f6d75e46ae7bfc7c8a37e3e3812aa8e3.ppt
- Количество слайдов: 32
 An Overall View of Standardization May 26, 2004 Indira Hewlett, Ph. D. CBER/FDA
An Overall View of Standardization May 26, 2004 Indira Hewlett, Ph. D. CBER/FDA
 NAT and blood screening u NAT assays are used to screen blood and plasma donations for multiple viruses u NAT can reduce viral burden in plasma for further manufacturing u NAT can reduce virus transmission by blood through early detection of viremic window period donations
NAT and blood screening u NAT assays are used to screen blood and plasma donations for multiple viruses u NAT can reduce viral burden in plasma for further manufacturing u NAT can reduce virus transmission by blood through early detection of viremic window period donations
 Standardization of NAT u Multiple, different NAT technologies: PCR, TMA, b. DNA u Multiple, genetically diverse viral subtypes u Different pooling algorithms and pool sizes for minipool NAT : up to 512 for Source Plasma and 6 -24 for Whole blood u Varied sensitivity, specificity and reproducibility of NAT
Standardization of NAT u Multiple, different NAT technologies: PCR, TMA, b. DNA u Multiple, genetically diverse viral subtypes u Different pooling algorithms and pool sizes for minipool NAT : up to 512 for Source Plasma and 6 -24 for Whole blood u Varied sensitivity, specificity and reproducibility of NAT
 Standardization of NAT-con’t u Need analytical standards – Tools for quality control and quality assurance of NAT – Aid in licensing of investigational NAT and for post-market surveillance through lot release testing – Establish and monitor LOD and analytical sensitivity of NAT – Assure global standardization of NAT assays and reporting e. g. copies, IU/ml – Monitor laboratory proficiency
Standardization of NAT-con’t u Need analytical standards – Tools for quality control and quality assurance of NAT – Aid in licensing of investigational NAT and for post-market surveillance through lot release testing – Establish and monitor LOD and analytical sensitivity of NAT – Assure global standardization of NAT assays and reporting e. g. copies, IU/ml – Monitor laboratory proficiency
 Quality Assurance of NAT (QA) u Assay validation – sensitivity, specificity, precision – clinical specificity and sensitivity u Quality control testing of components, final test kit, instrumentation u Acceptance criteria, specifications u Good manufacturing and good laboratory practices
Quality Assurance of NAT (QA) u Assay validation – sensitivity, specificity, precision – clinical specificity and sensitivity u Quality control testing of components, final test kit, instrumentation u Acceptance criteria, specifications u Good manufacturing and good laboratory practices
