Biochemistry_02.ppt
- Количество слайдов: 45
 Amino Acids, Peptides, Proteins Learning goals: • Structure and naming of amino acids • Structure and properties of peptides • Ionization behavior of amino acids and peptides • Methods to characterize peptides and proteins
Amino Acids, Peptides, Proteins Learning goals: • Structure and naming of amino acids • Structure and properties of peptides • Ionization behavior of amino acids and peptides • Methods to characterize peptides and proteins
 Proteins: Main Agents of Biological Function • Catalysis – enolase (in the glycolytic pathway) – DNA polymerase (in DNA replication) • Transport – hemoglobin (transports O 2 in the blood) – lactose permease (transports lactose across the cell membrane) • Structure – collagen (connective tissue) – keratin (hair, nails, feathers, horns) • Motion – myosin (muscle tissue) – actin (muscle tissue, cell motility)
Proteins: Main Agents of Biological Function • Catalysis – enolase (in the glycolytic pathway) – DNA polymerase (in DNA replication) • Transport – hemoglobin (transports O 2 in the blood) – lactose permease (transports lactose across the cell membrane) • Structure – collagen (connective tissue) – keratin (hair, nails, feathers, horns) • Motion – myosin (muscle tissue) – actin (muscle tissue, cell motility)
 Amino Acids: Building Blocks of Protein • Proteins are linear heteropolymers of -amino acids • Amino acids have properties that are well-suited to carry out a variety of biological functions – – Capacity to polymerize Useful acid-base properties Varied physical properties Varied chemical functionality
Amino Acids: Building Blocks of Protein • Proteins are linear heteropolymers of -amino acids • Amino acids have properties that are well-suited to carry out a variety of biological functions – – Capacity to polymerize Useful acid-base properties Varied physical properties Varied chemical functionality
