4eae04ee99a17bf0df44d9606cc40575.ppt
- Количество слайдов: 107

An introduction to biological databases
Database or databank ? At the beginning, subtle distinctions were done between databases and databanks (in UK, but not in the USA), such as: « Database management programs for the gestion of databanks » From now on, the term « database » (db) is usually preferred
What is a database ? A collection of. . . structured searchable (index) updated periodically (release) cross-referenced (hyperlinks) -> table of contents -> new edition -> links with other db …data Includes also associated tools (software) necessary for db access, db updating, db information insertion, db information deletion…. Data storage management: flat files, relational databases…
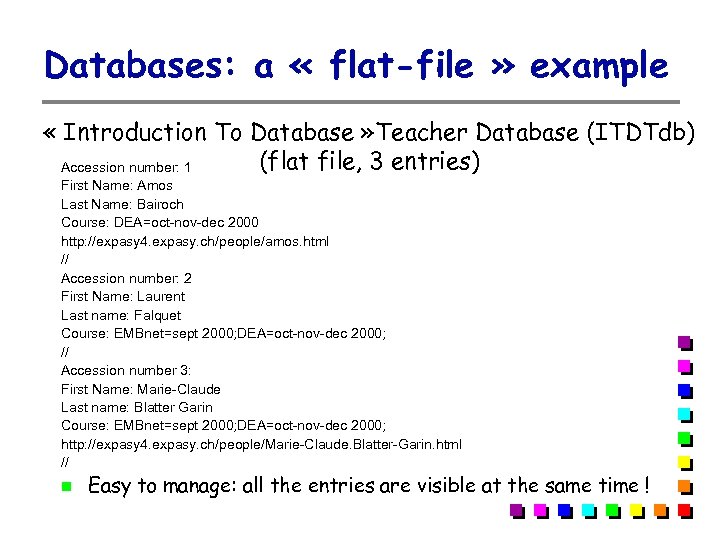
Databases: a « flat-file » example « Introduction To Database » Teacher Database (ITDTdb) (flat file, 3 entries) Accession number: 1 First Name: Amos Last Name: Bairoch Course: DEA=oct-nov-dec 2000 http: //expasy 4. expasy. ch/people/amos. html // Accession number: 2 First Name: Laurent Last name: Falquet Course: EMBnet=sept 2000; DEA=oct-nov-dec 2000; // Accession number 3: First Name: Marie-Claude Last name: Blatter Garin Course: EMBnet=sept 2000; DEA=oct-nov-dec 2000; http: //expasy 4. expasy. ch/people/Marie-Claude. Blatter-Garin. html // Easy to manage: all the entries are visible at the same time !
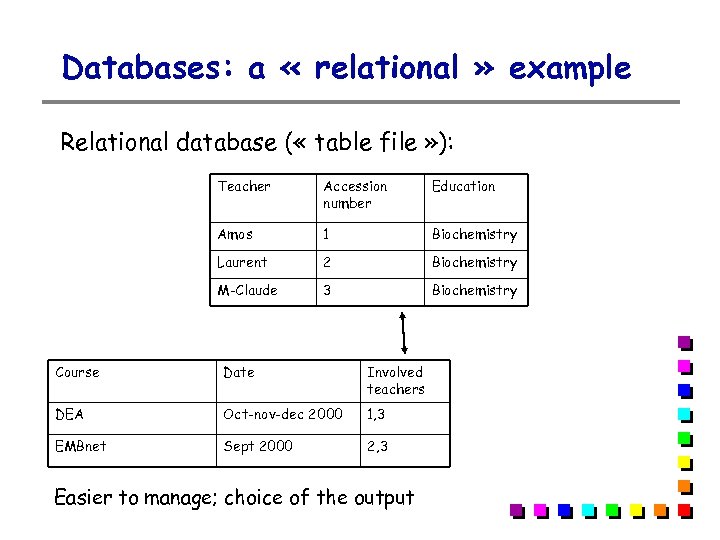
Databases: a « relational » example Relational database ( « table file » ): Teacher Accession number Education Amos 1 Biochemistry Laurent 2 Biochemistry M-Claude 3 Biochemistry Course Date Involved teachers DEA Oct-nov-dec 2000 1, 3 EMBnet Sept 2000 2, 3 Easier to manage; choice of the output
Why biological databases ? Explosive growth in biological data Data (sequences, 3 D structures, 2 D gel analysis, MS analysis, Microarrays…. ) are no longer published in a conventional manner, but directly submitted to databases Essential tools for biological research, as classical publications used to be !
Some statistics More than 1000 different databases Variable size: <100 Kb to >10 Gb DNA: > 10 Gb Protein: 1 Gb 3 D structure: 5 Gb Other: smaller Update frequency: daily to annually Generally accessible through the web (free!? ) Amos’ links: www. expasy. org/alinks. html Google: http: //www. google. com
Biological databases Some databases in the field of molecular biology… AATDB, Ace. Db, ACUTS, ADB, AFDB, AGIS, AMSdb, ARR, As. Db, BBDB, BCGD, Beanref, Biolmage, Bio. Mag. Res. Bank, BIOMDB, BLOCKS, Bov. GBASE, BOVMAP, BSORF, BTKbase, CANSITE, Carb. Bank, CARBHYD, CATH, CAZY, CCDC, CD 4 OLbase, CGAP, Chick. GBASE, Colibri, COPE, Cotton. DB, CSNDB, CUTG, Cyano. Base, db. CFC, db. EST, db. STS, DDBJ, DGP, Dicty. Db, Picty_c. DB, DIP, DOGS, DOMO, DPD, DPlnteract, ECDC, ECGC, EC 02 DBASE, Eco. Cyc, Eco. Gene, EMBL, EMD db, ENZYME, EPD, Epo. DB, ESTHER, Fly. Base, Fly. View, GCRDB, GENATLAS, Genbank, Gene. Cards, Genline, Gen. Link, GENOTK, Gen. Prot. EC, GIFTS, GPCRDB, GRAP, GRBase, g. RNAsdb, GRR, GSDB, HAEMB, HAMSTERS, HEART-2 DPAGE, HEXAdb, HGMD, HIDB, HIDC, Hl. Vdb, Hot. Molec. Base, HOVERGEN, HPDB, HSC-2 DPAGE, ICN, ICTVDB, IL 2 RGbase, IMGT, Kabat, KDNA, KEGG, Klotho, LGIC, MAD, Maize. Db, MDB, Medline, Mendel, MEROPS, MGDB, MGI, MHCPEP 5 Micado, Mito. Dat, MITOMAP, MJDB, Mmt. DB, Mol-R-Us, MPDB, MRR, Mut. Base, Myc. DB, NRSub, 0 -lyc. Base, OMIA, OMIM, OPD, ORDB, OWL, PAHdb, Pat. Base, PDB, PDD, Pfam, Phospho. Base, Pig. BASE, PIR, PKR, PMD, PPDB, PRESAGE, PRINTS, Pro. Dom, Prolysis, PROSITE, PROTOMAP, Rat. MAP, RDP, REBASE, RGP, SBASE, SCOP, Seq. Anai. Ref, SGD, SGP, Sheep. Map, Soybase, SPAD, SRNA db, SRPDB, STACK, Sty. Gene, Sub 2 D, Subti. List, SWISS-2 DPAGE, SWISS-3 DIMAGE, SWISSMODEL Repository, SWISS-PROT, Tel. DB, TGN, tm. RDB, TOPS, TRANSFAC, TRR, Uni. Gene, URNADB, V BASE, VDRR, Vector. DB, WDCM, WIT, Worm. Pep, YEPD, YPM, etc. . . . !!!!
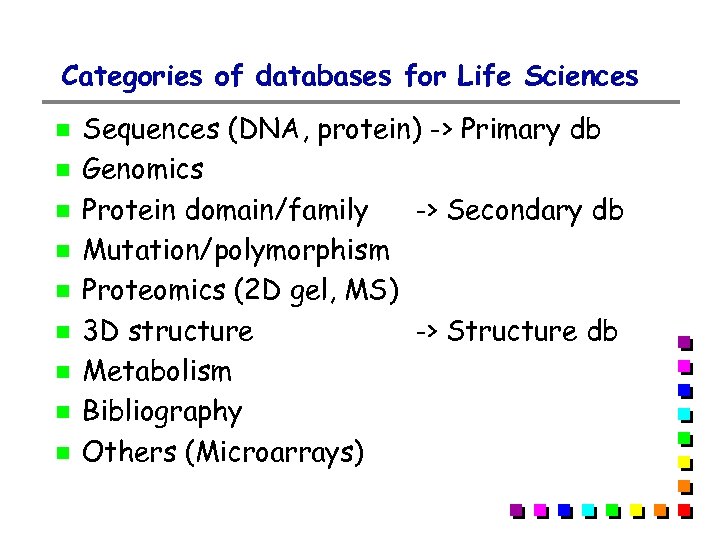
Categories of databases for Life Sciences Sequences (DNA, protein) -> Primary db Genomics Protein domain/family -> Secondary db Mutation/polymorphism Proteomics (2 D gel, MS) 3 D structure -> Structure db Metabolism Bibliography Others (Microarrays)
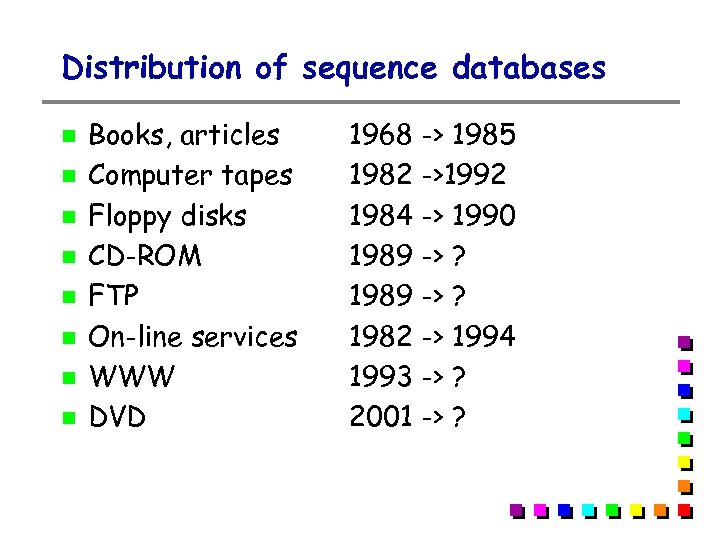
Distribution of sequence databases Books, articles Computer tapes Floppy disks CD-ROM FTP On-line services WWW DVD 1968 -> 1985 1982 ->1992 1984 -> 1990 1989 -> ? 1982 -> 1994 1993 -> ? 2001 -> ?
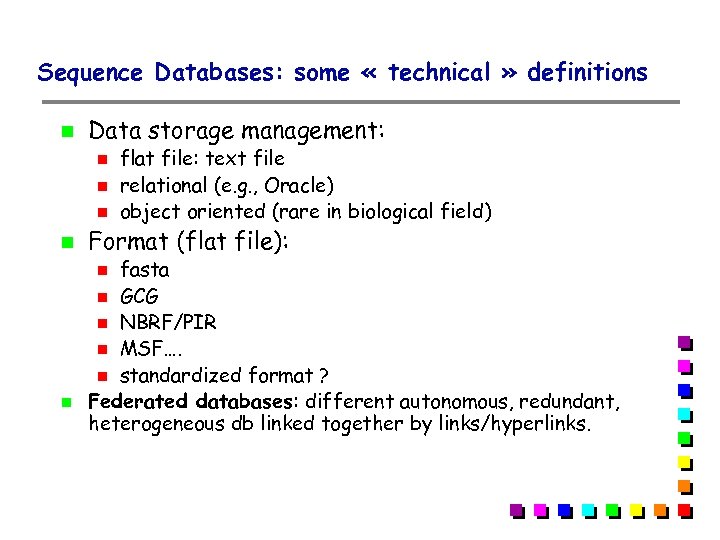
Sequence Databases: some « technical » definitions Data storage management: Format (flat file): fasta GCG NBRF/PIR MSF…. standardized format ? Federated databases: different autonomous, redundant, heterogeneous db linked together by links/hyperlinks. flat file: text file relational (e. g. , Oracle) object oriented (rare in biological field)
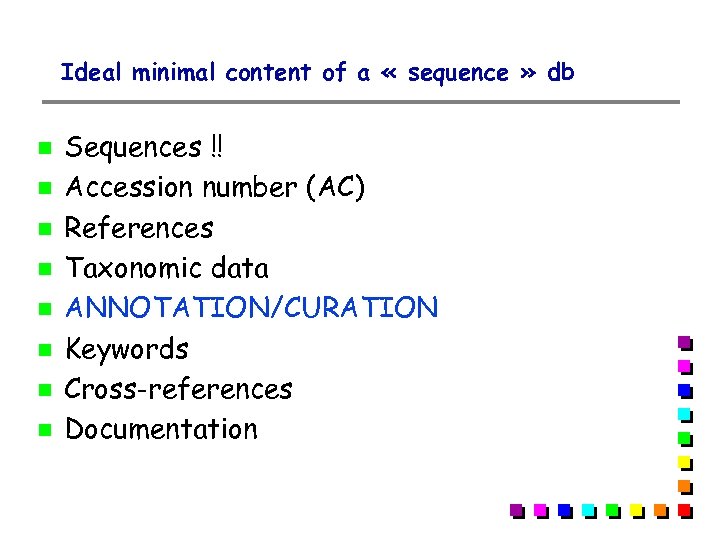
Ideal minimal content of a « sequence » db Sequences !! Accession number (AC) References Taxonomic data ANNOTATION/CURATION Keywords Cross-references Documentation
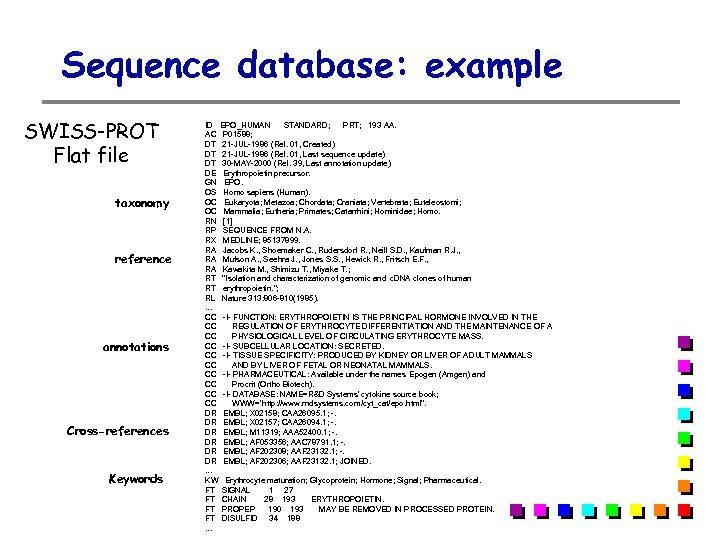