 QA Issues - I u Standardize assays using reference reagents and to compare values between laboratories u Ensure quality control of components and final test kit using panels and reference standards u Monitor operator proficiency using proficiency panels and training programs
QA Issues - I u Standardize assays using reference reagents and to compare values between laboratories u Ensure quality control of components and final test kit using panels and reference standards u Monitor operator proficiency using proficiency panels and training programs
 QA Issues -II u Sample preparation including collection, storage and extraction u Manufacturing consistency of primers, probes and enzymes u Performance of controls, calibrators and quantitation standards u Specimen and kit stability u Instrument and software validation
QA Issues -II u Sample preparation including collection, storage and extraction u Manufacturing consistency of primers, probes and enzymes u Performance of controls, calibrators and quantitation standards u Specimen and kit stability u Instrument and software validation
 QA: Controls and Quantitation Standards u Potency of specimen controls u Purity, identity and potency of synthetic oligo based internal controls or quantitation standards (e. g. transcripts) u Stability conditions of controls u Low and medium copy number kit controls, at least one close to LLOD u Quantitation standard close to LLOQ u Acceptance criteria, specifications
QA: Controls and Quantitation Standards u Potency of specimen controls u Purity, identity and potency of synthetic oligo based internal controls or quantitation standards (e. g. transcripts) u Stability conditions of controls u Low and medium copy number kit controls, at least one close to LLOD u Quantitation standard close to LLOQ u Acceptance criteria, specifications
 Current status of NAT u Donor screening NAT assays licensed for HIV-1 and HCV u Investigational NAT for West Nile virus (WNV) since July 2003 u HBV NAT in IND phase u In-process quality control NAT for Parvo virus B 19, Hepatitis A virus (HAV)
Current status of NAT u Donor screening NAT assays licensed for HIV-1 and HCV u Investigational NAT for West Nile virus (WNV) since July 2003 u HBV NAT in IND phase u In-process quality control NAT for Parvo virus B 19, Hepatitis A virus (HAV)
 Current status of CBER NAT standards u HIV-1, HBV and HCV NAT panels currently available for lot release of licensed NAT u HIV-1 subtype panel in final stages of formulation u HIV-2 panel in initial stages of testing inhouse u WNV NAT panel in final stages of testing