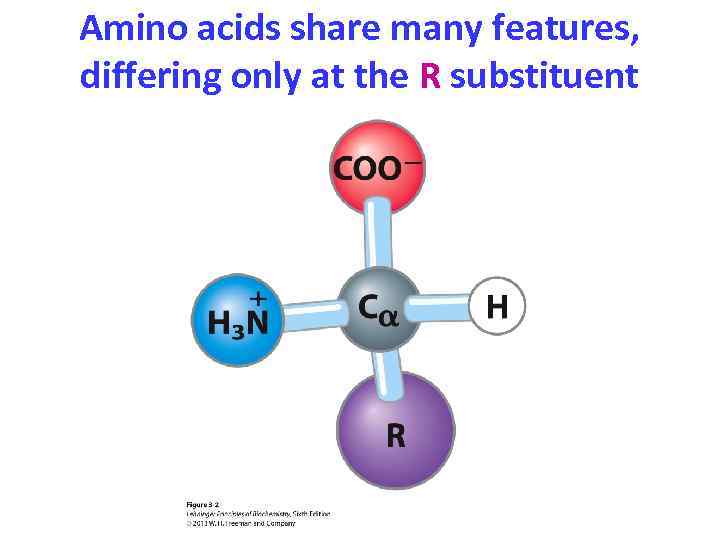 Amino acids share many features, differing only at the R substituent
Amino acids share many features, differing only at the R substituent
 Most -amino acids are chiral • The -carbon always has four substituents and is tetrahedral • All (except proline) have: – an acidic carboxyl group – a basic amino group – an -hydrogen connected to the -carbon • The fourth substituent (R) is unique – In glycine, the fourth substituent is also hydrogen
Most -amino acids are chiral • The -carbon always has four substituents and is tetrahedral • All (except proline) have: – an acidic carboxyl group – a basic amino group – an -hydrogen connected to the -carbon • The fourth substituent (R) is unique – In glycine, the fourth substituent is also hydrogen
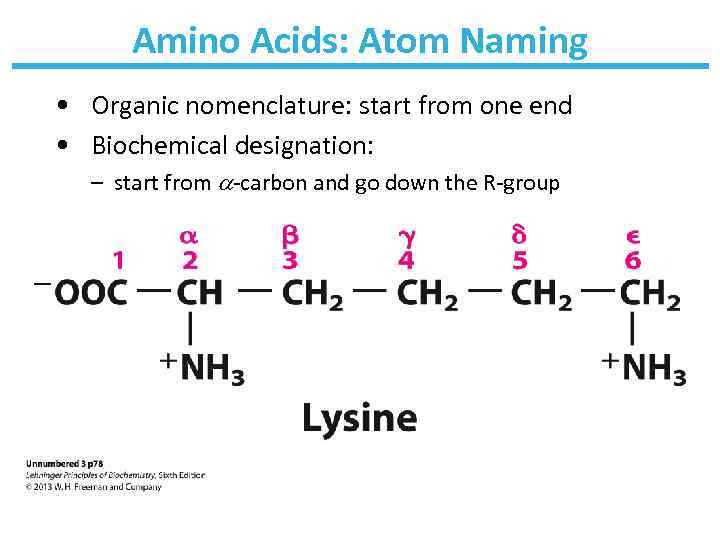 Amino Acids: Atom Naming • Organic nomenclature: start from one end • Biochemical designation: – start from -carbon and go down the R-group
Amino Acids: Atom Naming • Organic nomenclature: start from one end • Biochemical designation: – start from -carbon and go down the R-group
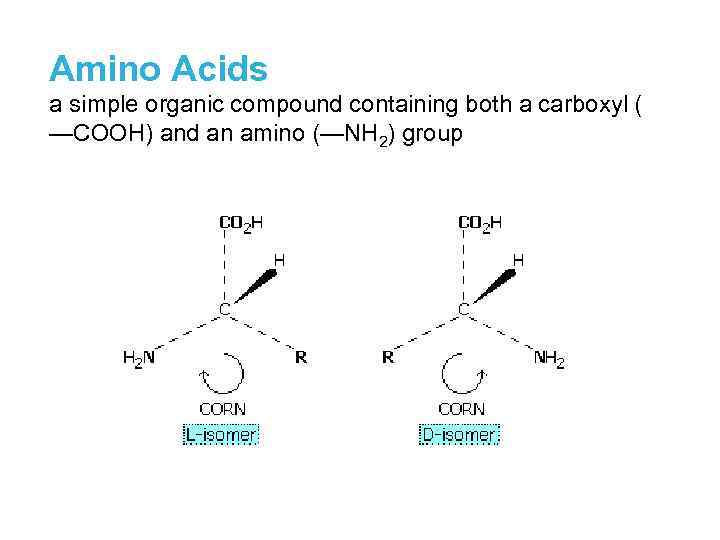 Amino Acids a simple organic compound containing both a carboxyl ( —COOH) and an amino (—NH 2) group
Amino Acids a simple organic compound containing both a carboxyl ( —COOH) and an amino (—NH 2) group
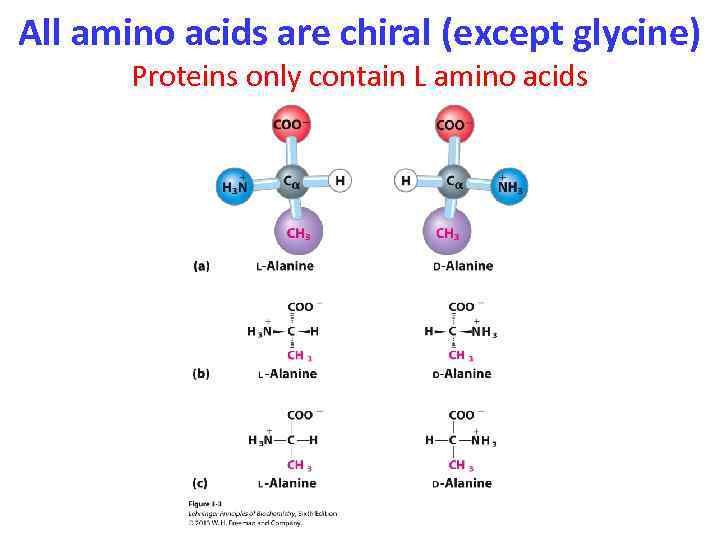 All amino acids are chiral (except glycine) Proteins only contain L amino acids
All amino acids are chiral (except glycine) Proteins only contain L amino acids
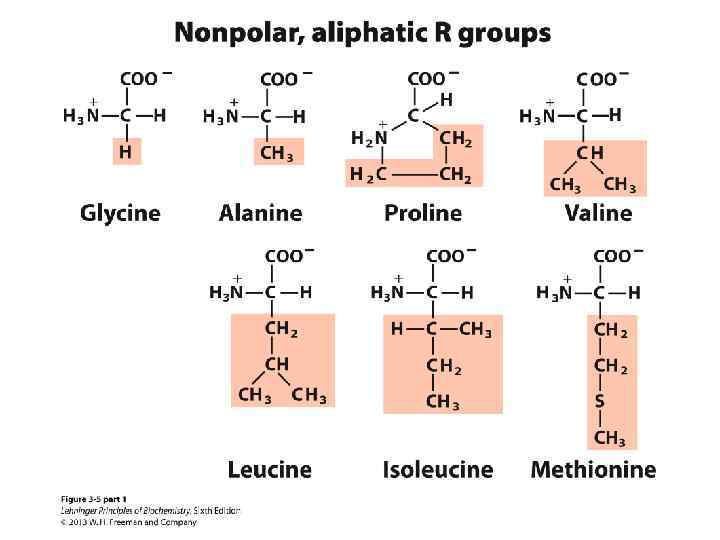
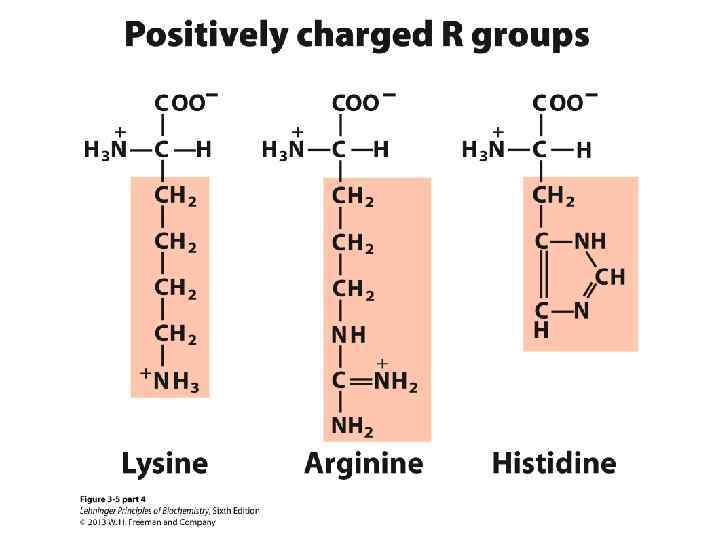
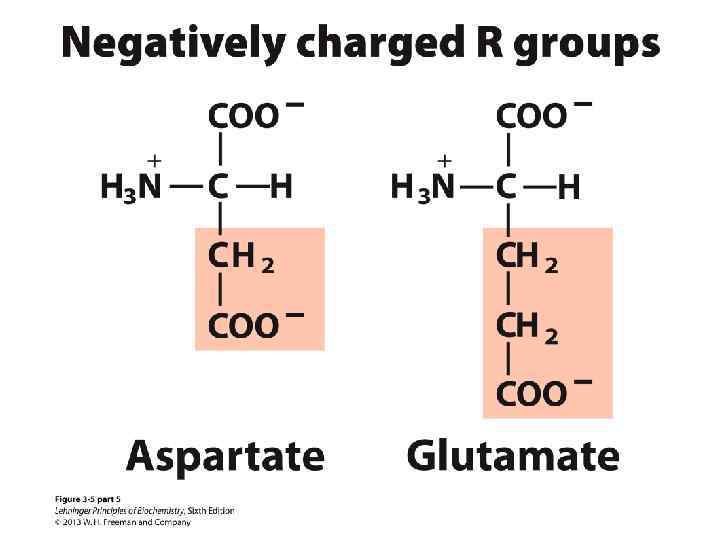
 Amino Acids: Classification Common amino acids can be placed in five basic groups depending on their R substituents: • Nonpolar, aliphatic (7) • Aromatic (3) • Polar, uncharged (5) • Positively charged (3) • Negatively charged (2)