Sequence database: example SWISS-PROT Flat file taxonomy reference annotations Cross-references Keywords ID EPO_HUMAN STANDARD; PRT; 193 AA. AC P 01588; DT 21 -JUL-1986 (Rel. 01, Created) DT 21 -JUL-1986 (Rel. 01, Last sequence update) DT 30 -MAY-2000 (Rel. 39, Last annotation update) DE Erythropoietin precursor. GN EPO. OS Homo sapiens (Human). OC Eukaryota; Metazoa; Chordata; Craniata; Vertebrata; Euteleostomi; OC Mammalia; Eutheria; Primates; Catarrhini; Hominidae; Homo. RN [1] RP SEQUENCE FROM N. A. RX MEDLINE; 85137899. RA Jacobs K. , Shoemaker C. , Rudersdorf R. , Neill S. D. , Kaufman R. J. , RA Mufson A. , Seehra J. , Jones S. S. , Hewick R. , Fritsch E. F. , RA Kawakita M. , Shimizu T. , Miyake T. ; RT "Isolation and characterization of genomic and c. DNA clones of human RT erythropoietin. "; RL Nature 313: 806 -810(1985). . CC -!- FUNCTION: ERYTHROPOIETIN IS THE PRINCIPAL HORMONE INVOLVED IN THE CC REGULATION OF ERYTHROCYTE DIFFERENTIATION AND THE MAINTENANCE OF A CC PHYSIOLOGICAL LEVEL OF CIRCULATING ERYTHROCYTE MASS. CC -!- SUBCELLULAR LOCATION: SECRETED. CC -!- TISSUE SPECIFICITY: PRODUCED BY KIDNEY OR LIVER OF ADULT MAMMALS CC AND BY LIVER OF FETAL OR NEONATAL MAMMALS. CC -!- PHARMACEUTICAL: Available under the names Epogen (Amgen) and CC Procrit (Ortho Biotech). CC -!- DATABASE: NAME=R&D Systems' cytokine source book; CC WWW="http: //www. rndsystems. com/cyt_cat/epo. html". DR EMBL; X 02158; CAA 26095. 1; -. DR EMBL; X 02157; CAA 26094. 1; -. DR EMBL; M 11319; AAA 52400. 1; -. DR EMBL; AF 053356; AAC 78791. 1; -. DR EMBL; AF 202308; AAF 23132. 1; -. DR EMBL; AF 202306; AAF 23132. 1; JOINED. . KW Erythrocyte maturation; Glycoprotein; Hormone; Signal; Pharmaceutical. FT SIGNAL 1 27 FT CHAIN 28 193 ERYTHROPOIETIN. FT PROPEP 190 193 MAY BE REMOVED IN PROCESSED PROTEIN. FT DISULFID 34 188. . .
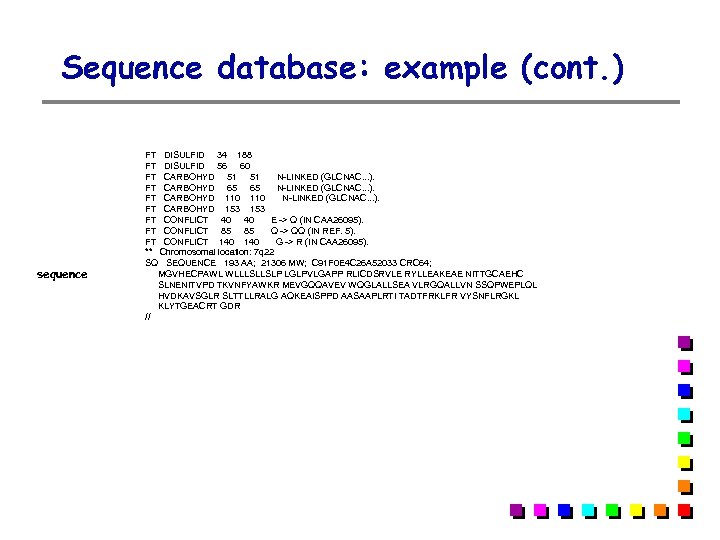
Sequence database: example (cont. ) sequence FT DISULFID 34 188 FT DISULFID 56 60 FT CARBOHYD 51 51 N-LINKED (GLCNAC. . . ). FT CARBOHYD 65 65 N-LINKED (GLCNAC. . . ). FT CARBOHYD 110 N-LINKED (GLCNAC. . . ). FT CARBOHYD 153 FT CONFLICT 40 40 E -> Q (IN CAA 26095). FT CONFLICT 85 85 Q -> QQ (IN REF. 5). FT CONFLICT 140 G -> R (IN CAA 26095). ** Chromosomal location: 7 q 22 SQ SEQUENCE 193 AA; 21306 MW; C 91 F 0 E 4 C 26 A 52033 CRC 64; MGVHECPAWL WLLLSLLSLP LGLPVLGAPP RLICDSRVLE RYLLEAKEAE NITTGCAEHC SLNENITVPD TKVNFYAWKR MEVGQQAVEV WQGLALLSEA VLRGQALLVN SSQPWEPLQL HVDKAVSGLR SLTTLLRALG AQKEAISPPD AASAAPLRTI TADTFRKLFR VYSNFLRGKL KLYTGEACRT GDR //
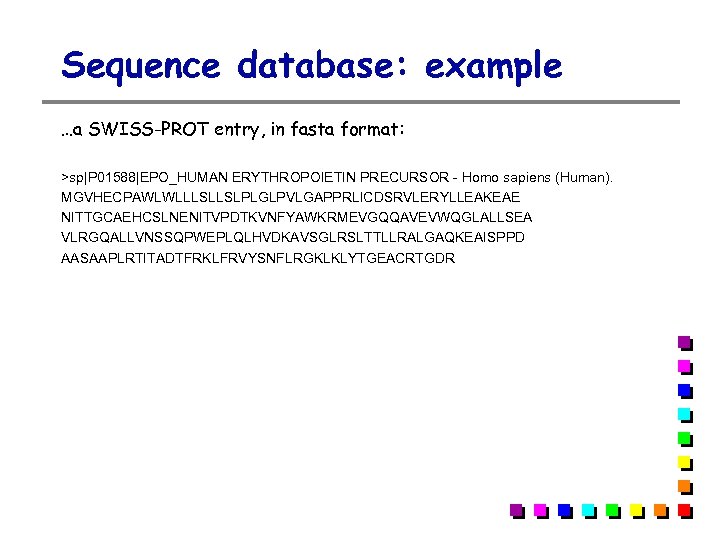
Sequence database: example …a SWISS-PROT entry, in fasta format: >sp|P 01588|EPO_HUMAN ERYTHROPOIETIN PRECURSOR - Homo sapiens (Human). MGVHECPAWLWLLLSLLSLPLGLPVLGAPPRLICDSRVLERYLLEAKEAE NITTGCAEHCSLNENITVPDTKVNFYAWKRMEVGQQAVEVWQGLALLSEA VLRGQALLVNSSQPWEPLQLHVDKAVSGLRSLTTLLRALGAQKEAISPPD AASAAPLRTITADTFRKLFRVYSNFLRGKLKLYTGEACRTGDR
Databases 1: nucleotide sequence The main DNA sequence db are EMBL (Europe)/Gen. Bank (USA) /DDBJ (Japan) There also specialized databases for the different types of RNAs (i. e. t. RNA, r. RNA, tm RNA, u. RNA, etc…) 3 D structure (DNA and RNA) Others: Aberrant splicing db; Eucaryotic promoter db (EPD); RNA editing sites, Multimedia Telomere Resource ……
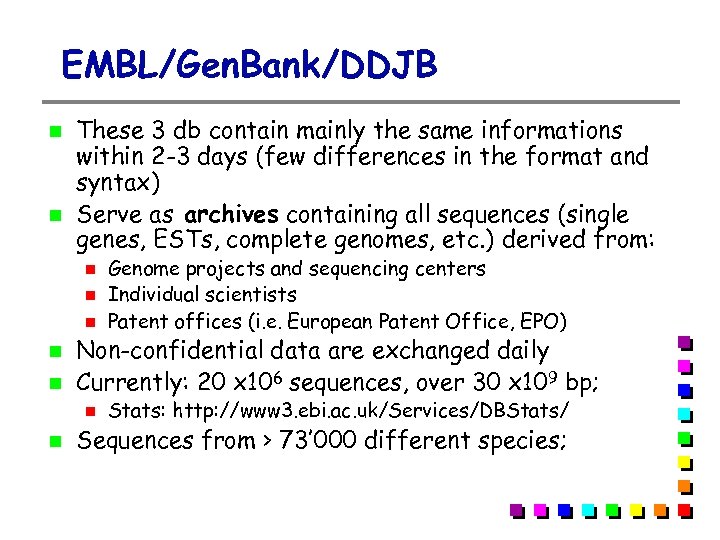
EMBL/Gen. Bank/DDJB These 3 db contain mainly the same informations within 2 -3 days (few differences in the format and syntax) Serve as archives containing all sequences (single genes, ESTs, complete genomes, etc. ) derived from: Non-confidential data are exchanged daily Currently: 20 x 106 sequences, over 30 x 109 bp; Genome projects and sequencing centers Individual scientists Patent offices (i. e. European Patent Office, EPO) Stats: http: //www 3. ebi. ac. uk/Services/DBStats/ Sequences from > 73’ 000 different species;
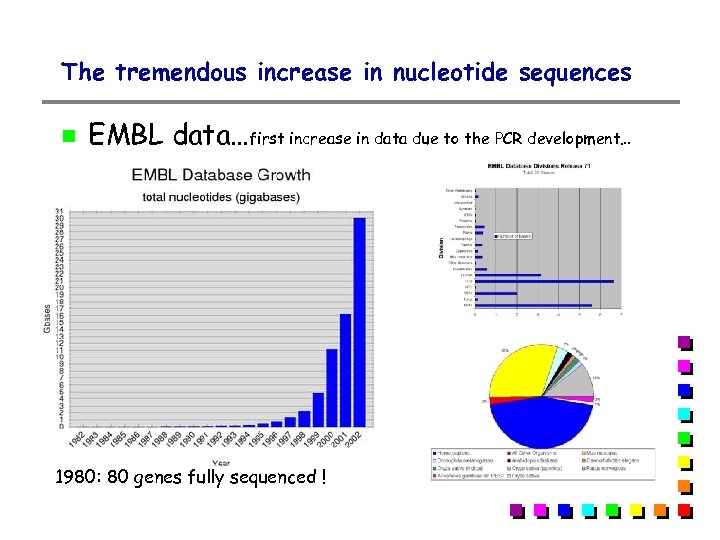
The tremendous increase in nucleotide sequences EMBL data…first increase in data due to the PCR development… 1980: 80 genes fully sequenced !
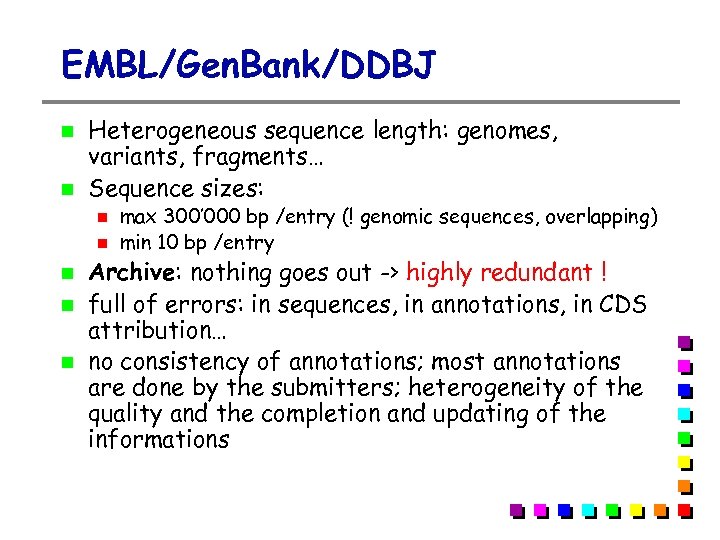
EMBL/Gen. Bank/DDBJ Heterogeneous sequence length: genomes, variants, fragments… Sequence sizes: max 300’ 000 bp /entry (! genomic sequences, overlapping) min 10 bp /entry Archive: nothing goes out -> highly redundant ! full of errors: in sequences, in annotations, in CDS attribution… no consistency of annotations; most annotations are done by the submitters; heterogeneity of the quality and the completion and updating of the informations
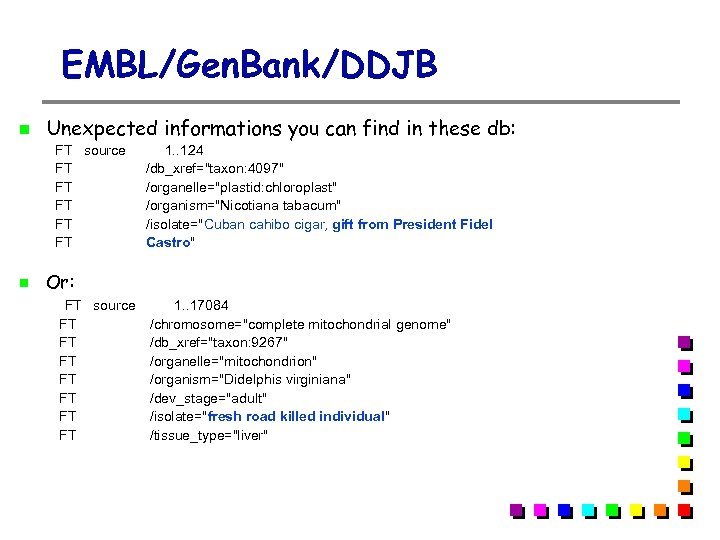
EMBL/Gen. Bank/DDJB Unexpected informations you can find in these db: FT source 1. . 124 FT /db_xref="taxon: 4097" FT /organelle="plastid: chloroplast" FT /organism="Nicotiana tabacum" FT /isolate="Cuban cahibo cigar, gift from President Fidel FT Castro" Or: FT source 1. . 17084 FT /chromosome="complete mitochondrial genome" FT /db_xref="taxon: 9267" FT /organelle="mitochondrion" FT /organism="Didelphis virginiana" FT /dev_stage="adult" FT /isolate="fresh road killed individual" FT /tissue_type="liver"
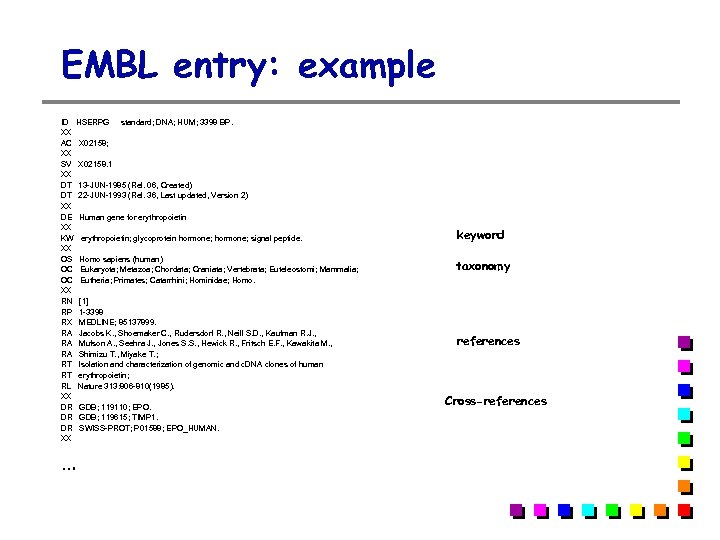
EMBL entry: example ID HSERPG standard; DNA; HUM; 3398 BP. XX AC X 02158; XX SV X 02158. 1 XX DT 13 -JUN-1985 (Rel. 06, Created) DT 22 -JUN-1993 (Rel. 36, Last updated, Version 2) XX DE Human gene for erythropoietin XX KW erythropoietin; glycoprotein hormone; signal peptide. XX OS Homo sapiens (human) OC Eukaryota; Metazoa; Chordata; Craniata; Vertebrata; Euteleostomi; Mammalia; OC Eutheria; Primates; Catarrhini; Hominidae; Homo. XX RN [1] RP 1 -3398 RX MEDLINE; 85137899. RA Jacobs K. , Shoemaker C. , Rudersdorf R. , Neill S. D. , Kaufman R. J. , RA Mufson A. , Seehra J. , Jones S. S. , Hewick R. , Fritsch E. F. , Kawakita M. , RA Shimizu T. , Miyake T. ; RT Isolation and characterization of genomic and c. DNA clones of human RT erythropoietin; RL Nature 313: 806 -810(1985). XX DR GDB; 119110; EPO. DR GDB; 119615; TIMP 1. DR SWISS-PROT; P 01588; EPO_HUMAN. XX … keyword taxonomy references Cross-references
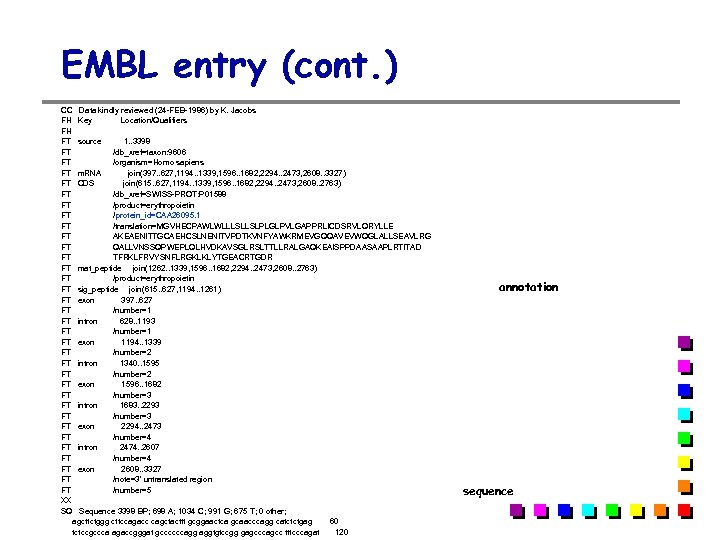
EMBL entry (cont. ) CC Data kindly reviewed (24 -FEB-1986) by K. Jacobs FH Key Location/Qualifiers FH FT source 1. . 3398 FT /db_xref=taxon: 9606 FT /organism=Homo sapiens FT m. RNA join(397. . 627, 1194. . 1339, 1596. . 1682, 2294. . 2473, 2608. . 3327) FT CDS join(615. . 627, 1194. . 1339, 1596. . 1682, 2294. . 2473, 2608. . 2763) FT /db_xref=SWISS-PROT: P 01588 FT /product=erythropoietin FT /protein_id=CAA 26095. 1 FT /translation=MGVHECPAWLWLLLSLLSLPLGLPVLGAPPRLICDSRVLQRYLLE FT AKEAENITTGCAEHCSLNENITVPDTKVNFYAWKRMEVGQQAVEVWQGLALLSEAVLRG FT QALLVNSSQPWEPLQLHVDKAVSGLRSLTTLLRALGAQKEAISPPDAASAAPLRTITAD FT TFRKLFRVYSNFLRGKLKLYTGEACRTGDR FT mat_peptide join(1262. . 1339, 1596. . 1682, 2294. . 2473, 2608. . 2763) FT /product=erythropoietin FT sig_peptide join(615. . 627, 1194. . 1261) FT exon 397. . 627 FT /number=1 FT intron 628. . 1193 FT /number=1 FT exon 1194. . 1339 FT /number=2 FT intron 1340. . 1595 FT /number=2 FT exon 1596. . 1682 FT /number=3 FT intron 1683. . 2293 FT /number=3 FT exon 2294. . 2473 FT /number=4 FT intron 2474. . 2607 FT /number=4 FT exon 2608. . 3327 FT /note=3' untranslated region FT /number=5 XX SQ Sequence 3398 BP; 698 A; 1034 C; 991 G; 675 T; 0 other; agcttctggg cttccagacc cagctacttt gcggaactca gcaacccagg catctctgag 60 tctccgccca agaccgggat gccccccagg aggtgtccgg gagcccagcc tttcccagat 120 annotation sequence
