Postdocs. See Lectures. Gerstein. Lab. org KY Yip, V Colonna, XJ Mu, … , 1000 Genomes, et al
MUSIC. gersteinlab. org A Harmanci, J Rozowsky github. com/gersteinlab/Mr. TADfinder K Yan, S Lou Fun. Seq. gersteinlab. org Y Fu, E Khurana, XJ Mu, Z Liu, S Lou, J Bedford, Jobs. gersteinlab. org Acknowledgments 42 - Hiring Postdocs. See Lectures. Gerstein. Lab. org KY Yip, V Colonna, XJ Mu, … , 1000 Genomes, et al
 43 - Lectures. Gerstein. Lab. org Extra
43 - Lectures. Gerstein. Lab. org Extra
 Info about content in this slide pack • General PERMISSIONS - This Presentation is copyright Mark Gerstein, Yale University, 2016. - Please read statement at www. gersteinlab. org/misc/permissions. html. - Feel free to use slides & images in the talk with PROPER acknowledgement (via citation to relevant papers or link to gersteinlab. org). Paper references in the talk are mostly from Papers. Gerstein. Lab. org. 44 - Lectures. Gerstein. Lab. org • PHOTOS & IMAGES. For thoughts on the source and permissions of many of the photos and clipped images in this presentation see streams. gerstein. info.
Info about content in this slide pack • General PERMISSIONS - This Presentation is copyright Mark Gerstein, Yale University, 2016. - Please read statement at www. gersteinlab. org/misc/permissions. html. - Feel free to use slides & images in the talk with PROPER acknowledgement (via citation to relevant papers or link to gersteinlab. org). Paper references in the talk are mostly from Papers. Gerstein. Lab. org. 44 - Lectures. Gerstein. Lab. org • PHOTOS & IMAGES. For thoughts on the source and permissions of many of the photos and clipped images in this presentation see streams. gerstein. info.