Amino Acids: Classification Common amino acids can be placed in five basic groups depending on their R substituents: • Nonpolar, aliphatic (7) • Aromatic (3) • Polar, uncharged (5) • Positively charged (3) • Negatively charged (2)
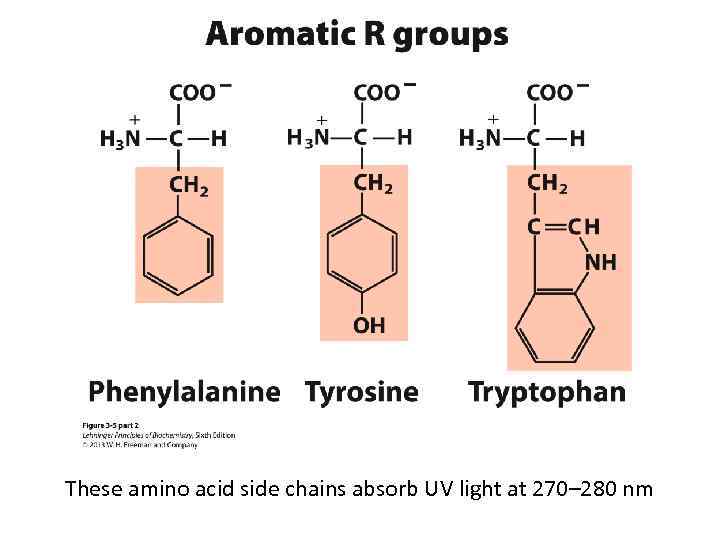 These amino acid side chains absorb UV light at 270– 280 nm
These amino acid side chains absorb UV light at 270– 280 nm
 These amino acids side chains can form hydrogen bonds. Cysteine can form disulfide bonds.
These amino acids side chains can form hydrogen bonds. Cysteine can form disulfide bonds.
 Uncommon Amino Acids in Proteins • Not incorporated by ribosomes except for Selenocysteine • Arise by post-translational modifications of proteins • Reversible modifications, especially phosphorylation, are important in regulation and signaling
Uncommon Amino Acids in Proteins • Not incorporated by ribosomes except for Selenocysteine • Arise by post-translational modifications of proteins • Reversible modifications, especially phosphorylation, are important in regulation and signaling
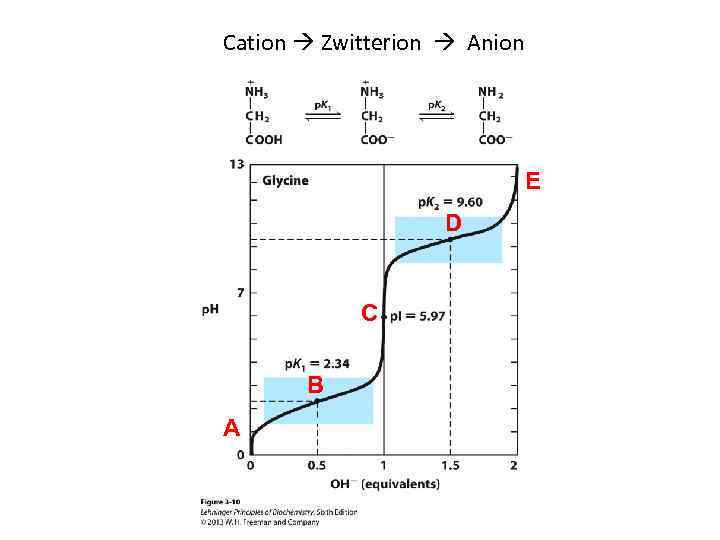
 Ionization of Amino Acids • At acidic p. H, the carboxyl group is protonated and the amino acid is in the cationic form. • At neutral p. H, the carboxyl group is deprotonated but the amino group is protonated. The net charge is zero; such ions are called Zwitterions. • At alkaline p. H, the amino group is neutral –NH 2 and the amino acid is in the anionic form.
Ionization of Amino Acids • At acidic p. H, the carboxyl group is protonated and the amino acid is in the cationic form. • At neutral p. H, the carboxyl group is deprotonated but the amino group is protonated. The net charge is zero; such ions are called Zwitterions. • At alkaline p. H, the amino group is neutral –NH 2 and the amino acid is in the anionic form.