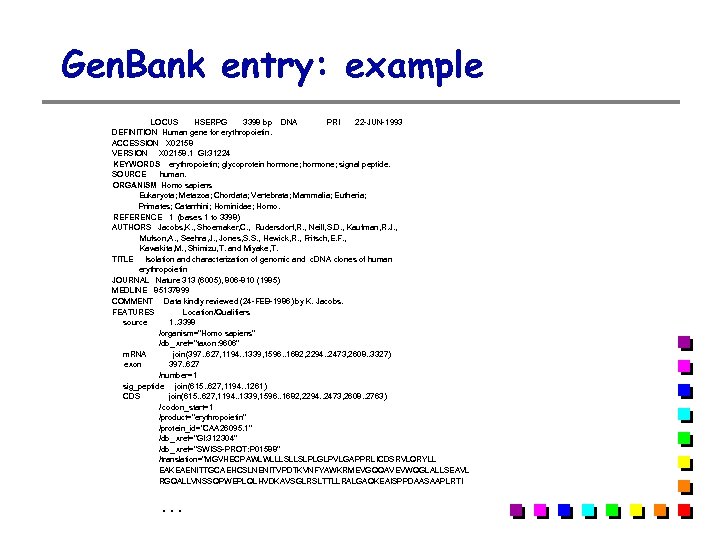
Gen. Bank entry: example LOCUS HSERPG 3398 bp DNA PRI 22 -JUN-1993 DEFINITION Human gene for erythropoietin. ACCESSION X 02158 VERSION X 02158. 1 GI: 31224 KEYWORDS erythropoietin; glycoprotein hormone; signal peptide. SOURCE human. ORGANISM Homo sapiens Eukaryota; Metazoa; Chordata; Vertebrata; Mammalia; Eutheria; Primates; Catarrhini; Hominidae; Homo. REFERENCE 1 (bases 1 to 3398) AUTHORS Jacobs, K. , Shoemaker, C. , Rudersdorf, R. , Neill, S. D. , Kaufman, R. J. , Mufson, A. , Seehra, J. , Jones, S. S. , Hewick, R. , Fritsch, E. F. , Kawakita, M. , Shimizu, T. and Miyake, T. TITLE Isolation and characterization of genomic and c. DNA clones of human erythropoietin JOURNAL Nature 313 (6005), 806 -810 (1985) MEDLINE 85137899 COMMENT Data kindly reviewed (24 -FEB-1986) by K. Jacobs. FEATURES Location/Qualifiers source 1. . 3398 /organism="Homo sapiens" /db_ xref="taxon: 9606" m. RNA join(397. . 627, 1194. . 1339, 1596. . 1682, 2294. . 2473, 2608. . 3327) exon 397. . 627 /number=1 sig_peptide join(615. . 627, 1194. . 1261) CDS join(615. . 627, 1194. . 1339, 1596. . 1682, 2294. . 2473, 2608. . 2763) / codon_start=1 /product="erythropoietin" /protein_id="CAA 26095. 1" /db_ xref="GI: 312304" /db_ xref="SWISS-PROT: P 01588" /translation="MGVHECPAWLWLLLSLLSLPLGLPVLGAPPRLICDSRVLQRYLL EAKEAENITTGCAEHCSLNENITVPDTKVNFYAWKRMEVGQQAVEVWQGLALLSEAVL RGQALLVNSSQPWEPLQLHVDKAVSGLRSLTTLLRALGAQKEAISPPDAASAAPLRTI …
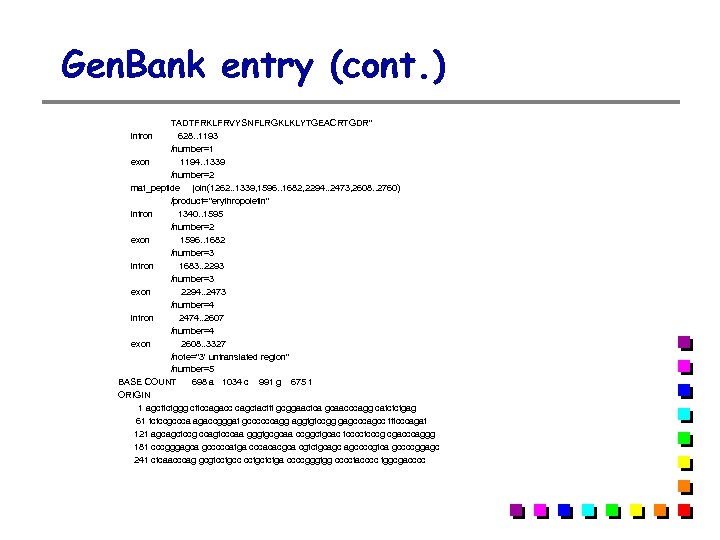
Gen. Bank entry (cont. ) TADTFRKLFRVYSNFLRGKLKLYTGEACRTGDR" intron 628. . 1193 /number=1 exon 1194. . 1339 /number=2 mat_peptide join(1262. . 1339, 1596. . 1682, 2294. . 2473, 2608. . 2760) /product="erythropoietin" intron 1340. . 1595 /number=2 exon 1596. . 1682 /number=3 intron 1683. . 2293 /number=3 exon 2294. . 2473 /number=4 intron 2474. . 2607 /number=4 exon 2608. . 3327 /note="3' untranslated region" /number=5 BASE COUNT 698 a 1034 c 991 g 675 t ORIGIN 1 agcttctggg cttccagacc cagctacttt gcggaactca gcaacccagg catctctgag 61 tctccgccca agaccgggat gccccccagg aggtgtccgg gagcccagcc tttcccagat 121 agcagctccg ccagtcccaa gggtgcgcaa ccggctgcac tcccctcccg cgacccaggg 181 cccgggagca gcccccatga cccacacgca cgtctgcagc agccccgtca gccccggagc 241 ctcaacccag gcgtcctgcc cctgctctga ccccgggtgg cccctacccc tggcgacccc
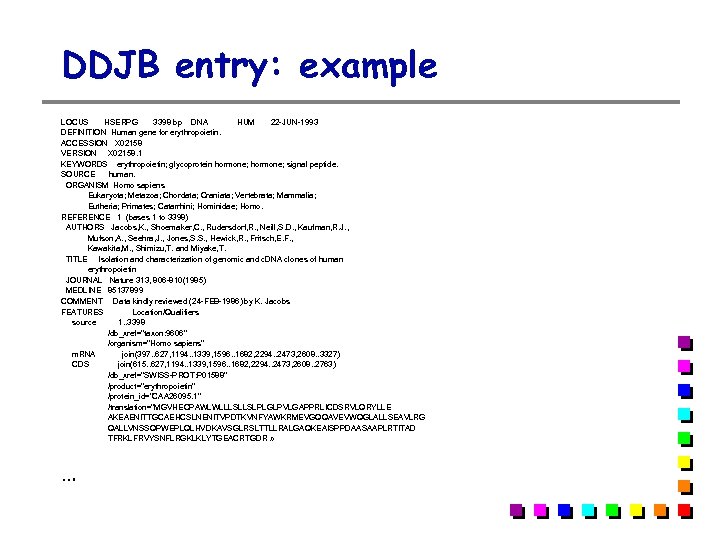
DDJB entry: example LOCUS HSERPG 3398 bp DNA HUM 22 -JUN-1993 DEFINITION Human gene for erythropoietin. ACCESSION X 02158 VERSION X 02158. 1 KEYWORDS erythropoietin; glycoprotein hormone; signal peptide. SOURCE human. ORGANISM Homo sapiens Eukaryota; Metazoa; Chordata; Craniata; Vertebrata; Mammalia; Eutheria; Primates; Catarrhini; Hominidae; Homo. REFERENCE 1 (bases 1 to 3398) AUTHORS Jacobs, K. , Shoemaker, C. , Rudersdorf, R. , Neill, S. D. , Kaufman, R. J. , Mufson, A. , Seehra, J. , Jones, S. S. , Hewick, R. , Fritsch, E. F. , Kawakita, M. , Shimizu, T. and Miyake, T. TITLE Isolation and characterization of genomic and c. DNA clones of human erythropoietin JOURNAL Nature 313, 806 -810(1985) MEDLINE 85137899 COMMENT Data kindly reviewed (24 -FEB-1986) by K. Jacobs FEATURES Location/Qualifiers source 1. . 3398 /db_xref="taxon: 9606" /organism="Homo sapiens" m. RNA join(397. . 627, 1194. . 1339, 1596. . 1682, 2294. . 2473, 2608. . 3327) CDS join(615. . 627, 1194. . 1339, 1596. . 1682, 2294. . 2473, 2608. . 2763) /db_xref="SWISS-PROT: P 01588" /product="erythropoietin" /protein_id="CAA 26095. 1" /translation="MGVHECPAWLWLLLSLLSLPLGLPVLGAPPRLICDSRVLQRYLLE AKEAENITTGCAEHCSLNENITVPDTKVNFYAWKRMEVGQQAVEVWQGLALLSEAVLRG QALLVNSSQPWEPLQLHVDKAVSGLRSLTTLLRALGAQKEAISPPDAASAAPLRTITAD TFRKLFRVYSNFLRGKLKLYTGEACRTGDR » …
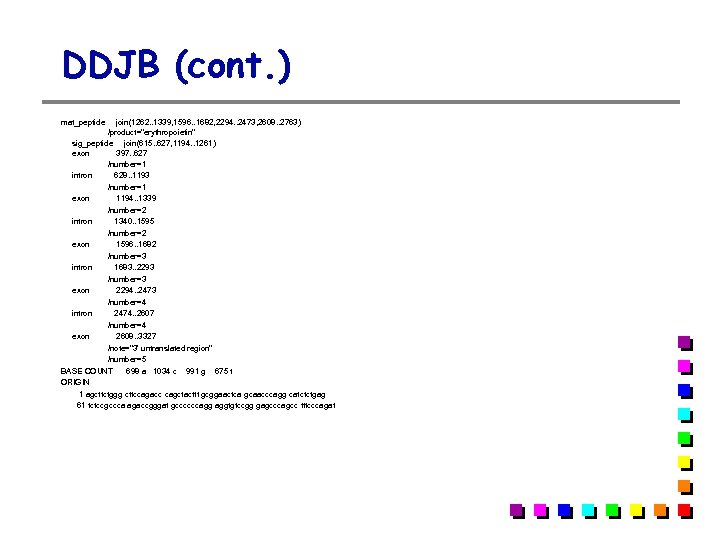
DDJB (cont. ) mat_peptide join(1262. . 1339, 1596. . 1682, 2294. . 2473, 2608. . 2763) /product="erythropoietin" sig_peptide join(615. . 627, 1194. . 1261) exon 397. . 627 /number=1 intron 628. . 1193 /number=1 exon 1194. . 1339 /number=2 intron 1340. . 1595 /number=2 exon 1596. . 1682 /number=3 intron 1683. . 2293 /number=3 exon 2294. . 2473 /number=4 intron 2474. . 2607 /number=4 exon 2608. . 3327 /note="3' untranslated region" /number=5 BASE COUNT 698 a 1034 c 991 g 675 t ORIGIN 1 agcttctggg cttccagacc cagctacttt gcggaactca gcaacccagg catctctgag 61 tctccgccca agaccgggat gccccccagg aggtgtccgg gagcccagcc tttcccagat
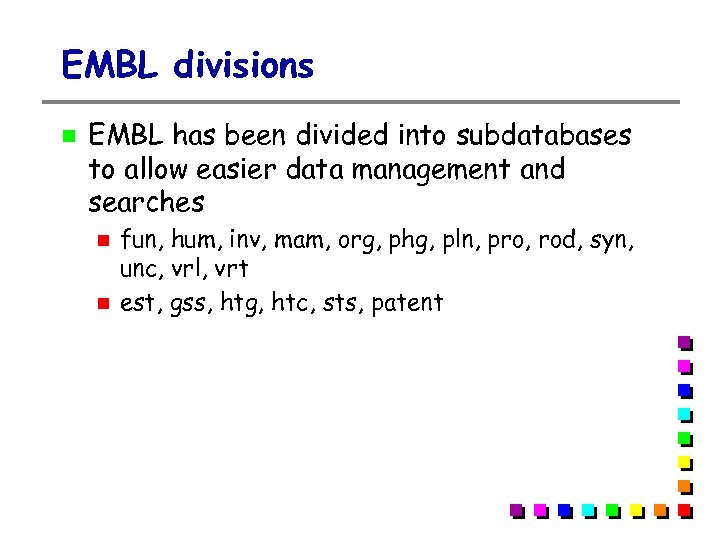
EMBL divisions EMBL has been divided into subdatabases to allow easier data management and searches fun, hum, inv, mam, org, phg, pln, pro, rod, syn, unc, vrl, vrt est, gss, htg, htc, sts, patent
EMBL: The Genome divisions http: //www. ebi. ac. uk/genomes/ Schizosaccharomyces pombe strain 972 h- complete genome
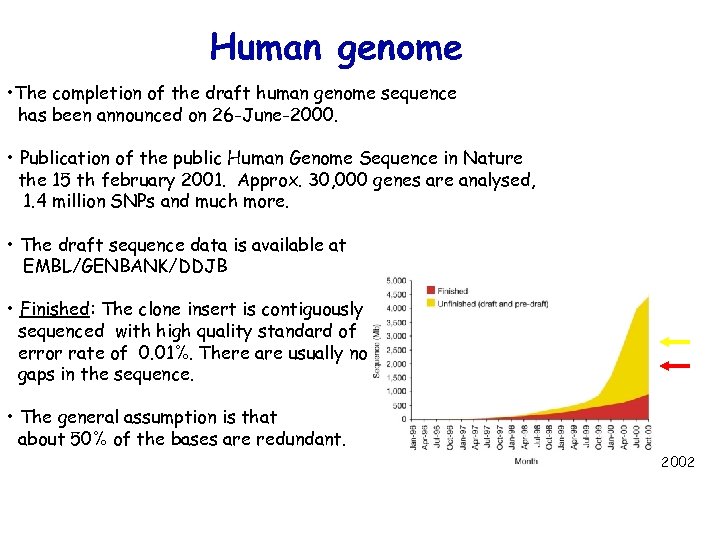
Human genome • The completion of the draft human genome sequence has been announced on 26 -June-2000. • Publication of the public Human Genome Sequence in Nature the 15 th february 2001. Approx. 30, 000 genes are analysed, 1. 4 million SNPs and much more. • The draft sequence data is available at EMBL/GENBANK/DDJB • Finished: The clone insert is contiguously sequenced with high quality standard of error rate of 0. 01%. There are usually no gaps in the sequence. • The general assumption is that about 50% of the bases are redundant. 2002
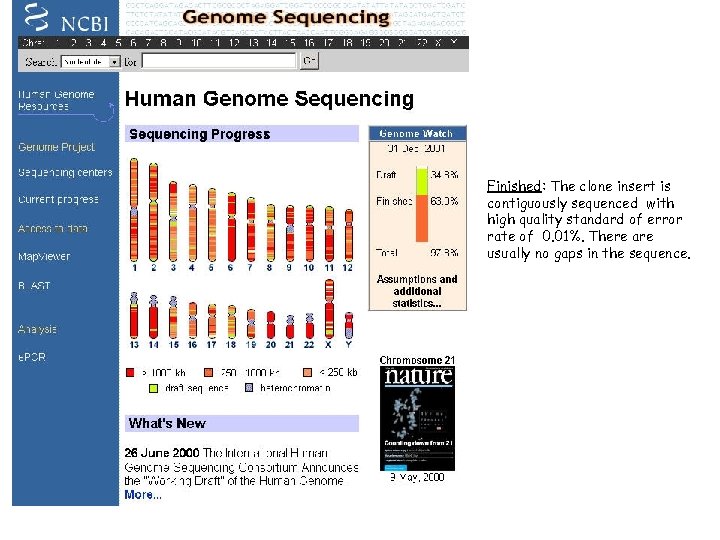
Finished: The clone insert is contiguously sequenced with high quality standard of error rate of 0. 01%. There are usually no gaps in the sequence.