Current status of CBER NAT standards u HIV-1, HBV and HCV NAT panels currently available for lot release of licensed NAT u HIV-1 subtype panel in final stages of formulation u HIV-2 panel in initial stages of testing inhouse u WNV NAT panel in final stages of testing
 CBER HIV-1 RNA Panel HIV-1 subtype B panel has 10 members – Cultured patient isolate, heat inactivated and diluted with defibrinated Ab-ve plasma – Gag, pol and env regions sequenced – Virus dilutions tested by 15 labs in collaborative study – 8 positives: 10, 50, 100, 500, 2500, 5000, 2. 5 x 104, and 2. 5 x 105 copies/m. L and 2 negatives – Current HIV-1 standard is 100 IU/ml for the pool test and 10, 000 IU/ml for the original donation
CBER HIV-1 RNA Panel HIV-1 subtype B panel has 10 members – Cultured patient isolate, heat inactivated and diluted with defibrinated Ab-ve plasma – Gag, pol and env regions sequenced – Virus dilutions tested by 15 labs in collaborative study – 8 positives: 10, 50, 100, 500, 2500, 5000, 2. 5 x 104, and 2. 5 x 105 copies/m. L and 2 negatives – Current HIV-1 standard is 100 IU/ml for the pool test and 10, 000 IU/ml for the original donation
 NIBSC/So. GAT Collaborative Study Calibration of HIV-1 Working reagents Candidate Preparation QC 105 (NRL, Australia) B 5 (CBER, USA) B 10 (CBER, USA) Pelispy (CLB, The Netherlands) PWS-1 (NIBSC, UK) PWS-3 (NIBSC, UK) IRC (Utrecht, The Netherlands) Log IU/m. L 4. 04 2. 21 3. 82 4. 43 3. 56 2. 72 4. 27 Ref: Davis et al (2003) J Virol Methods 107: 37 -44 12
NIBSC/So. GAT Collaborative Study Calibration of HIV-1 Working reagents Candidate Preparation QC 105 (NRL, Australia) B 5 (CBER, USA) B 10 (CBER, USA) Pelispy (CLB, The Netherlands) PWS-1 (NIBSC, UK) PWS-3 (NIBSC, UK) IRC (Utrecht, The Netherlands) Log IU/m. L 4. 04 2. 21 3. 82 4. 43 3. 56 2. 72 4. 27 Ref: Davis et al (2003) J Virol Methods 107: 37 -44 12
 CBER HIV-1 RNA Panel -con’t u HIV-1 subtype panel – 7 subtypes of HIV-1 group M: A, B, C, D, E, F, G ; group N, and group O » Pilot-scale prototype panels were tested in collaborative study involving 5 NAT manufacturers at various dilutions » Data analyzed at FDA and consensus values assigned to viral stocks » Full-scale final panel currently under formulation
CBER HIV-1 RNA Panel -con’t u HIV-1 subtype panel – 7 subtypes of HIV-1 group M: A, B, C, D, E, F, G ; group N, and group O » Pilot-scale prototype panels were tested in collaborative study involving 5 NAT manufacturers at various dilutions » Data analyzed at FDA and consensus values assigned to viral stocks » Full-scale final panel currently under formulation