 Cation Zwitterion Anion E D C B A
Cation Zwitterion Anion E D C B A
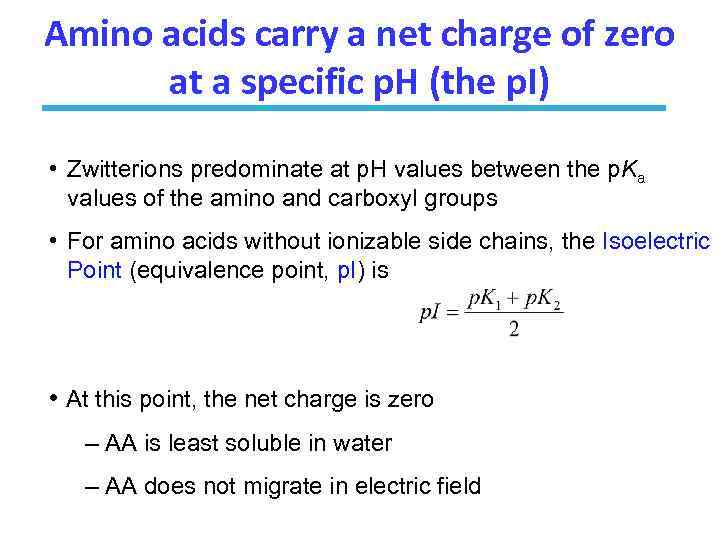 Amino acids carry a net charge of zero at a specific p. H (the p. I) • Zwitterions predominate at p. H values between the p. Ka values of the amino and carboxyl groups • For amino acids without ionizable side chains, the Isoelectric Point (equivalence point, p. I) is • At this point, the net charge is zero – AA is least soluble in water – AA does not migrate in electric field
Amino acids carry a net charge of zero at a specific p. H (the p. I) • Zwitterions predominate at p. H values between the p. Ka values of the amino and carboxyl groups • For amino acids without ionizable side chains, the Isoelectric Point (equivalence point, p. I) is • At this point, the net charge is zero – AA is least soluble in water – AA does not migrate in electric field
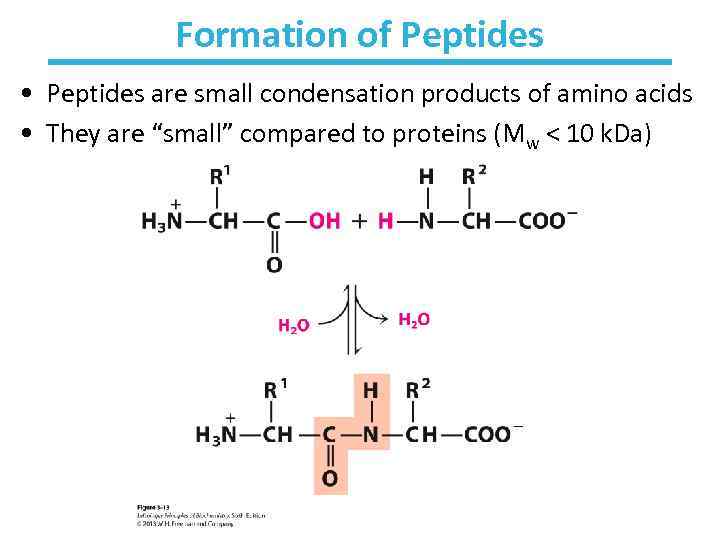 Formation of Peptides • Peptides are small condensation products of amino acids • They are “small” compared to proteins (Mw < 10 k. Da)
Formation of Peptides • Peptides are small condensation products of amino acids • They are “small” compared to proteins (Mw < 10 k. Da)
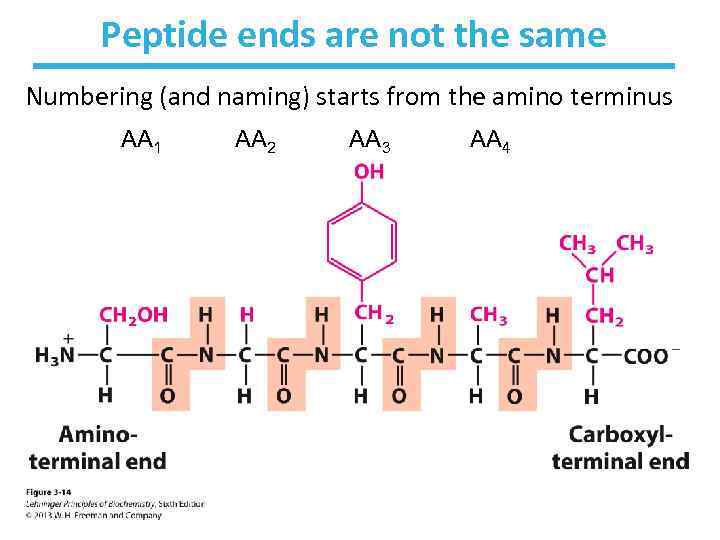 Peptide ends are not the same Numbering (and naming) starts from the amino terminus AA 1 AA 5 AA 2 AA 3 AA 4
Peptide ends are not the same Numbering (and naming) starts from the amino terminus AA 1 AA 5 AA 2 AA 3 AA 4
 Peptides: A Variety of Functions • Hormones and pheromones – insulin (think sugar) – oxytocin (think childbirth) – sex-peptide (think fruit fly mating) • Neuropeptides – substance P (pain mediator) • Antibiotics – polymyxin B (for Gram – bacteria) – bacitracin (for Gram + bacteria) • Protection, e. g. , toxins – amanitin (mushrooms) – conotoxin (cone snails) – chlorotoxin (scorpions)
Peptides: A Variety of Functions • Hormones and pheromones – insulin (think sugar) – oxytocin (think childbirth) – sex-peptide (think fruit fly mating) • Neuropeptides – substance P (pain mediator) • Antibiotics – polymyxin B (for Gram – bacteria) – bacitracin (for Gram + bacteria) • Protection, e. g. , toxins – amanitin (mushrooms) – conotoxin (cone snails) – chlorotoxin (scorpions)
 Proteins are: • Polypeptides (covalently linked -amino acids) + possibly: • cofactors functional non-amino acid component metal ions or organic molecules • coenzymes organic cofactors NAD+ in lactate dehydrogenase • prosthetic groups covalently attached cofactors heme in myoglobin • other modifications
Proteins are: • Polypeptides (covalently linked -amino acids) + possibly: • cofactors functional non-amino acid component metal ions or organic molecules • coenzymes organic cofactors NAD+ in lactate dehydrogenase • prosthetic groups covalently attached cofactors heme in myoglobin • other modifications
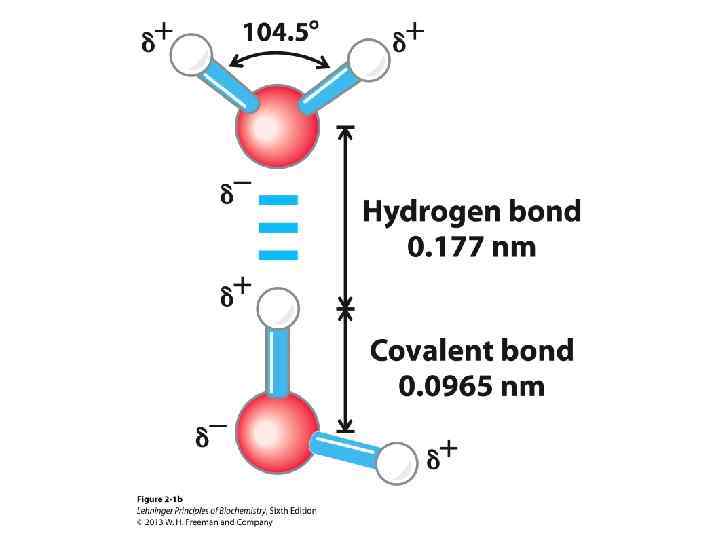
 Hydrogen Bonds • Strong dipole-dipole or charge-dipole interaction that arises between an acid (proton donor) and a base (proton acceptor) • Typically 4– 6 k. J/mol for bonds with neutral atoms, and 6– 10 k. J/mol for bonds with one charged atom • Typically involves two electronegative atoms (frequently nitrogen and oxygen) • Hydrogen bonds are strongest when the bonded molecules are oriented to maximize electrostatic interaction • Ideally the three atoms involved are in a line
Hydrogen Bonds • Strong dipole-dipole or charge-dipole interaction that arises between an acid (proton donor) and a base (proton acceptor) • Typically 4– 6 k. J/mol for bonds with neutral atoms, and 6– 10 k. J/mol for bonds with one charged atom • Typically involves two electronegative atoms (frequently nitrogen and oxygen) • Hydrogen bonds are strongest when the bonded molecules are oriented to maximize electrostatic interaction • Ideally the three atoms involved are in a line
 What to Study about Peptides and Proteins What is its sequence and composition? What is its three-dimensional structure? How does it find its native fold? How does it achieve its biochemical role? How is its function regulated? How does it interacts with other macromolecules? How is it related to other proteins? Where is it localized within the cell? What are its physico-chemical properties?