Nucleotid databases and « associated » genomic projects/databases Problem: Redundancy = makes Blasts searches of the complete databases useless for detecting anything behond the closest homologs. Solutions: • assemblies of genomic sequence data (contigs) and corresponding RNA and protein sequences -> dataset of genomic contigs, RNAs and proteins • annotation of genes, RNAs, proteins, variation (SNPs), STS markers, gene prediction, nomenclature and chromosomal location. • compute connexions to other resources (cross-references) Examples: Ref. Seq/Locus link (drosophila, human, mouse, rat and zebrafish), TIGR (microbes and plants), Ens. EMBL (Eukaryota)…
Locus. Link / Ref. Seq Erythropoitin receptor
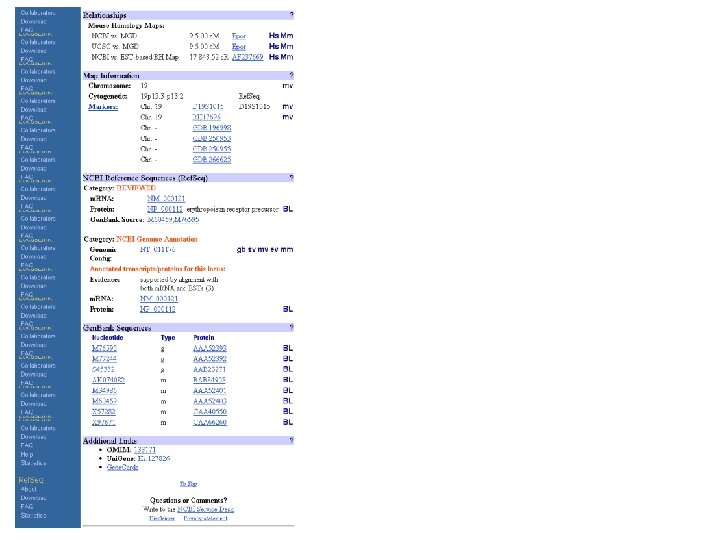
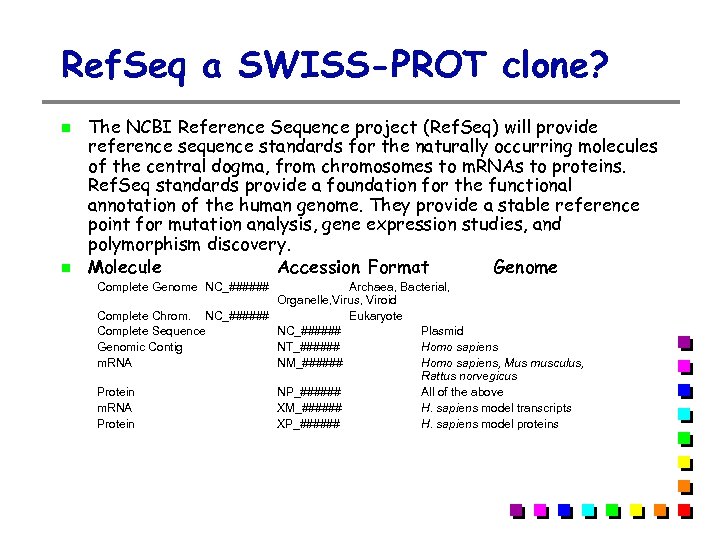
Ref. Seq a SWISS-PROT clone? The NCBI Reference Sequence project (Ref. Seq) will provide reference sequence standards for the naturally occurring molecules of the central dogma, from chromosomes to m. RNAs to proteins. Ref. Seq standards provide a foundation for the functional annotation of the human genome. They provide a stable reference point for mutation analysis, gene expression studies, and polymorphism discovery. Molecule Accession Format Genome Complete Genome NC_###### Archaea, Bacterial, Organelle, Virus, Viroid Complete Chrom. NC_###### Eukaryote Complete Sequence NC_###### Plasmid Genomic Contig NT_###### Homo sapiens m. RNA NM_###### Homo sapiens, Mus musculus, Rattus norvegicus Protein NP_###### All of the above m. RNA XM_###### H. sapiens model transcripts Protein XP_###### H. sapiens model proteins
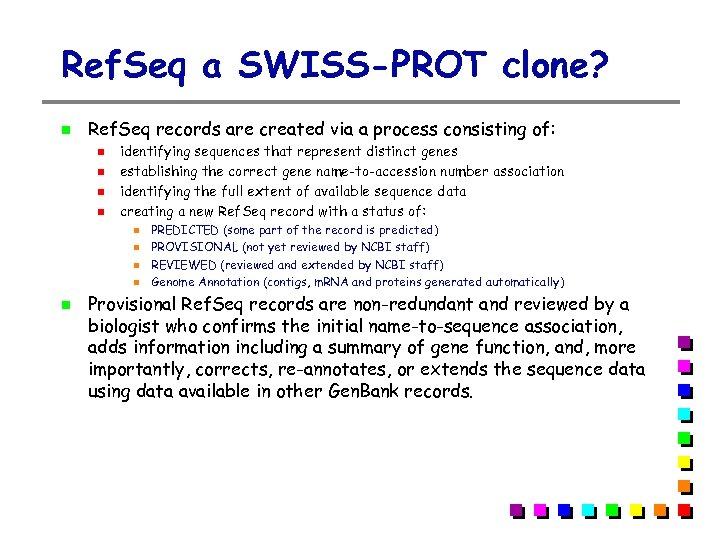
Ref. Seq a SWISS-PROT clone? Ref. Seq records are created via a process consisting of: identifying sequences that represent distinct genes establishing the correct gene name-to-accession number association identifying the full extent of available sequence data creating a new Ref. Seq record with a status of: PREDICTED (some part of the record is predicted) PROVISIONAL (not yet reviewed by NCBI staff) REVIEWED (reviewed and extended by NCBI staff) Genome Annotation (contigs, m. RNA and proteins generated automatically) Provisional Ref. Seq records are non-redundant and reviewed by a biologist who confirms the initial name-to-sequence association, adds information including a summary of gene function, and, more importantly, corrects, re-annotates, or extends the sequence data using data available in other Gen. Bank records.
ESTs and Unigene is an ongoing effort at NCBI to cluster EST sequences with traditional gene sequences For each cluster, there is a lot of additional information included Unigene is regularly rebuilt. Therefore, cluster identifiers are not stable gene indices Species: Human, Mouse, Rat, Cow, Zebrafish, and recently also Frog, Cress, Rice, Barley, Maize, Wheat
Databases 2: genomics Contain information on genes, gene location (mapping), gene nomenclature and links to sequence databases; usually no sequence! Exist for most organisms important for life science research; species specific. Examples: MIM, GDB (human), MGD (mouse), Fly. Base (Drosophila), SGD (yeast), Maize. DB (maize), Subti. List (B. subtilis), etc. ; Format: generally relational (Oracle, Sy. Base or Ace. Db).
MIM OMIM™: Online Mendelian Inheritance in Man a catalog of human genes and genetic disorders contains a summary of literature, pictures, and reference information. It also contains numerous links to articles and sequence information.