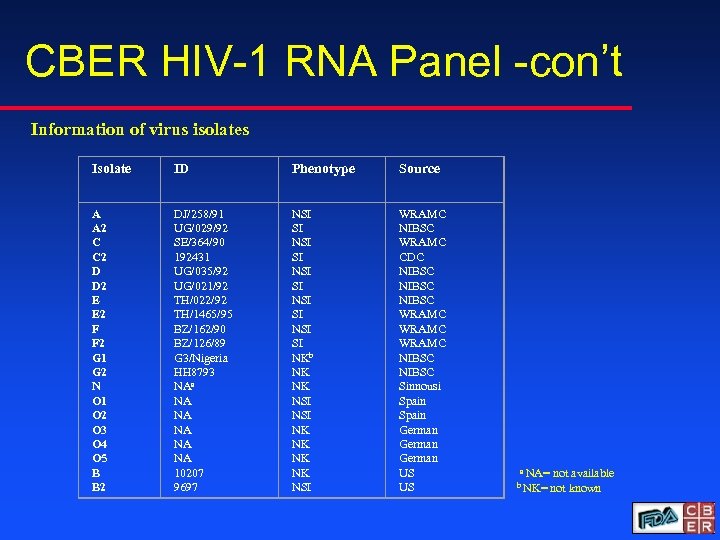 CBER HIV-1 RNA Panel -con’t Information of virus isolates Isolate ID Phenotype Source A A 2 C C 2 D D 2 E E 2 F F 2 G 1 G 2 N O 1 O 2 O 3 O 4 O 5 B B 2 DJ/258/91 UG/029/92 SE/364/90 192431 UG/035/92 UG/021/92 TH/022/92 TH/1465/95 BZ/162/90 BZ/126/89 G 3/Nigeria HH 8793 NAa NA NA NA 10207 9697 NSI SI NSI SI NKb NK NK NSI WRAMC NIBSC WRAMC CDC NIBSC WRAMC NIBSC Sinnousi Spain German US US a NA= not available b NK= not known
CBER HIV-1 RNA Panel -con’t Information of virus isolates Isolate ID Phenotype Source A A 2 C C 2 D D 2 E E 2 F F 2 G 1 G 2 N O 1 O 2 O 3 O 4 O 5 B B 2 DJ/258/91 UG/029/92 SE/364/90 192431 UG/035/92 UG/021/92 TH/022/92 TH/1465/95 BZ/162/90 BZ/126/89 G 3/Nigeria HH 8793 NAa NA NA NA 10207 9697 NSI SI NSI SI NKb NK NK NSI WRAMC NIBSC WRAMC CDC NIBSC WRAMC NIBSC Sinnousi Spain German US US a NA= not available b NK= not known
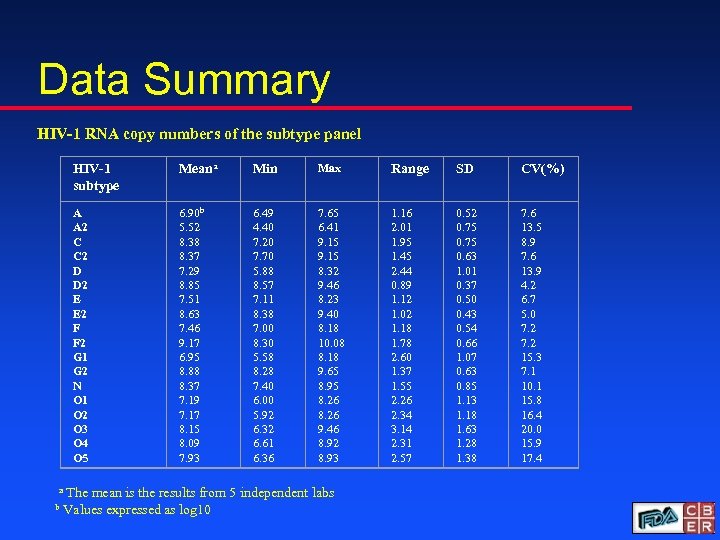 Data Summary HIV-1 RNA copy numbers of the subtype panel HIV-1 subtype Meana Min Max Range SD CV(%) A A 2 C C 2 D D 2 E E 2 F F 2 G 1 G 2 N O 1 O 2 O 3 O 4 O 5 6. 90 b 5. 52 8. 38 8. 37 7. 29 8. 85 7. 51 8. 63 7. 46 9. 17 6. 95 8. 88 8. 37 7. 19 7. 17 8. 15 8. 09 7. 93 6. 49 4. 40 7. 20 7. 70 5. 88 8. 57 7. 11 8. 38 7. 00 8. 30 5. 58 8. 28 7. 40 6. 00 5. 92 6. 32 6. 61 6. 36 7. 65 6. 41 9. 15 8. 32 9. 46 8. 23 9. 40 8. 18 10. 08 8. 18 9. 65 8. 95 8. 26 9. 46 8. 92 8. 93 1. 16 2. 01 1. 95 1. 45 2. 44 0. 89 1. 12 1. 02 1. 18 1. 78 2. 60 1. 37 1. 55 2. 26 2. 34 3. 14 2. 31 2. 57 0. 52 0. 75 0. 63 1. 01 0. 37 0. 50 0. 43 0. 54 0. 66 1. 07 0. 63 0. 85 1. 13 1. 18 1. 63 1. 28 1. 38 7. 6 13. 5 8. 9 7. 6 13. 9 4. 2 6. 7 5. 0 7. 2 15. 3 7. 1 10. 1 15. 8 16. 4 20. 0 15. 9 17. 4 a The mean is the results from 5 independent labs b Values expressed as log 10
Data Summary HIV-1 RNA copy numbers of the subtype panel HIV-1 subtype Meana Min Max Range SD CV(%) A A 2 C C 2 D D 2 E E 2 F F 2 G 1 G 2 N O 1 O 2 O 3 O 4 O 5 6. 90 b 5. 52 8. 38 8. 37 7. 29 8. 85 7. 51 8. 63 7. 46 9. 17 6. 95 8. 88 8. 37 7. 19 7. 17 8. 15 8. 09 7. 93 6. 49 4. 40 7. 20 7. 70 5. 88 8. 57 7. 11 8. 38 7. 00 8. 30 5. 58 8. 28 7. 40 6. 00 5. 92 6. 32 6. 61 6. 36 7. 65 6. 41 9. 15 8. 32 9. 46 8. 23 9. 40 8. 18 10. 08 8. 18 9. 65 8. 95 8. 26 9. 46 8. 92 8. 93 1. 16 2. 01 1. 95 1. 45 2. 44 0. 89 1. 12 1. 02 1. 18 1. 78 2. 60 1. 37 1. 55 2. 26 2. 34 3. 14 2. 31 2. 57 0. 52 0. 75 0. 63 1. 01 0. 37 0. 50 0. 43 0. 54 0. 66 1. 07 0. 63 0. 85 1. 13 1. 18 1. 63 1. 28 1. 38 7. 6 13. 5 8. 9 7. 6 13. 9 4. 2 6. 7 5. 0 7. 2 15. 3 7. 1 10. 1 15. 8 16. 4 20. 0 15. 9 17. 4 a The mean is the results from 5 independent labs b Values expressed as log 10