What to Study about Peptides and Proteins What is its sequence and composition? What is its three-dimensional structure? How does it find its native fold? How does it achieve its biochemical role? How is its function regulated? How does it interacts with other macromolecules? How is it related to other proteins? Where is it localized within the cell? What are its physico-chemical properties?
 A mixture of proteins can be separated • Separation relies on differences in physical and chemical properties – – – Charge Size Affinity for a ligand Solubility Hydrophobicity Thermal stability • Chromatography is commonly used for preparative separation
A mixture of proteins can be separated • Separation relies on differences in physical and chemical properties – – – Charge Size Affinity for a ligand Solubility Hydrophobicity Thermal stability • Chromatography is commonly used for preparative separation
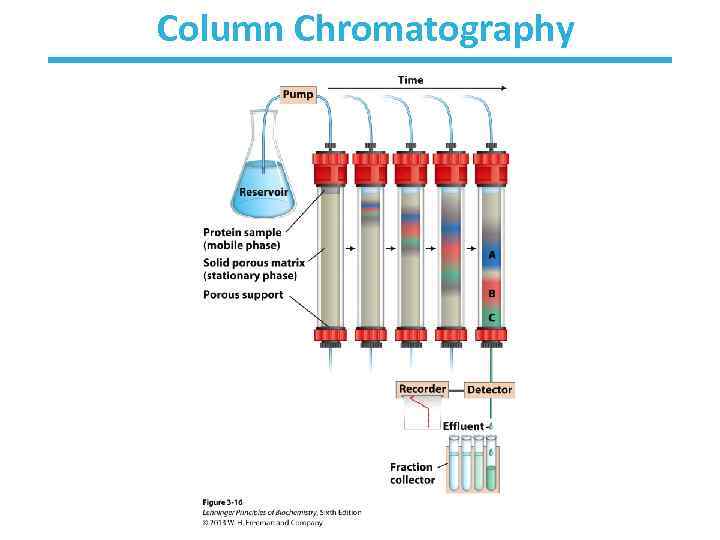 Column Chromatography
Column Chromatography
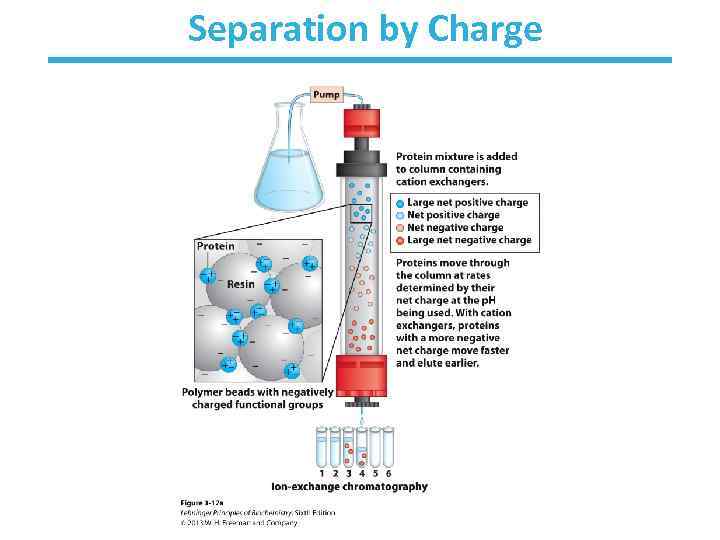 Separation by Charge
Separation by Charge
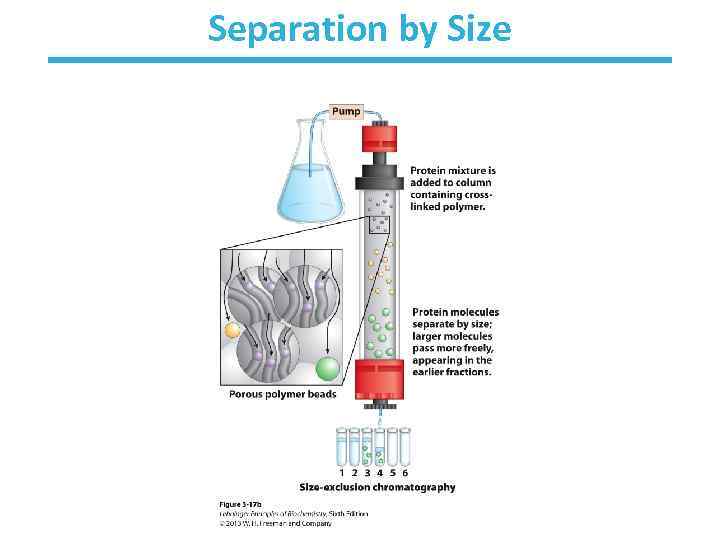 Separation by Size
Separation by Size
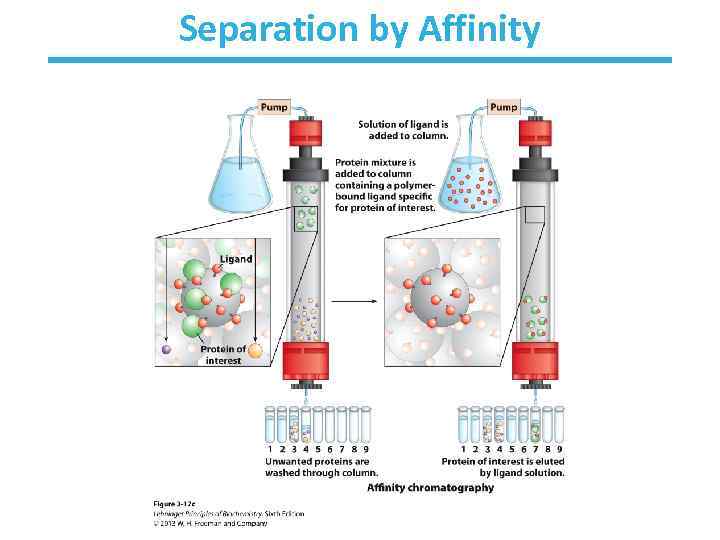 Separation by Affinity
Separation by Affinity
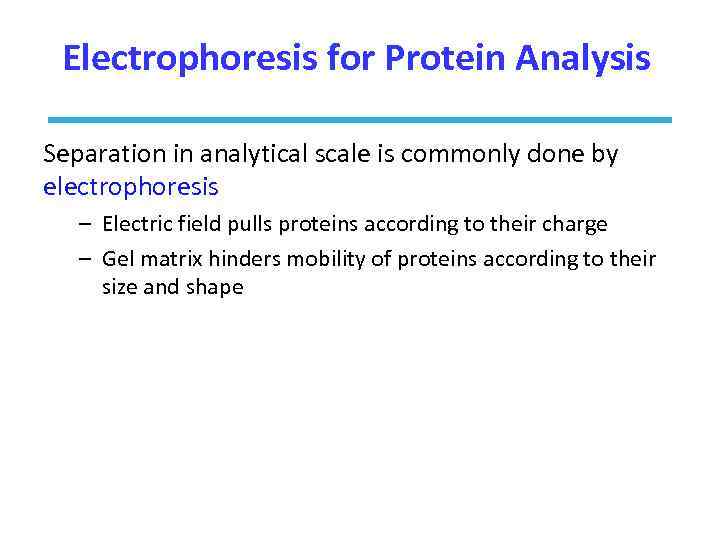 Electrophoresis for Protein Analysis Separation in analytical scale is commonly done by electrophoresis – Electric field pulls proteins according to their charge – Gel matrix hinders mobility of proteins according to their size and shape