MIM OMIM™: Online Mendelian Inheritance in Man catalog of human genes and genetic disorders contains a summary of literature and reference information. It also contains links to publications and sequence information.


Genecard an electronic encyclopedia of biological and medical information based on intelligent knowledge navigation technology
http: //www. genelynx. org/
Collections of hyperlinks for each human gene
Ensembl Contains all the human genome DNA sequences currently available in the public domain. Automated annotation: by using different software tools, features are identified in the DNA sequences: Genes (known or predicted) Single nucleotide polymorphisms (SNPs) Repeats Homologies Created and maintained by the EBI and the Sanger Center (UK) www. ensembl. org
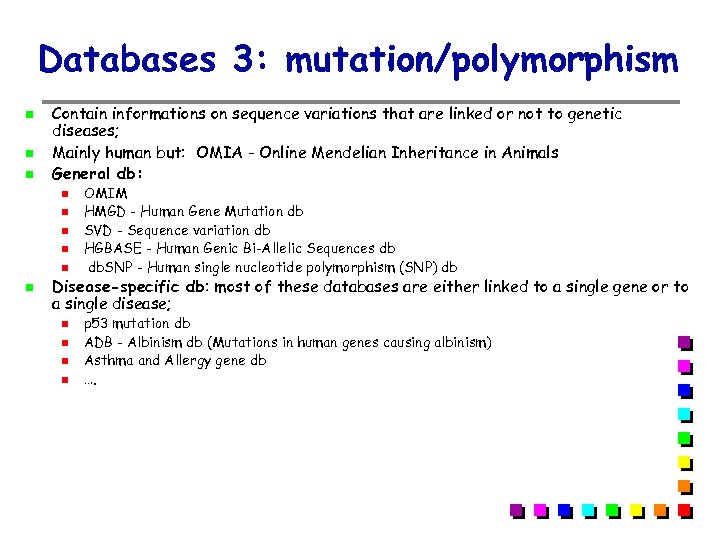
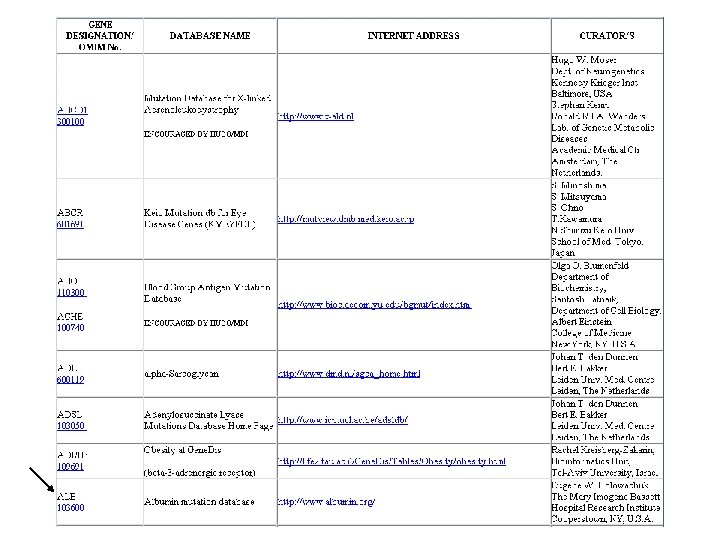
Databases 3: mutation/polymorphism Contain informations on sequence variations that are linked or not to genetic diseases; Mainly human but: OMIA - Online Mendelian Inheritance in Animals General db: OMIM HMGD - Human Gene Mutation db SVD - Sequence variation db HGBASE - Human Genic Bi-Allelic Sequences db db. SNP - Human single nucleotide polymorphism (SNP) db Disease-specific db: most of these databases are either linked to a single gene or to a single disease; p 53 mutation db ADB - Albinism db (Mutations in human genes causing albinism) Asthma and Allergy gene db ….
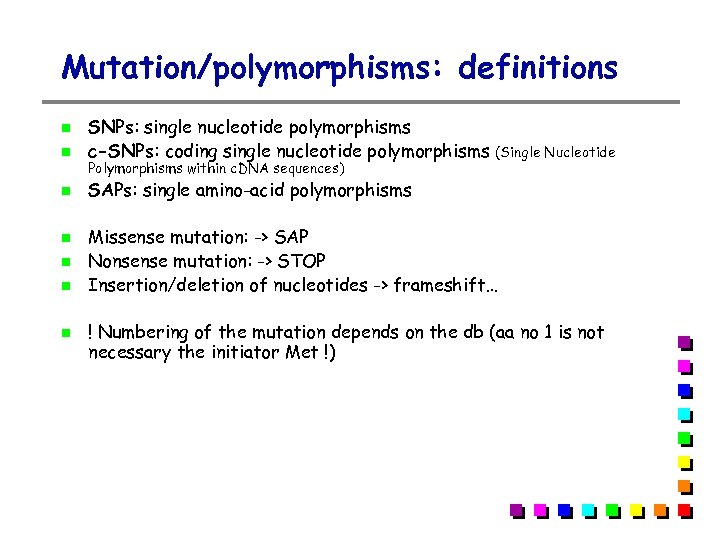
Mutation/polymorphisms: definitions SNPs: single nucleotide polymorphisms c-SNPs: coding single nucleotide polymorphisms SAPs: single amino-acid polymorphisms Polymorphisms within c. DNA sequences) (Single Nucleotide Missense mutation: -> SAP Nonsense mutation: -> STOP Insertion/deletion of nucleotides -> frameshift… ! Numbering of the mutation depends on the db (aa no 1 is not necessary the initiator Met !)
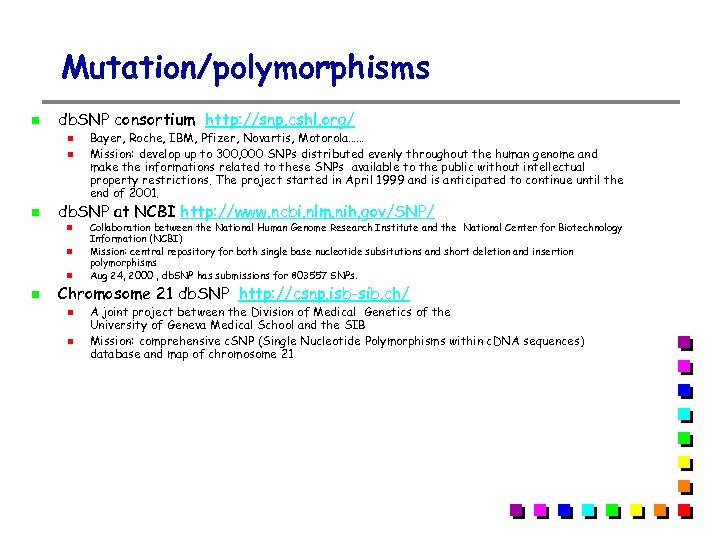
Mutation/polymorphisms db. SNP consortium http: //snp. cshl. org/ db. SNP at NCBI http: //www. ncbi. nlm. nih. gov/SNP/ Bayer, Roche, IBM, Pfizer, Novartis, Motorola…… Mission: develop up to 300, 000 SNPs distributed evenly throughout the human genome and make the informations related to these SNPs available to the public without intellectual property restrictions. The project started in April 1999 and is anticipated to continue until the end of 2001. Collaboration between the National Human Genome Research Institute and the National Center for Biotechnology Information (NCBI) Mission: central repository for both single base nucleotide subsitutions and short deletion and insertion polymorphisms Aug 24, 2000 , db. SNP has submissions for 803557 SNPs. Chromosome 21 db. SNP http: //csnp. isb-sib. ch/ A joint project between the Division of Medical Genetics of the University of Geneva Medical School and the SIB Mission: comprehensive c. SNP (Single Nucleotide Polymorphisms within c. DNA sequences) database and map of chromosome 21
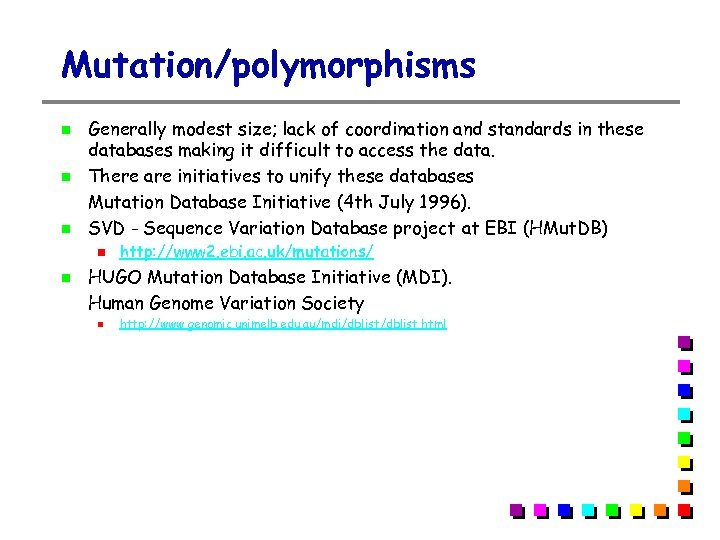
Mutation/polymorphisms Generally modest size; lack of coordination and standards in these databases making it difficult to access the data. There are initiatives to unify these databases Mutation Database Initiative (4 th July 1996). SVD - Sequence Variation Database project at EBI (HMut. DB) http: //www 2. ebi. ac. uk/mutations/ HUGO Mutation Database Initiative (MDI). Human Genome Variation Society http: //www. genomic. unimelb. edu. au/mdi/dblist. html