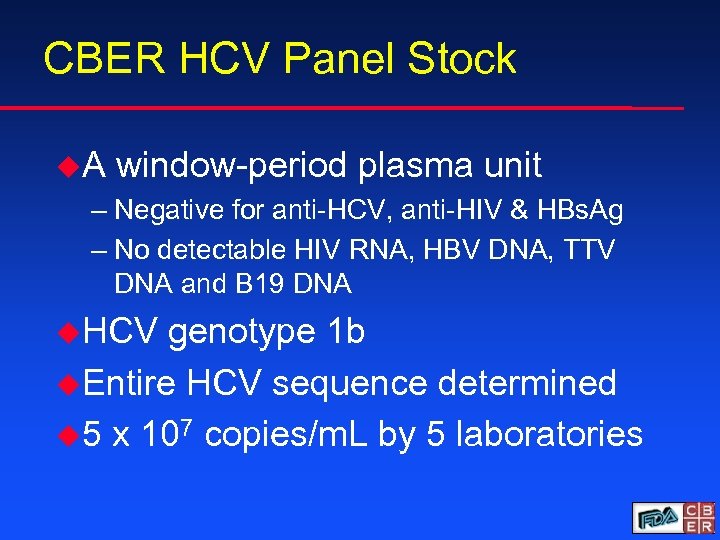 CBER HCV Panel Stock u. A window-period plasma unit – Negative for anti-HCV, anti-HIV & HBs. Ag – No detectable HIV RNA, HBV DNA, TTV DNA and B 19 DNA u. HCV genotype 1 b u. Entire HCV sequence determined u 5 x 107 copies/m. L by 5 laboratories
CBER HCV Panel Stock u. A window-period plasma unit – Negative for anti-HCV, anti-HIV & HBs. Ag – No detectable HIV RNA, HBV DNA, TTV DNA and B 19 DNA u. HCV genotype 1 b u. Entire HCV sequence determined u 5 x 107 copies/m. L by 5 laboratories
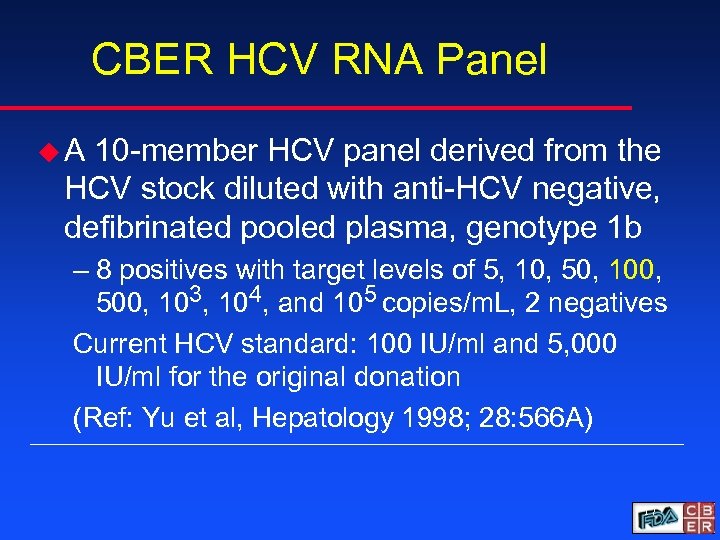 CBER HCV RNA Panel u. A 10 -member HCV panel derived from the HCV stock diluted with anti-HCV negative, defibrinated pooled plasma, genotype 1 b – 8 positives with target levels of 5, 10, 50, 100, 500, 103, 104, and 105 copies/m. L, 2 negatives Current HCV standard: 100 IU/ml and 5, 000 IU/ml for the original donation (Ref: Yu et al, Hepatology 1998; 28: 566 A) 17
CBER HCV RNA Panel u. A 10 -member HCV panel derived from the HCV stock diluted with anti-HCV negative, defibrinated pooled plasma, genotype 1 b – 8 positives with target levels of 5, 10, 50, 100, 500, 103, 104, and 105 copies/m. L, 2 negatives Current HCV standard: 100 IU/ml and 5, 000 IU/ml for the original donation (Ref: Yu et al, Hepatology 1998; 28: 566 A) 17
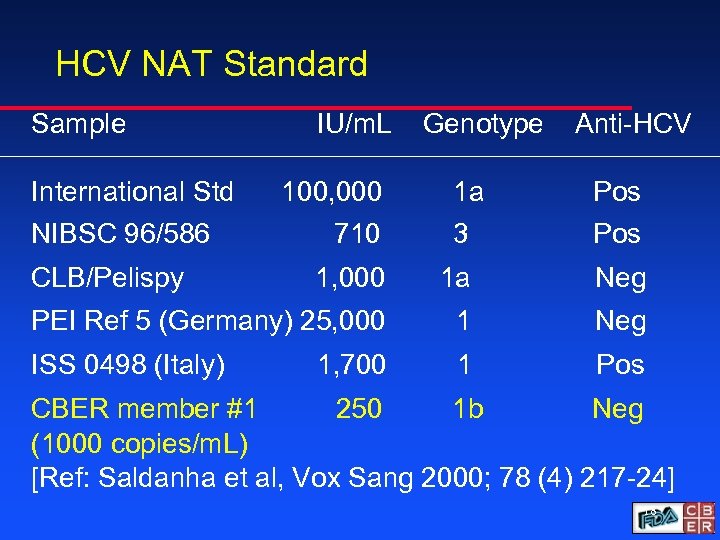 HCV NAT Standard Sample International Std NIBSC 96/586 CLB/Pelispy IU/m. L 100, 000 710 Genotype 1 a 3 Anti-HCV Pos 1, 000 1 a Neg PEI Ref 5 (Germany) 25, 000 1 Neg ISS 0498 (Italy) 1 Pos 1, 700 CBER member #1 250 1 b Neg (1000 copies/m. L) [Ref: Saldanha et al, Vox Sang 2000; 78 (4) 217 -24] 18
HCV NAT Standard Sample International Std NIBSC 96/586 CLB/Pelispy IU/m. L 100, 000 710 Genotype 1 a 3 Anti-HCV Pos 1, 000 1 a Neg PEI Ref 5 (Germany) 25, 000 1 Neg ISS 0498 (Italy) 1 Pos 1, 700 CBER member #1 250 1 b Neg (1000 copies/m. L) [Ref: Saldanha et al, Vox Sang 2000; 78 (4) 217 -24] 18
 Parvovirus B 19 NAT as an In-Process Control u Require validation as an analytical test and approve it under relevant product’s license u Proposed limit: <104 IU of B 19 DNA per m. L in all manufacturing pools – B 19 transmissions associated with S/D Treated Pooled Plasma in a phase 4 study in healthy donors » <104 GE/m. L in non-transmitting lots – Viral neutralization by anti-B 19 in pools – Viral clearance by manufacturing procedures
Parvovirus B 19 NAT as an In-Process Control u Require validation as an analytical test and approve it under relevant product’s license u Proposed limit: <104 IU of B 19 DNA per m. L in all manufacturing pools – B 19 transmissions associated with S/D Treated Pooled Plasma in a phase 4 study in healthy donors » <104 GE/m. L in non-transmitting lots – Viral neutralization by anti-B 19 in pools – Viral clearance by manufacturing procedures