Electrophoresis for Protein Analysis Separation in analytical scale is commonly done by electrophoresis – Electric field pulls proteins according to their charge – Gel matrix hinders mobility of proteins according to their size and shape
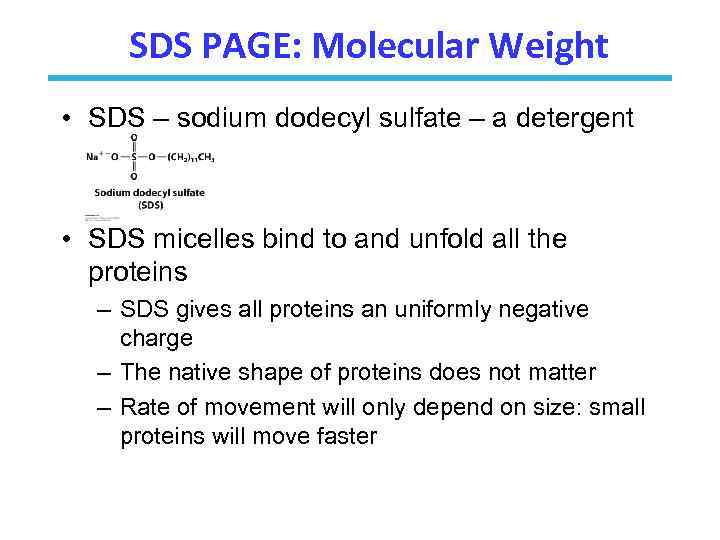 SDS PAGE: Molecular Weight • SDS – sodium dodecyl sulfate – a detergent • SDS micelles bind to and unfold all the proteins – SDS gives all proteins an uniformly negative charge – The native shape of proteins does not matter – Rate of movement will only depend on size: small proteins will move faster
SDS PAGE: Molecular Weight • SDS – sodium dodecyl sulfate – a detergent • SDS micelles bind to and unfold all the proteins – SDS gives all proteins an uniformly negative charge – The native shape of proteins does not matter – Rate of movement will only depend on size: small proteins will move faster
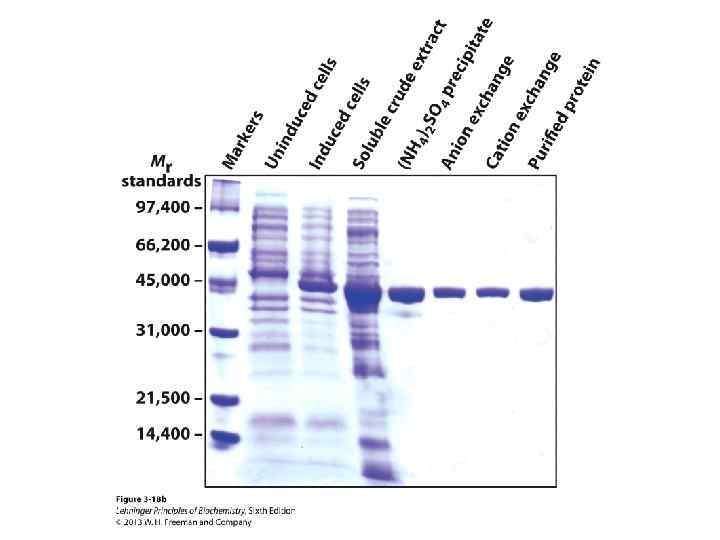
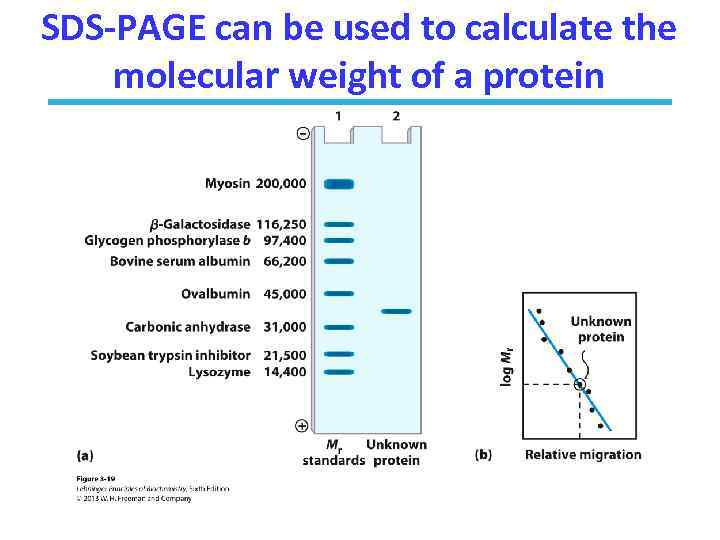 SDS-PAGE can be used to calculate the molecular weight of a protein
SDS-PAGE can be used to calculate the molecular weight of a protein
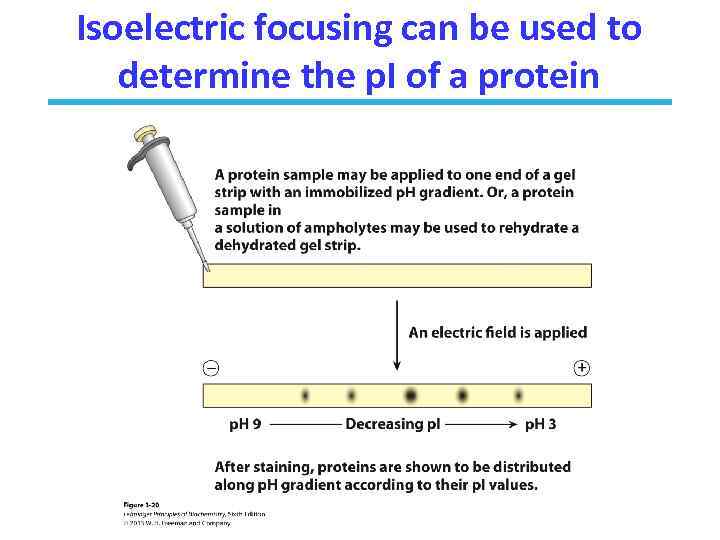 Isoelectric focusing can be used to determine the p. I of a protein
Isoelectric focusing can be used to determine the p. I of a protein
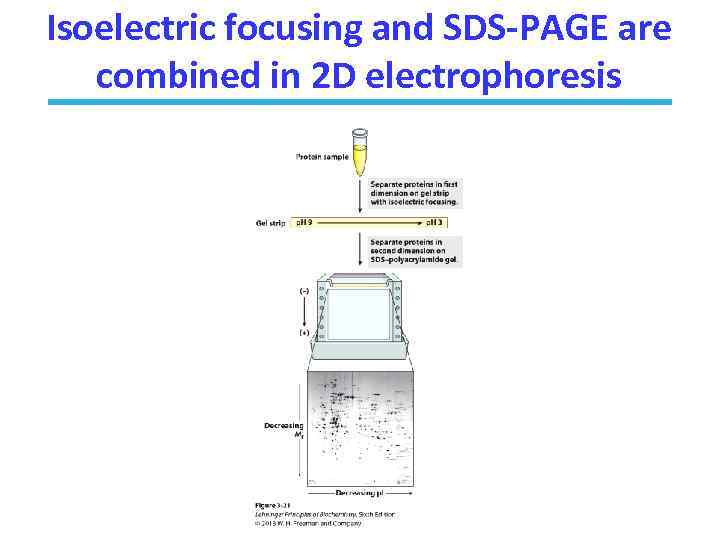 Isoelectric focusing and SDS-PAGE are combined in 2 D electrophoresis
Isoelectric focusing and SDS-PAGE are combined in 2 D electrophoresis
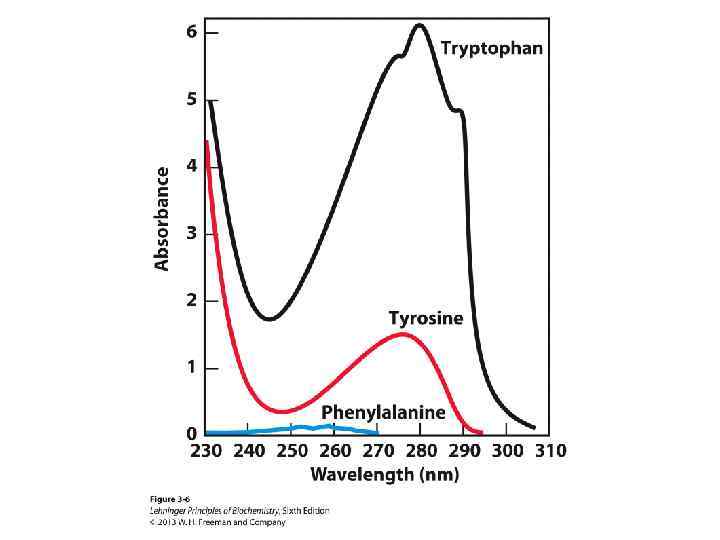
 Spectroscopic Detection of Aromatic Amino Acids • The aromatic amino acids absorb light in the UV region • Proteins typically have UV absorbance maxima around 275– 280 nm • Tryptophan and tyrosine are the strongest chromophores • Concentration can be determined by UV-visible spectrophotometry using Beers law: A = ·c·l