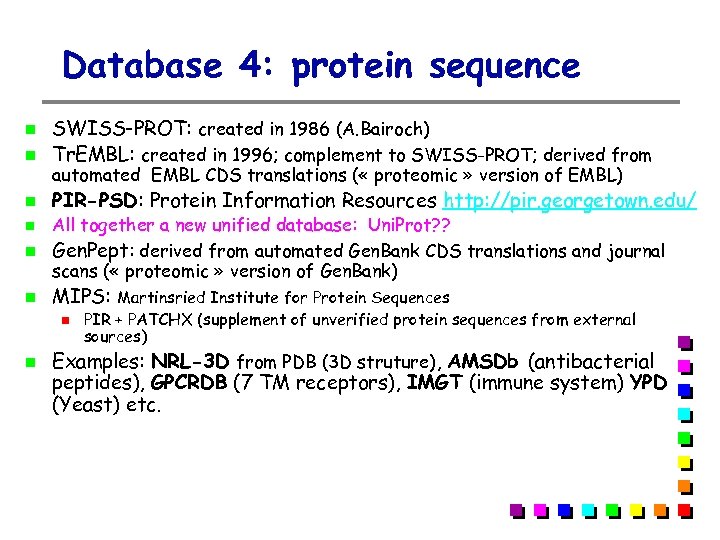
Database 4: protein sequence SWISS-PROT: created in 1986 (A. Bairoch) Tr. EMBL: created in 1996; complement to SWISS-PROT; derived from PIR-PSD: Protein Information Resources http: //pir. georgetown. edu/ automated EMBL CDS translations ( « proteomic » version of EMBL) All together a new unified database: Uni. Prot? ? Gen. Pept: derived from automated Gen. Bank CDS translations and journal scans ( « proteomic » version of Gen. Bank) MIPS: Martinsried Institute for Protein Sequences PIR + PATCHX (supplement of unverified protein sequences from external sources) Examples: NRL-3 D from PDB (3 D struture), AMSDb (antibacterial peptides), GPCRDB (7 TM receptors), IMGT (immune system) YPD (Yeast) etc.
SWISS-PROT Collaboration between the SIB (CH) and EMBL/EBI (UK) Annotated (manually), non-redundant, crossreferenced, documented protein sequence database. ~113 ’ 000 sequences from more than 6’ 800 different species; 70 ’ 000 references (publications); 550 ’ 000 cross-references (databases); ~200 Mb of annotations. Weekly releases; available from about 50 servers across the world, the main source being Ex. PASy
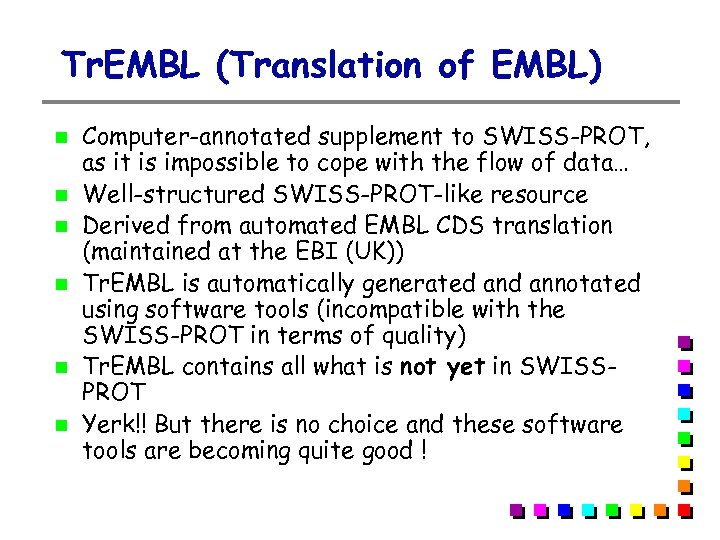
Tr. EMBL (Translation of EMBL) Computer-annotated supplement to SWISS-PROT, as it is impossible to cope with the flow of data… Well-structured SWISS-PROT-like resource Derived from automated EMBL CDS translation (maintained at the EBI (UK)) Tr. EMBL is automatically generated annotated using software tools (incompatible with the SWISS-PROT in terms of quality) Tr. EMBL contains all what is not yet in SWISSPROT Yerk!! But there is no choice and these software tools are becoming quite good !
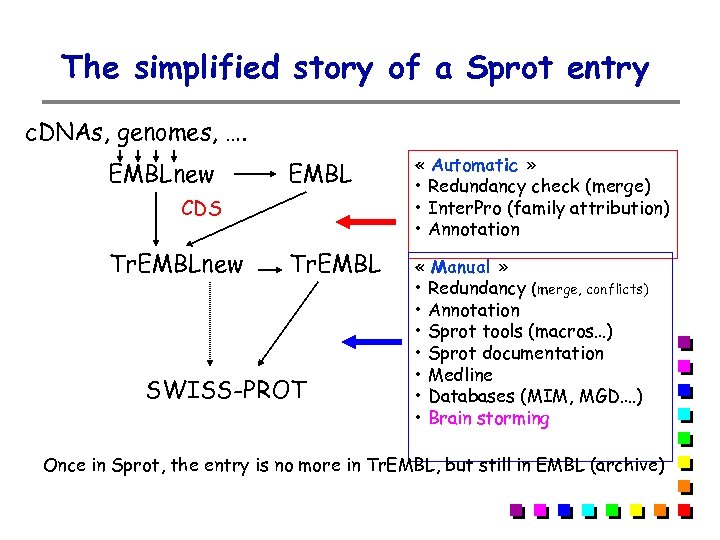
The simplified story of a Sprot entry c. DNAs, genomes, …. EMBLnew EMBL « Automatic » • Redundancy check (merge) • Inter. Pro (family attribution) • Annotation Tr. EMBL « Manual » • Redundancy (merge, conflicts) • Annotation • Sprot tools (macros…) • Sprot documentation • Medline • Databases (MIM, MGD…. ) • Brain storming CDS Tr. EMBLnew SWISS-PROT Once in Sprot, the entry is no more in Tr. EMBL, but still in EMBL (archive)
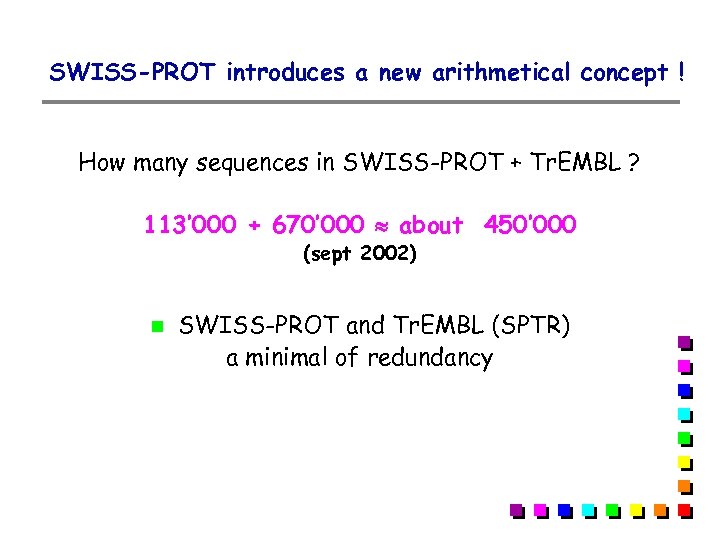
SWISS-PROT introduces a new arithmetical concept ! How many sequences in SWISS-PROT + Tr. EMBL ? 113’ 000 + 670’ 000 about 450’ 000 (sept 2002) SWISS-PROT and Tr. EMBL (SPTR) a minimal of redundancy
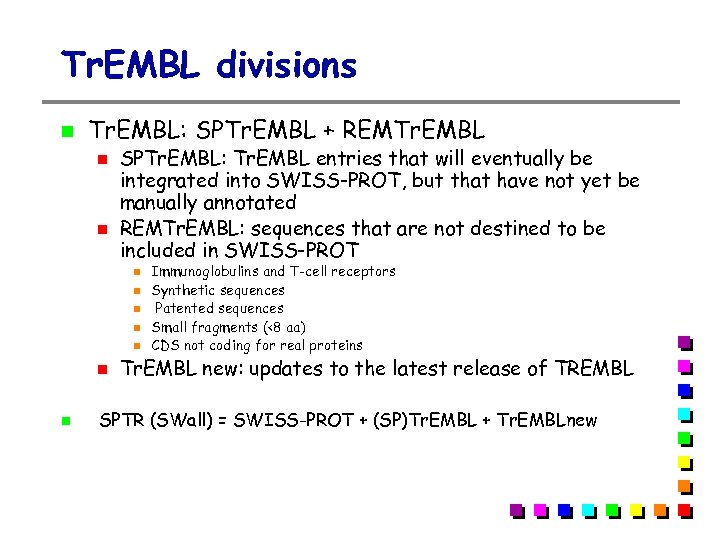
Tr. EMBL divisions Tr. EMBL: SPTr. EMBL + REMTr. EMBL SPTr. EMBL: Tr. EMBL entries that will eventually be integrated into SWISS-PROT, but that have not yet be manually annotated REMTr. EMBL: sequences that are not destined to be included in SWISS-PROT Immunoglobulins and T-cell receptors Synthetic sequences Patented sequences Small fragments (<8 aa) CDS not coding for real proteins Tr. EMBL new: updates to the latest release of TREMBL SPTR (SWall) = SWISS-PROT + (SP)Tr. EMBL + Tr. EMBLnew
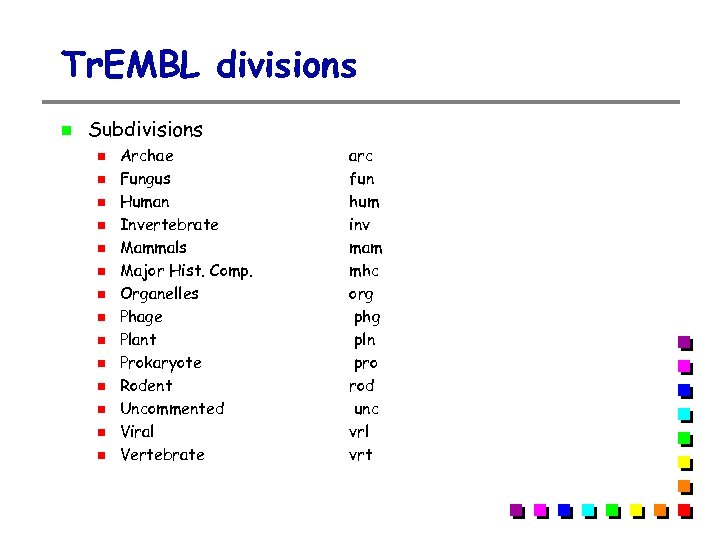
Tr. EMBL divisions Subdivisions Archae Fungus Human Invertebrate Mammals Major Hist. Comp. Organelles Phage Plant Prokaryote Rodent Uncommented Viral Vertebrate arc fun hum inv mam mhc org phg pln pro rod unc vrl vrt
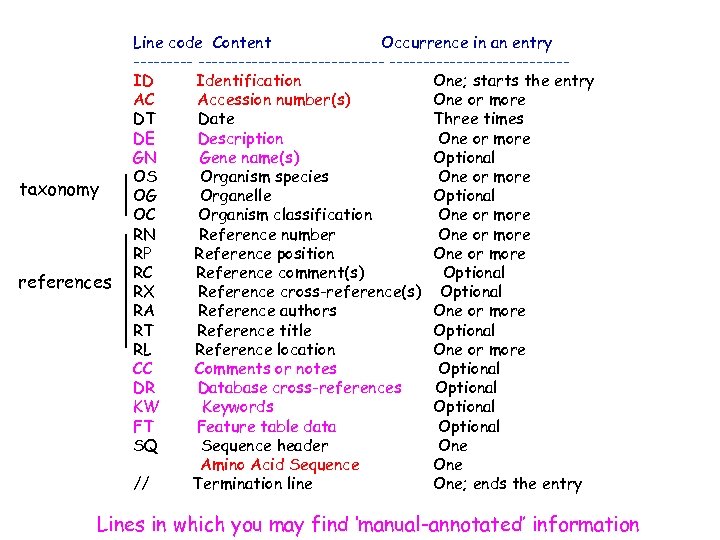
taxonomy references Line code Content Occurrence in an entry -------------------ID Identification One; starts the entry AC Accession number(s) One or more DT Date Three times DE Description One or more GN Gene name(s) Optional OS Organism species One or more OG Organelle Optional OC Organism classification One or more RN Reference number One or more RP Reference position One or more RC Reference comment(s) Optional RX Reference cross-reference(s) Optional RA Reference authors One or more RT Reference title Optional RL Reference location One or more CC Comments or notes Optional DR Database cross-references Optional KW Keywords Optional FT Feature table data Optional SQ Sequence header One Amino Acid Sequence One // Termination line One; ends the entry Lines in which you may find ‘manual-annotated’ information

a Swiss-Prot entry… overview Entry name Accession number sequence

Protein name Gene name Taxonomy

References

Comments

Cross-references

Keywords

Feature table (sequence description)

Tr. EMBL: example Original Tr. EMBL entry which has been integrated into the SWISS-PROT EPO_HUMAN entry and thus which is not found in Tr. EMBL anymore.

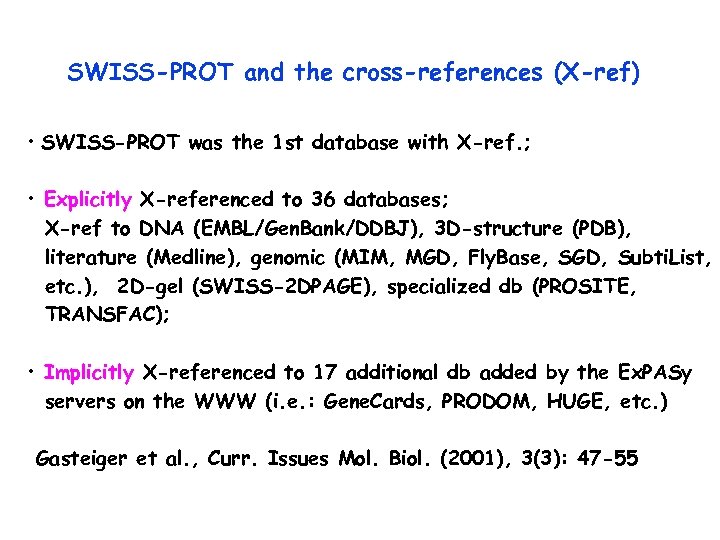
SWISS-PROT and the cross-references (X-ref) • SWISS-PROT was the 1 st database with X-ref. ; • Explicitly X-referenced to 36 databases; X-ref to DNA (EMBL/Gen. Bank/DDBJ), 3 D-structure (PDB), literature (Medline), genomic (MIM, MGD, Fly. Base, SGD, Subti. List, etc. ), 2 D-gel (SWISS-2 DPAGE), specialized db (PROSITE, TRANSFAC); • Implicitly X-referenced to 17 additional db added by the Ex. PASy servers on the WWW (i. e. : Gene. Cards, PRODOM, HUGE, etc. ) Gasteiger et al. , Curr. Issues Mol. Biol. (2001), 3(3): 47 -55
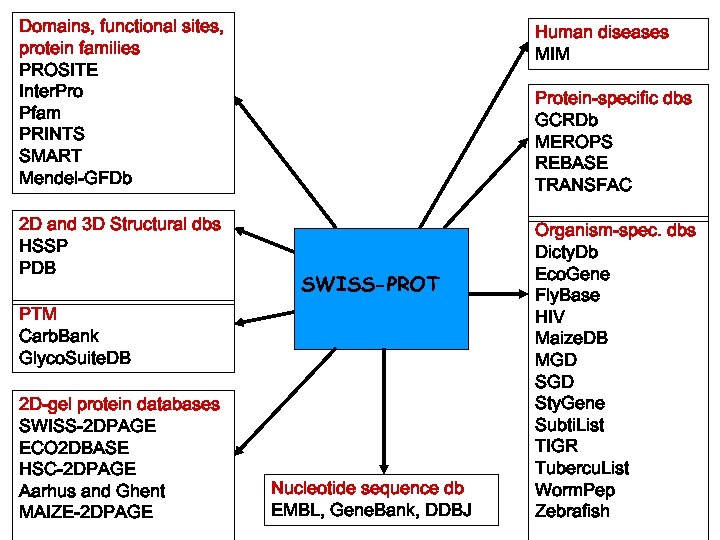
Domains, functional sites, protein families PROSITE Inter. Pro Pfam PRINTS SMART Mendel-GFDb Human diseases MIM 2 D and 3 D Structural dbs HSSP PDB Organism-spec. dbs Dicty. Db Eco. Gene Fly. Base HIV Maize. DB MGD Sty. Gene Subti. List TIGR Tubercu. List Worm. Pep Zebrafish Protein-specific dbs GCRDb MEROPS REBASE TRANSFAC SWISS-PROT PTM Carb. Bank Glyco. Suite. DB 2 D-gel protein databases SWISS-2 DPAGE ECO 2 DBASE HSC-2 DPAGE Aarhus and Ghent MAIZE-2 DPAGE Nucleotide sequence db EMBL, Gene. Bank, DDBJ
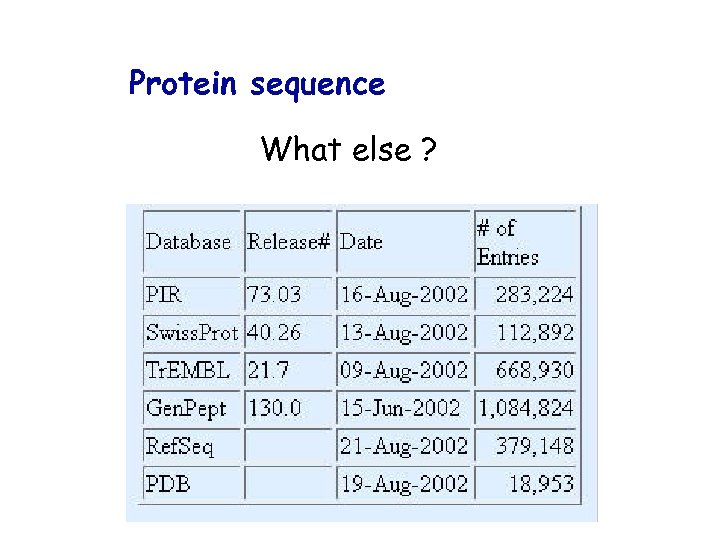
Protein sequence What else ?
http: //pir. georgetown. edu/