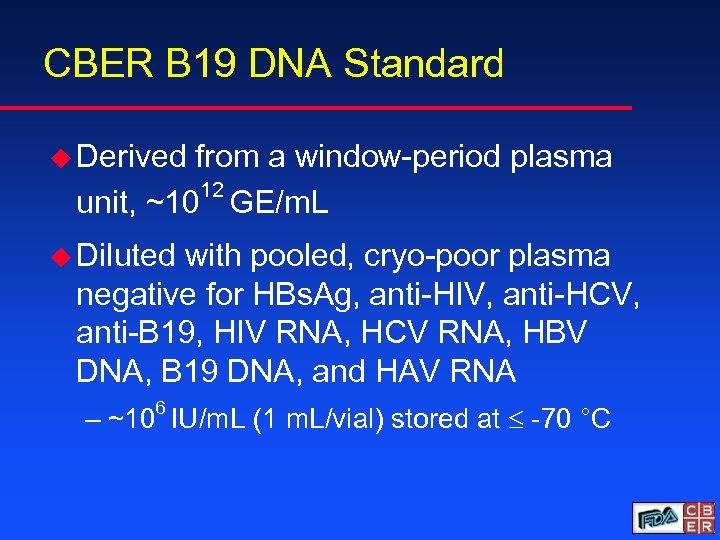 CBER B 19 DNA Standard u Derived from a window-period plasma 12 unit, ~10 GE/m. L u Diluted with pooled, cryo-poor plasma negative for HBs. Ag, anti-HIV, anti-HCV, anti-B 19, HIV RNA, HCV RNA, HBV DNA, B 19 DNA, and HAV RNA – ~106 IU/m. L (1 m. L/vial) stored at -70 °C
CBER B 19 DNA Standard u Derived from a window-period plasma 12 unit, ~10 GE/m. L u Diluted with pooled, cryo-poor plasma negative for HBs. Ag, anti-HIV, anti-HCV, anti-B 19, HIV RNA, HCV RNA, HBV DNA, B 19 DNA, and HAV RNA – ~106 IU/m. L (1 m. L/vial) stored at -70 °C
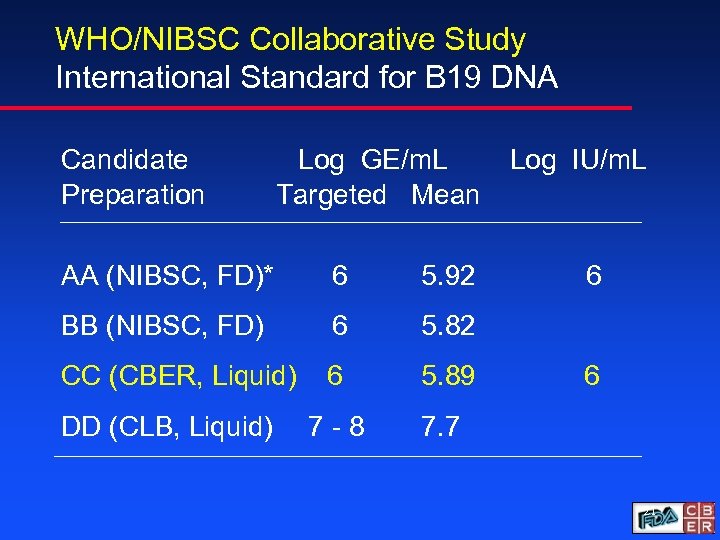 WHO/NIBSC Collaborative Study International Standard for B 19 DNA Candidate Preparation Log GE/m. L Targeted Mean AA (NIBSC, FD)* 6 5. 92 BB (NIBSC, FD) 6 5. 82 CC (CBER, Liquid) 6 5. 89 7 -8 Log IU/m. L 7. 7 DD (CLB, Liquid) 6 6 21
WHO/NIBSC Collaborative Study International Standard for B 19 DNA Candidate Preparation Log GE/m. L Targeted Mean AA (NIBSC, FD)* 6 5. 92 BB (NIBSC, FD) 6 5. 82 CC (CBER, Liquid) 6 5. 89 7 -8 Log IU/m. L 7. 7 DD (CLB, Liquid) 6 6 21
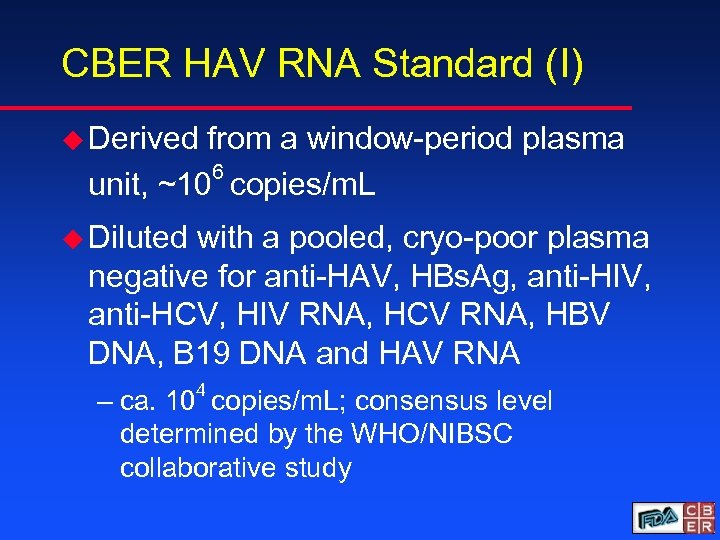 CBER HAV RNA Standard (I) u Derived from a window-period plasma 6 unit, ~10 copies/m. L u Diluted with a pooled, cryo-poor plasma negative for anti-HAV, HBs. Ag, anti-HIV, anti-HCV, HIV RNA, HCV RNA, HBV DNA, B 19 DNA and HAV RNA 4 – ca. 10 copies/m. L; consensus level determined by the WHO/NIBSC collaborative study
CBER HAV RNA Standard (I) u Derived from a window-period plasma 6 unit, ~10 copies/m. L u Diluted with a pooled, cryo-poor plasma negative for anti-HAV, HBs. Ag, anti-HIV, anti-HCV, HIV RNA, HCV RNA, HBV DNA, B 19 DNA and HAV RNA 4 – ca. 10 copies/m. L; consensus level determined by the WHO/NIBSC collaborative study
 WHO/NIBSC Collaborative Study International Standard for HAV RNA Candidate Preparation Log GE/m. L AA (NIBSC, FD)* 5. 29 BB (NIBSC, FD) 5. 07 CC (CLB, Liquid) 4. 99 DD (ISS, Liquid) 5. 40 EE (CBER, Liquid) Log IU/m. L 5 4. 08 Ref: Saldanha et al, WHO ECBS Report, Feb 2003. 23
WHO/NIBSC Collaborative Study International Standard for HAV RNA Candidate Preparation Log GE/m. L AA (NIBSC, FD)* 5. 29 BB (NIBSC, FD) 5. 07 CC (CLB, Liquid) 4. 99 DD (ISS, Liquid) 5. 40 EE (CBER, Liquid) Log IU/m. L 5 4. 08 Ref: Saldanha et al, WHO ECBS Report, Feb 2003. 23
 HBV NAT panel u CBER HBV DNA panel derived from a window period specimen genotype A, serotype adw 2 u Panel members are 0, 10 and 100 copies/ml u Panel tested by 3 NAT manufacturers