Spectroscopic Detection of Aromatic Amino Acids • The aromatic amino acids absorb light in the UV region • Proteins typically have UV absorbance maxima around 275– 280 nm • Tryptophan and tyrosine are the strongest chromophores • Concentration can be determined by UV-visible spectrophotometry using Beers law: A = ·c·l
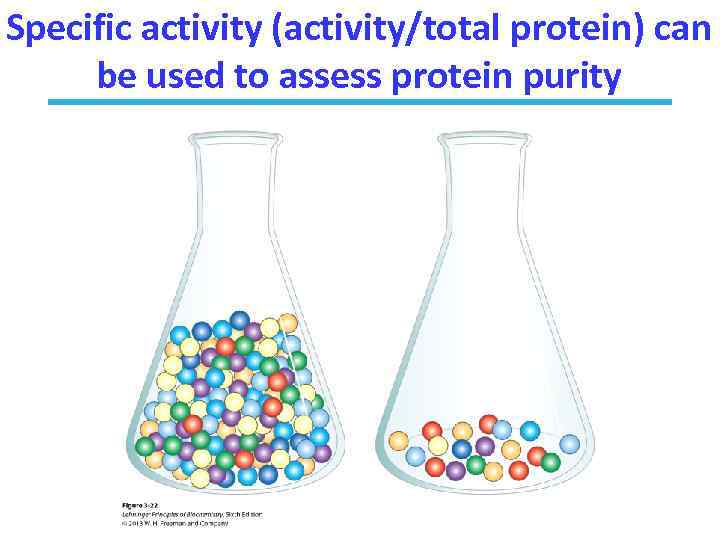 Specific activity (activity/total protein) can be used to assess protein purity
Specific activity (activity/total protein) can be used to assess protein purity
 Protein Sequencing • It is essential to further biochemical analysis that we know the sequence of the protein we are studying • Actual sequence generally determined from DNA sequence • Edman Degradation (Classical method) – Successive rounds of N-terminal modification, cleavage, and identification – Can be used to identify protein with known sequence • Mass Spectrometry (Modern method) – MALDI MS and ESI MS can precisely identify the mass of a peptide, and thus the amino acid sequence – Can be used to determine post-translational modifications
Protein Sequencing • It is essential to further biochemical analysis that we know the sequence of the protein we are studying • Actual sequence generally determined from DNA sequence • Edman Degradation (Classical method) – Successive rounds of N-terminal modification, cleavage, and identification – Can be used to identify protein with known sequence • Mass Spectrometry (Modern method) – MALDI MS and ESI MS can precisely identify the mass of a peptide, and thus the amino acid sequence – Can be used to determine post-translational modifications