PIR-PSD: example « well annotated »
Gen. Pept (translation of Gen. Bank) Gen. Pept is a protein database translated from the last release of Gen. Bank (+ journal scans) The current release has > 1 million entries In contrast to Tr. EMBL, keeps all protein sequences including small fragments (< 8 aa), immunoglobulins…. Redundancy: > 20 entries for human EPO
When Amos dreams…
Database 5: protein domain/family Contains biologically significant « pattern / profiles/ HMM » formulated in such a way that, with appropriate computional tools, it can rapidly and reliably determine to which known family of proteins (if any) a new sequence belongs to -> tools to identify what is the function of uncharacterized proteins translated from genomic or c. DNA sequences ( « functional diagnostic » )
Database 4: protein domain/family Contains biologically significant « pattern / profiles/ HMM » formulated in such a way that, with appropriate computional tools, it can rapidly and reliably determine to which known family of proteins (if any) a new sequence belongs to -> tools to identify what is the function of uncharacterized proteins translated from genomic or c. DNA sequences ( « functional diagnostic » )
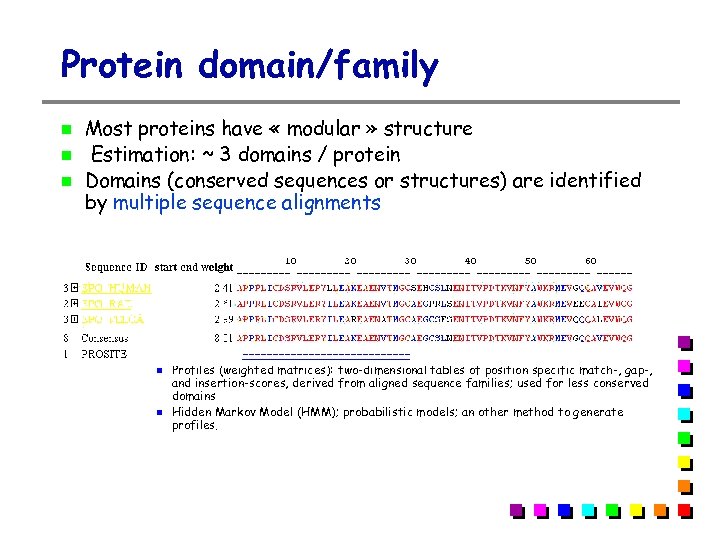
Protein domain/family Most proteins have « modular » structure Estimation: ~ 3 domains / protein Domains (conserved sequences or structures) are identified by multiple sequence alignments Domains can be defined by different methods: Pattern (regular expression); used for very conserved domains Profiles (weighted matrices): two-dimensional tables of position specific match-, gap-, and insertion-scores, derived from aligned sequence families; used for less conserved domains Hidden Markov Model (HMM); probabilistic models; an other method to generate profiles.
Protein domain/family db Secondary databases are the fruit of analyses of the sequences found in the primary sequence db Either manually curated (i. e. PROSITE, Pfam, etc. ) or automatically generated (i. e. Pro. Dom, DOMO) Some depend on the method used to detect if a protein belongs to a particular domain/family (patterns, profiles, HMM, PSI-BLAST)
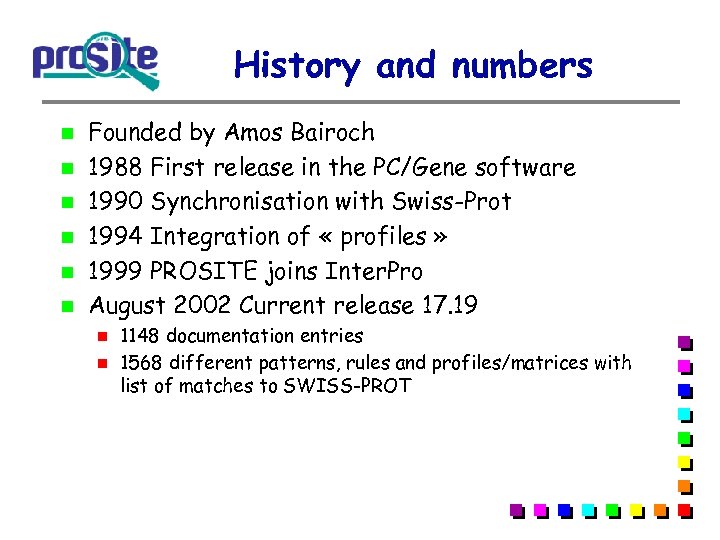
History and numbers Founded by Amos Bairoch 1988 First release in the PC/Gene software 1990 Synchronisation with Swiss-Prot 1994 Integration of « profiles » 1999 PROSITE joins Inter. Pro August 2002 Current release 17. 19 1148 documentation entries 1568 different patterns, rules and profiles/matrices with list of matches to SWISS-PROT
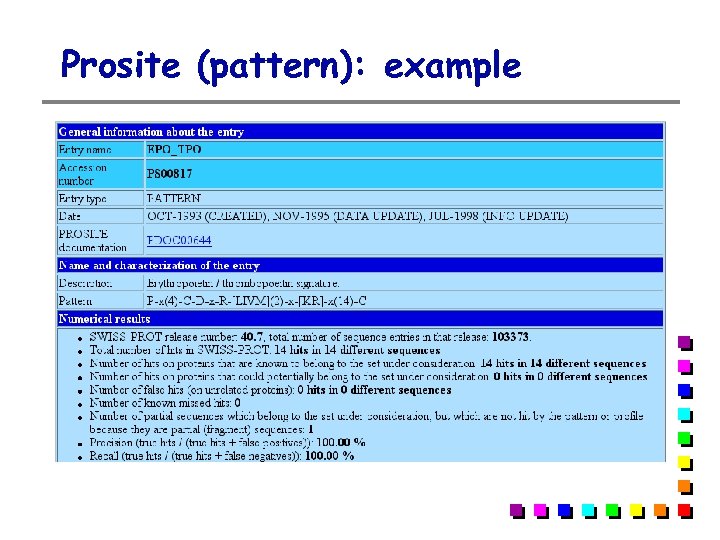
Prosite (pattern): example
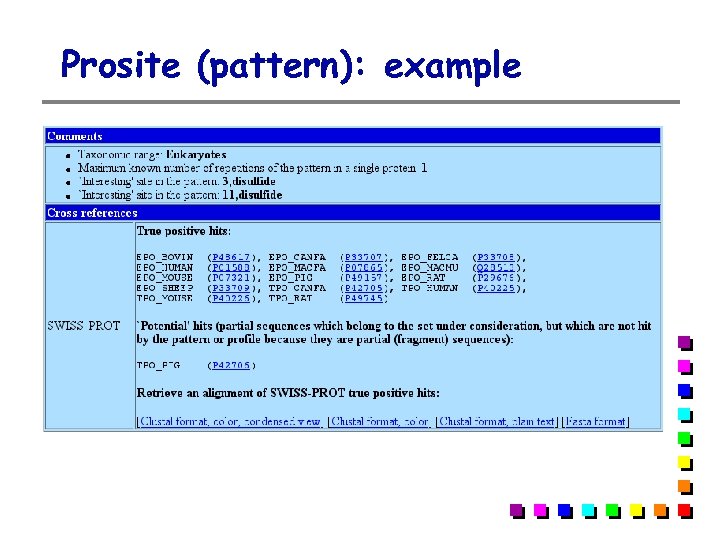
Prosite (pattern): example
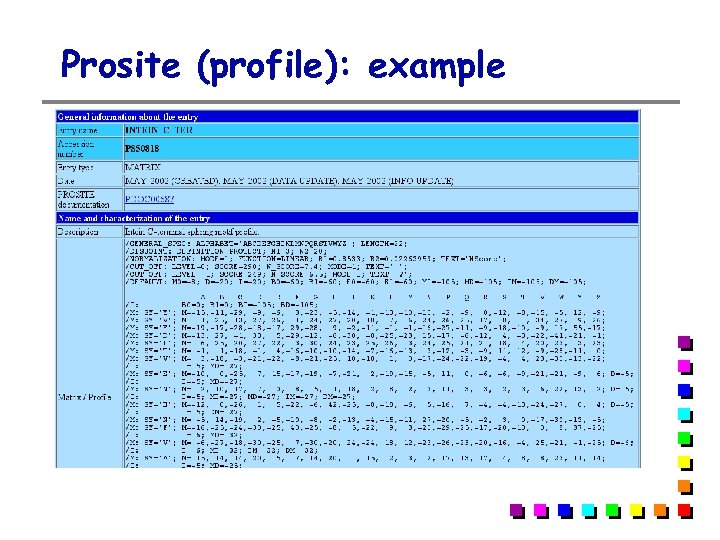
Prosite (profile): example
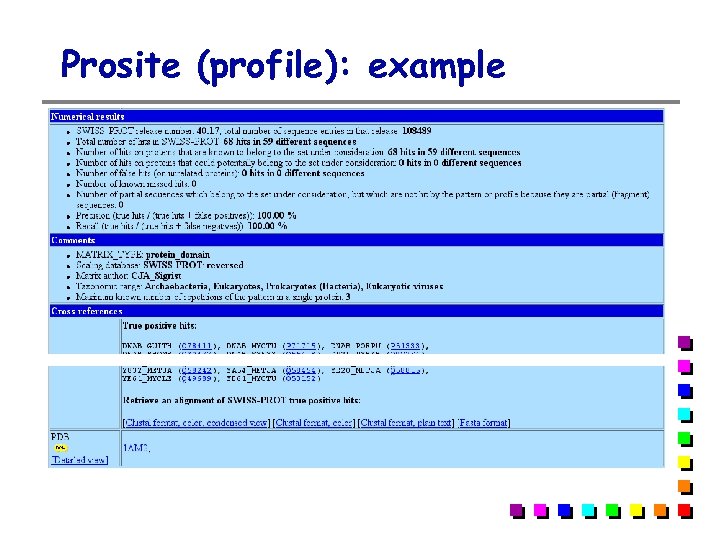
Prosite (profile): example
Protein domain/family db PROSITE Pro. Dom PRINTS Pfam SMART TIGRfam Patterns / Profiles Aligned motifs (PSI-BLAST) (Pfam B) Aligned motifs HMM (Hidden Markov Models) HMM DOMO BLOCKS CDD(CDART) Aligned motifs (PSI-BLAST) PSI-BLAST(PSSM) of Pfam and SMART I n t e r p r o

Inter. Pro: www. ebi. ac. uk/interpro
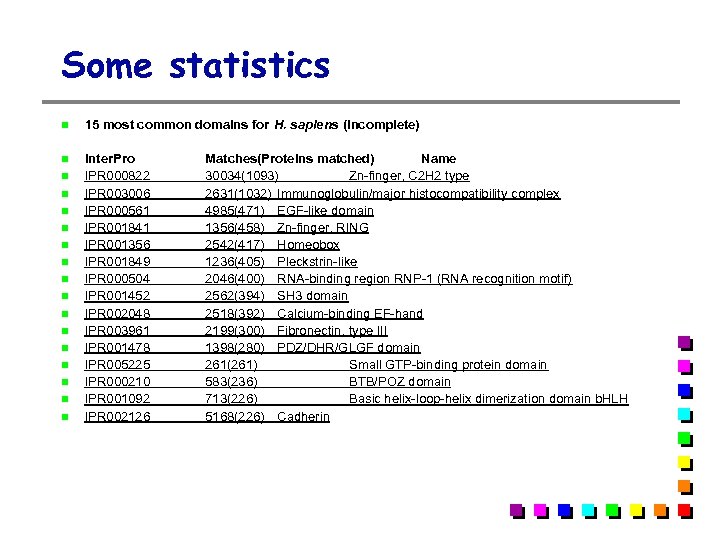
Some statistics 15 most common domains for H. sapiens (Incomplete) Inter. Pro IPR 000822 IPR 003006 IPR 000561 IPR 001841 IPR 001356 IPR 001849 IPR 000504 IPR 001452 IPR 002048 IPR 003961 IPR 001478 IPR 005225 IPR 000210 IPR 001092 IPR 002126 Matches(Proteins matched) Name 30034(1093) Zn-finger, C 2 H 2 type 2631(1032) Immunoglobulin/major histocompatibility complex 4985(471) EGF-like domain 1356(458) Zn-finger, RING 2542(417) Homeobox 1236(405) Pleckstrin-like 2046(400) RNA-binding region RNP-1 (RNA recognition motif) 2562(394) SH 3 domain 2518(392) Calcium-binding EF-hand 2199(300) Fibronectin, type III 1398(280) PDZ/DHR/GLGF domain 261(261) Small GTP-binding protein domain 583(236) BTB/POZ domain 713(226) Basic helix-loop-helix dimerization domain b. HLH 5168(226) Cadherin