HBV NAT panel u CBER HBV DNA panel derived from a window period specimen genotype A, serotype adw 2 u Panel members are 0, 10 and 100 copies/ml u Panel tested by 3 NAT manufacturers
 WNV testing u The 2002 outbreak indicated transmission of WNV by blood transfusion u All reported cases of transmission by blood transfusion occurred during the acute, viremic phase u NAT is the most appropriate strategy to interdict infectious donations u NAT on pooled donations implemented first since platforms for HIV/HCV NAT are already licensed u Need for standards to monitor sensitivity of WNV NAT assays
WNV testing u The 2002 outbreak indicated transmission of WNV by blood transfusion u All reported cases of transmission by blood transfusion occurred during the acute, viremic phase u NAT is the most appropriate strategy to interdict infectious donations u NAT on pooled donations implemented first since platforms for HIV/HCV NAT are already licensed u Need for standards to monitor sensitivity of WNV NAT assays
 Need for Viral Titer Standardization u Lack of consensus for viral titer – Viral titer defined in plaque forming units (PFU) – Broad range of viral particles per PFU (1 – 1000 virions) – Need for correlation of RNA copies with PFU – Non-infectious particles (defective) may be detected by PCR but not by infectivity assays u Copy number determination necessary to define analytical sensitivity and infectivity
Need for Viral Titer Standardization u Lack of consensus for viral titer – Viral titer defined in plaque forming units (PFU) – Broad range of viral particles per PFU (1 – 1000 virions) – Need for correlation of RNA copies with PFU – Non-infectious particles (defective) may be detected by PCR but not by infectivity assays u Copy number determination necessary to define analytical sensitivity and infectivity
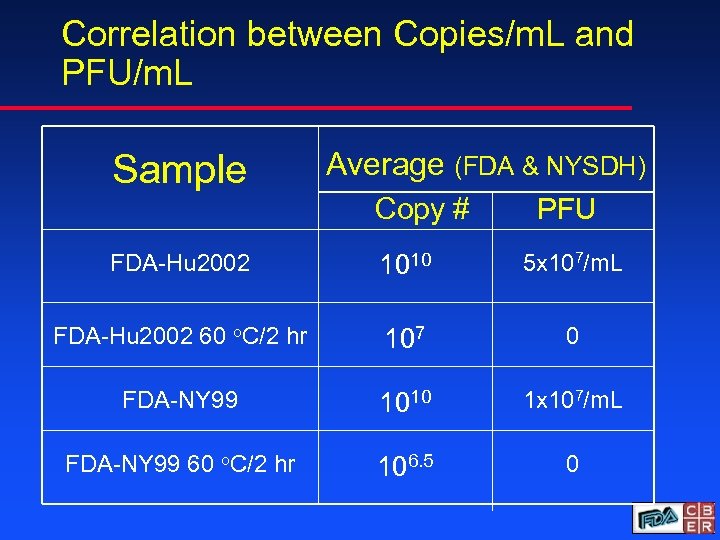 Correlation between Copies/m. L and PFU/m. L Sample Average (FDA & NYSDH) Copy # PFU FDA-Hu 2002 1010 5 x 107/m. L FDA-Hu 2002 60 o. C/2 hr 107 0 FDA-NY 99 1010 1 x 107/m. L FDA-NY 99 60 o. C/2 hr 106. 5 0
Correlation between Copies/m. L and PFU/m. L Sample Average (FDA & NYSDH) Copy # PFU FDA-Hu 2002 1010 5 x 107/m. L FDA-Hu 2002 60 o. C/2 hr 107 0 FDA-NY 99 1010 1 x 107/m. L FDA-NY 99 60 o. C/2 hr 106. 5 0
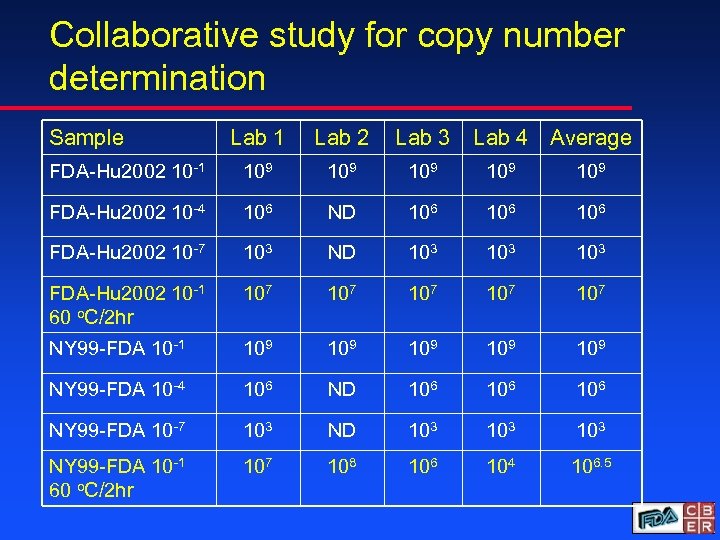 Collaborative study for copy number determination Sample Lab 1 Lab 2 Lab 3 Lab 4 Average FDA-Hu 2002 10 -1 109 109 109 FDA-Hu 2002 10 -4 106 ND 106 106 FDA-Hu 2002 10 -7 103 ND 103 103 FDA-Hu 2002 10 -1 60 o. C/2 hr 107 107 107 NY 99 -FDA 10 -1 109 109 109 NY 99 -FDA 10 -4 106 ND 106 106 NY 99 -FDA 10 -7 103 ND 103 103 NY 99 -FDA 10 -1 60 o. C/2 hr 107 108 106 104 106. 5