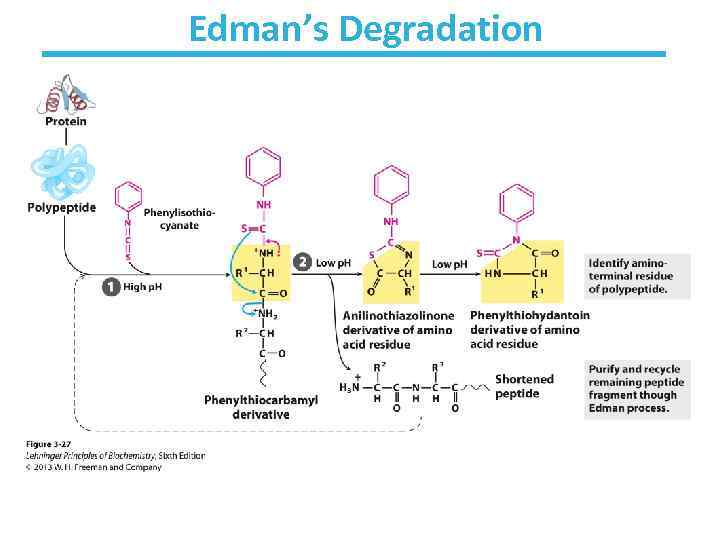 Edman’s Degradation
Edman’s Degradation
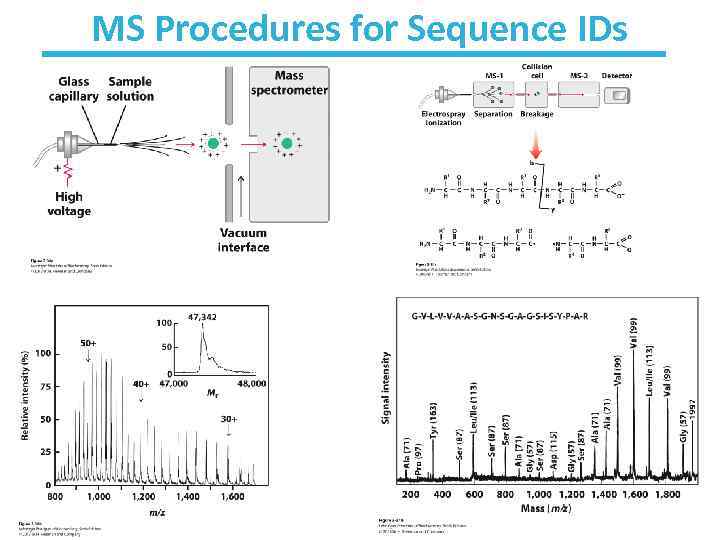 MS Procedures for Sequence IDs
MS Procedures for Sequence IDs
 Protein Sequences as Clues to Evolutionary Relationships • Sequences of homologous proteins from a wide range of species can be aligned analyzed for differences • Differences indicate evolutionary divergences • Analysis of multiple protein families can indicate evolutionary relationships between organisms, ultimately the history of life on Earth
Protein Sequences as Clues to Evolutionary Relationships • Sequences of homologous proteins from a wide range of species can be aligned analyzed for differences • Differences indicate evolutionary divergences • Analysis of multiple protein families can indicate evolutionary relationships between organisms, ultimately the history of life on Earth
 Chapter : Summary In this chapter, we learned about: • The many biological functions of peptides and proteins • The structures and names of amino acids found in proteins • The ionization properties of amino acids and peptides • The methods for separation and analysis of proteins
Chapter : Summary In this chapter, we learned about: • The many biological functions of peptides and proteins • The structures and names of amino acids found in proteins • The ionization properties of amino acids and peptides • The methods for separation and analysis of proteins