Inter. Pro example
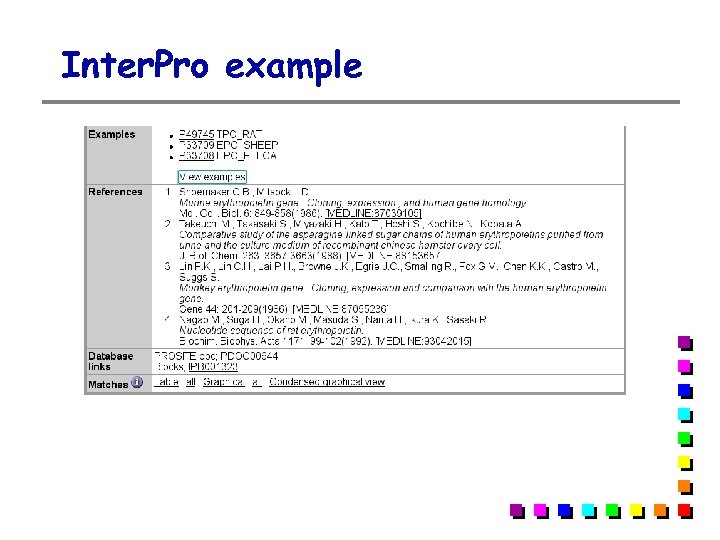
Inter. Pro example
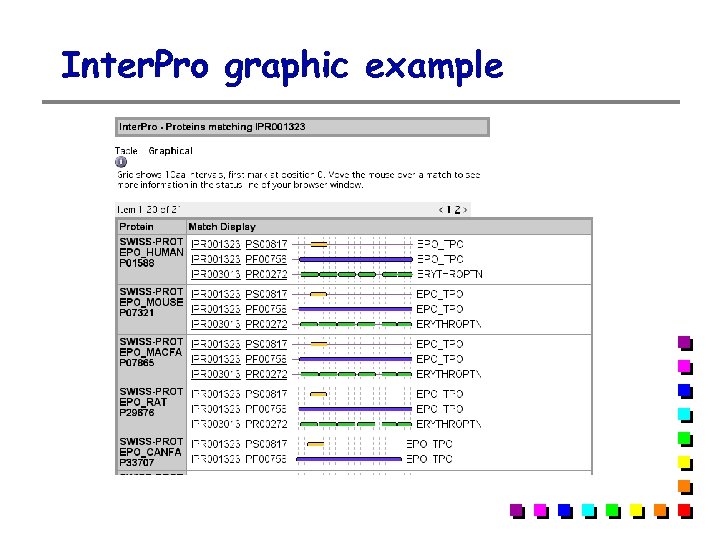
Inter. Pro graphic example
Databases 6: proteomics Contain informations obtained by 2 D-PAGE: master images of the gels and description of identified proteins Examples: SWISS-2 DPAGE, ECO 2 DBASE, Maize 2 DPAGE, Sub 2 D, Cyano 2 DBase, etc. Format: composed of image and text files Most 2 D-PAGE databases are “federated” and use SWISS-PROT as a master index There is currently no protein Mass Spectrometry (MS) database (not for long…)
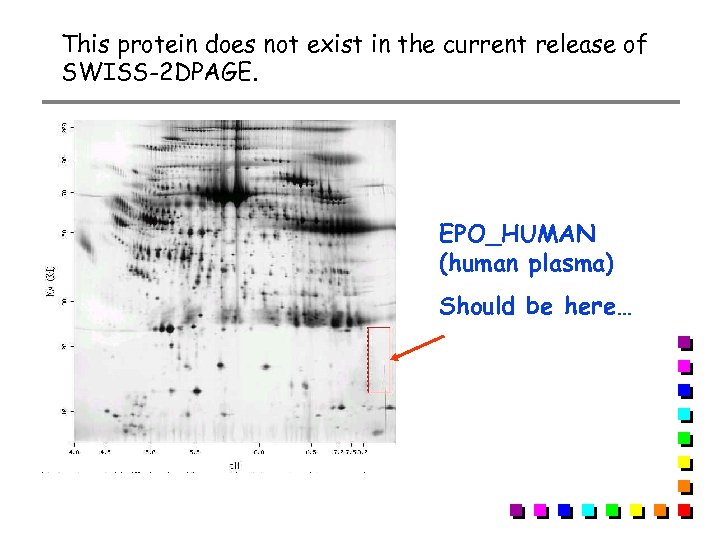
This protein does not exist in the current release of SWISS-2 DPAGE. EPO_HUMAN (human plasma) Should be here…
Databases 7: 3 D structure Contain the spatial coordinates of macromolecules whose 3 D structure has been obtained by X-ray or NMR studies Proteins represent more than 90% of available structures (others are DNA, RNA, sugars, virus, complex protein/DNA…) RCSB or PDB (Protein Data Bank), CATH and SCOP (structural classification of proteins (according to the secondary structures)), BMRB (Bio. Mag. Res. Bank; NMR results) DSSP: Database of Secondary Structure Assignments. HSSP: Homology-derived secondary structure of proteins. FSSP: Fold Classification based on Structure-Structure Assignments. SWISS-MODEL: Homology-derived 3 D structure db
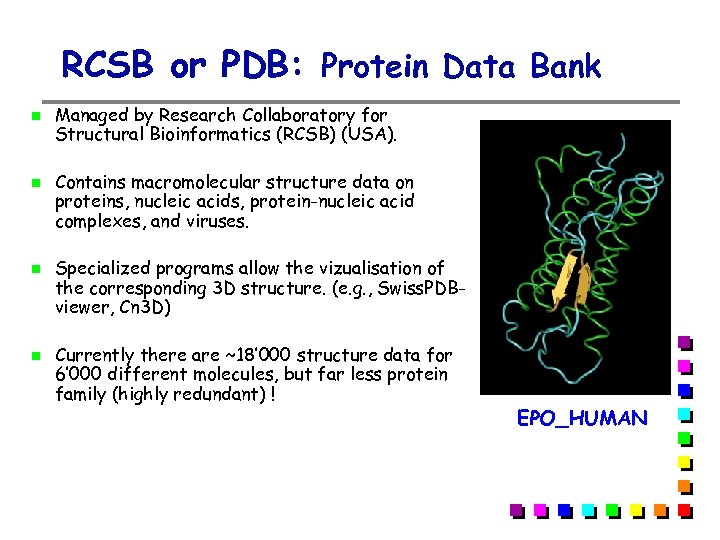
RCSB or PDB: Protein Data Bank Managed by Research Collaboratory for Structural Bioinformatics (RCSB) (USA). Contains macromolecular structure data on proteins, nucleic acids, protein-nucleic acid complexes, and viruses. Specialized programs allow the vizualisation of the corresponding 3 D structure. (e. g. , Swiss. PDBviewer, Cn 3 D) Currently there are ~18’ 000 structure data for 6’ 000 different molecules, but far less protein family (highly redundant) ! EPO_HUMAN
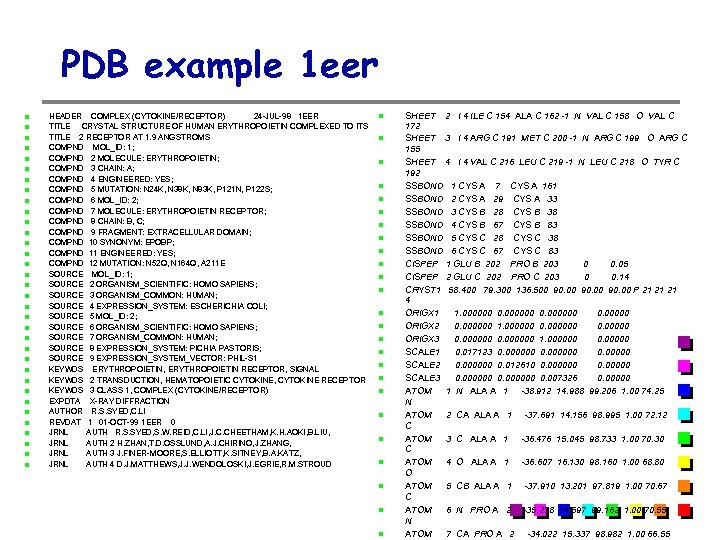
PDB example 1 eer HEADER COMPLEX (CYTOKINE/RECEPTOR) 24 -JUL-98 1 EER TITLE CRYSTAL STRUCTURE OF HUMAN ERYTHROPOIETIN COMPLEXED TO ITS TITLE 2 RECEPTOR AT 1. 9 ANGSTROMS COMPND MOL_ID: 1; COMPND 2 MOLECULE: ERYTHROPOIETIN; COMPND 3 CHAIN: A; COMPND 4 ENGINEERED: YES; COMPND 5 MUTATION: N 24 K, N 38 K, N 83 K, P 121 N, P 122 S; COMPND 6 MOL_ID: 2; COMPND 7 MOLECULE: ERYTHROPOIETIN RECEPTOR; COMPND 8 CHAIN: B, C; COMPND 9 FRAGMENT: EXTRACELLULAR DOMAIN; COMPND 10 SYNONYM: EPOBP; COMPND 11 ENGINEERED: YES; COMPND 12 MUTATION: N 52 Q, N 164 Q, A 211 E SOURCE MOL_ID: 1; SOURCE 2 ORGANISM_SCIENTIFIC: HOMO SAPIENS; SOURCE 3 ORGANISM_COMMON: HUMAN; SOURCE 4 EXPRESSION_SYSTEM: ESCHERICHIA COLI; SOURCE 5 MOL_ID: 2; SOURCE 6 ORGANISM_SCIENTIFIC: HOMO SAPIENS; SOURCE 7 ORGANISM_COMMON: HUMAN; SOURCE 8 EXPRESSION_SYSTEM: PICHIA PASTORIS; SOURCE 9 EXPRESSION_SYSTEM_VECTOR: PHIL-S 1 KEYWDS ERYTHROPOIETIN, ERYTHROPOIETIN RECEPTOR, SIGNAL KEYWDS 2 TRANSDUCTION, HEMATOPOIETIC CYTOKINE, CYTOKINE RECEPTOR KEYWDS 3 CLASS 1, COMPLEX (CYTOKINE/RECEPTOR) EXPDTA X-RAY DIFFRACTION AUTHOR R. S. SYED, C. LI REVDAT 1 01 -OCT-99 1 EER 0 JRNL AUTH R. S. SYED, S. W. REID, C. LI, J. C. CHEETHAM, K. H. AOKI, B. LIU, JRNL AUTH 2 H. ZHAN, T. D. OSSLUND, A. J. CHIRINO, J. ZHANG, JRNL AUTH 3 J. FINER-MOORE, S. ELLIOTT, K. SITNEY, B. A. KATZ, JRNL AUTH 4 D. J. MATTHEWS, J. J. WENDOLOSKI, J. EGRIE, R. M. STROUD SHEET 2 I 4 ILE C 154 ALA C 162 -1 N VAL C 158 O VAL C 172 SHEET 3 I 4 ARG C 191 MET C 200 -1 N ARG C 199 O ARG C 155 SHEET 4 I 4 VAL C 216 LEU C 219 -1 N LEU C 218 O TYR C 192 SSBOND 1 CYS A 7 CYS A 161 SSBOND 2 CYS A 29 CYS A 33 SSBOND 3 CYS B 28 CYS B 38 SSBOND 4 CYS B 67 CYS B 83 SSBOND 5 CYS C 28 CYS C 38 SSBOND 6 CYS C 67 CYS C 83 CISPEP 1 GLU B 202 PRO B 203 0 0. 05 CISPEP 2 GLU C 202 PRO C 203 0 0. 14 CRYST 1 58. 400 79. 300 136. 500 90. 00 P 21 21 21 4 ORIGX 1 1. 000000 0. 00000 ORIGX 2 0. 000000 1. 000000 0. 00000 ORIGX 3 0. 000000 1. 000000 0. 00000 SCALE 1 0. 017123 0. 000000 0. 00000 SCALE 2 0. 000000 0. 012610 0. 00000 SCALE 3 0. 000000 0. 007326 0. 00000 ATOM 1 N ALA A 1 -38. 912 14. 988 99. 206 1. 00 74. 25 N ATOM 2 CA ALA A 1 -37. 691 14. 156 98. 995 1. 00 72. 12 C ATOM 3 C ALA A 1 -36. 476 15. 045 98. 733 1. 00 70. 30 C ATOM 4 O ALA A 1 -36. 607 16. 130 98. 160 1. 00 68. 80 O ATOM 5 CB ALA A 1 -37. 910 13. 201 97. 819 1. 00 70. 67 C ATOM 6 N PRO A 2 -35. 278 14. 597 99. 162 1. 00 70. 55 N ATOM 7 CA PRO A 2 -34. 022 15. 337 98. 982 1. 00 66. 55
Databases 8: metabolic Contain informations that describe enzymes, biochemical reactions and metabolic pathways; ENZYME and BRENDA: nomenclature databases that store informations on enzyme names and reactions; Metabolic databases: Eco. Cyc (specialized on Escherichia coli), KEGG, EMP/WIT; Usualy these databases are tightly coupled with query software that allows the user to visualise reaction schemes.
Databases 9: bibliographic Bibliographic reference databases contain citations and abstract informations of published life science articles; Example: Medline Other more specialized databases also exist (example: Agricola).

Medline MEDLINE covers the fields of medicine, nursing, dentistry, veterinary medicine, the health care system, and the preclinical sciences more than 4, 600 biomedical journals published in the United States and 70 other countries Contains over 11 million citations since 1966 until now Contains links to biological db and to some journals New records are added to Pre. MEDLINE daily! Many papers not dealing with human are not in Medline ! Before 1970, keeps only the first 10 authors ! Not all journals have citations since 1966 !
Medline/Pubmed Pub. Med is developed by the National Center for Biotechnology Information (NCBI) Pub. Med provides access to bibliographic information such as MEDLINE, Pre. MEDLINE, Health. STAR, and to integrated molecular biology databases (composite db) PMID: 10923642 (Pub. Med ID) UI: 20378145 (Medline ID)
Databases 10: others There are many databases that cannot be classified in the categories listed previously; Examples: Re. Base (restriction enzymes), TRANSFAC (transcription factors), Carb. Bank, Glyco. Suite. DB (linked sugars), Protein-protein interactions db (DIP, Pro. Net, BIND, MINT), Protease db (MEROPS), biotechnology patents db, etc. ; As well as many other resources concerning any aspects of macromolecules and molecular biology.
Proliferation of databases What is the best db for sequence analysis ? Which does contain the highest quality data ? Which is the more comprehensive ? Which is the more up-to-date ? Which is the less redundant ? Which is the more indexed (allows complex queries) ? Which Web server does respond most quickly ? ……. ? ? ?
Some important practical remarks Databases: many errors (automated annotation) ! Not all db are available on all servers The update frequency is not the same for all servers; creation of db_new between releases (exemple: EMBLnew; Tr. EMBLnew…. ) Some servers add automatically useful crossreferences to an entry (implicit links) in addition to already existing links (explicit links)
Database retrieval tools Sequence Retrieval System (SRS, Europe) allows any flatfile db to be indexed to any other; allows to formulate queries across a wide range of different db types via a single interface, without any worry about data structure, query languages… Entrez (USA): less flexible than SRS but exploits the concept of « neighbouring » , which allows related articles in different db to be linked together, whether or not they are cross-referenced directly ATLAS: specific for macromolecular sequences db (i. e. NRL 3 D) ….
Before the introduction to databases… After the introduction to databases…
4eae04ee99a17bf0df44d9606cc40575.ppt