Collaborative study for copy number determination Sample Lab 1 Lab 2 Lab 3 Lab 4 Average FDA-Hu 2002 10 -1 109 109 109 FDA-Hu 2002 10 -4 106 ND 106 106 FDA-Hu 2002 10 -7 103 ND 103 103 FDA-Hu 2002 10 -1 60 o. C/2 hr 107 107 107 NY 99 -FDA 10 -1 109 109 109 NY 99 -FDA 10 -4 106 ND 106 106 NY 99 -FDA 10 -7 103 ND 103 103 NY 99 -FDA 10 -1 60 o. C/2 hr 107 108 106 104 106. 5
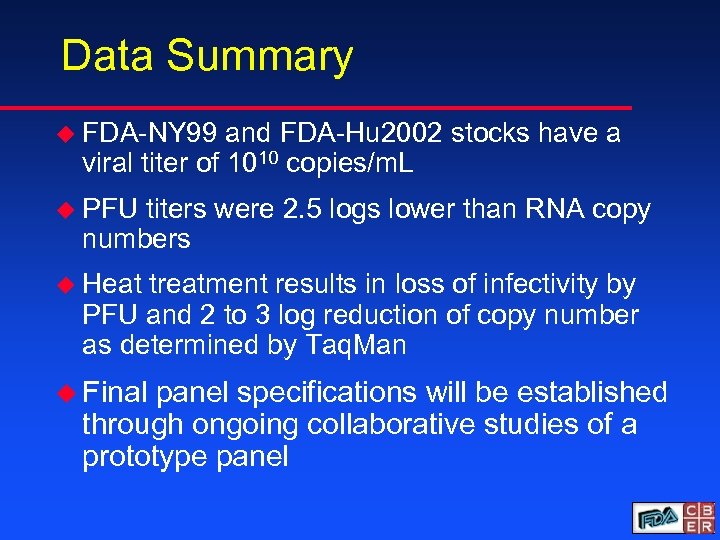 Data Summary u FDA-NY 99 and FDA-Hu 2002 stocks have a viral titer of 1010 copies/m. L u PFU titers were 2. 5 logs lower than RNA copy numbers u Heat treatment results in loss of infectivity by PFU and 2 to 3 log reduction of copy number as determined by Taq. Man u Final panel specifications will be established through ongoing collaborative studies of a prototype panel
Data Summary u FDA-NY 99 and FDA-Hu 2002 stocks have a viral titer of 1010 copies/m. L u PFU titers were 2. 5 logs lower than RNA copy numbers u Heat treatment results in loss of infectivity by PFU and 2 to 3 log reduction of copy number as determined by Taq. Man u Final panel specifications will be established through ongoing collaborative studies of a prototype panel
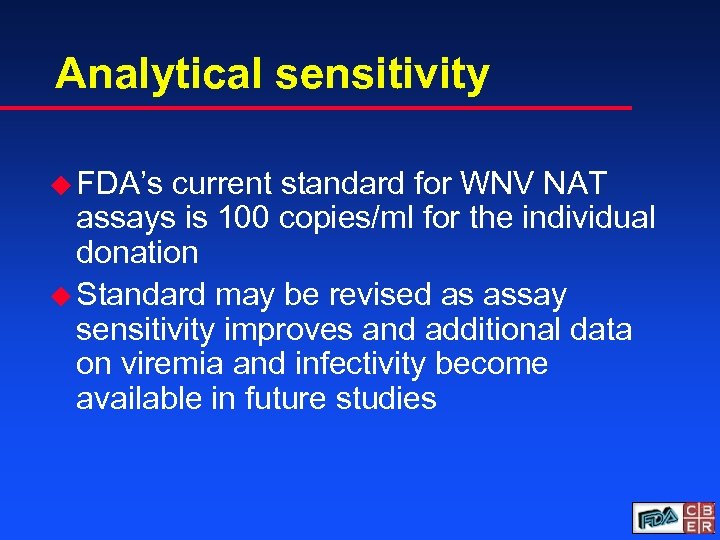 Analytical sensitivity u FDA’s current standard for WNV NAT assays is 100 copies/ml for the individual donation u Standard may be revised as assay sensitivity improves and additional data on viremia and infectivity become available in future studies
Analytical sensitivity u FDA’s current standard for WNV NAT assays is 100 copies/ml for the individual donation u Standard may be revised as assay sensitivity improves and additional data on viremia and infectivity become available in future studies
 Summary u u u FDA has established panels for HIV, HCV, HBV, B 19 and HAV and standards for licensing tests Panel for WNV under development Standards are useful for: -quality control and quality assurance of NAT - licensing tests and post-market surveillance -Standards useful for global harmonization of NAT assays
Summary u u u FDA has established panels for HIV, HCV, HBV, B 19 and HAV and standards for licensing tests Panel for WNV under development Standards are useful for: -quality control and quality assurance of NAT - licensing tests and post-market surveillance -Standards useful for global harmonization of NAT assays
 Acknowledgements CBER/FDA S. Lee M. Yu O. Wood M. Rios S. Kerby R. Taffs J. Hu R. Biswas WRAIR N. Michael ARC S. Stramer CDC R. Lanciotti NYDOH L. Kramer
Acknowledgements CBER/FDA S. Lee M. Yu O. Wood M. Rios S. Kerby R. Taffs J. Hu R. Biswas WRAIR N. Michael ARC S. Stramer CDC R. Lanciotti NYDOH L. Kramer


