8fa26c3f5d7532f9aa8e3bc56c35a09c.ppt
- Количество слайдов: 25
 Alternative Splicing A very short introduction (in plants)
Alternative Splicing A very short introduction (in plants)
 Alternative Splicing The exons and introns of a particular gene get shuffled to create multiple isoforms of a particular protein • First demonstrated in the late 1970’s in adenovirus • Fairly well characterized in animals (at least somewhat better than in plants) • Contributes to protein diversity • Affects m. RNA stability
Alternative Splicing The exons and introns of a particular gene get shuffled to create multiple isoforms of a particular protein • First demonstrated in the late 1970’s in adenovirus • Fairly well characterized in animals (at least somewhat better than in plants) • Contributes to protein diversity • Affects m. RNA stability
 One Gene / One Enzyme One Gene / One Polypeptide “One Gene / One set of connected transcripts” Ensembl- What is a gene, post-ENCODE? History and updated definition Genome Res. 2007. 17: 669 -681 1940’s ----------------------------------------2000’s
One Gene / One Enzyme One Gene / One Polypeptide “One Gene / One set of connected transcripts” Ensembl- What is a gene, post-ENCODE? History and updated definition Genome Res. 2007. 17: 669 -681 1940’s ----------------------------------------2000’s
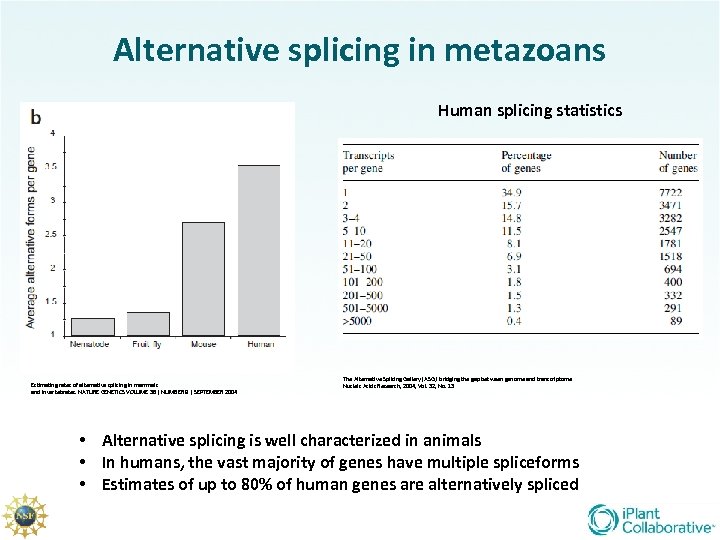 Alternative splicing in metazoans Human splicing statistics Estimating rates of alternative splicing in mammals and invertebrates. NATURE GENETICS VOLUME 36 | NUMBER 9 | SEPTEMBER 2004 The Alternative Splicing Gallery (ASG): bridging the gap between genome and transcriptome Nucleic Acids Research, 2004, Vol. 32, No. 13 • Alternative splicing is well characterized in animals • In humans, the vast majority of genes have multiple spliceforms • Estimates of up to 80% of human genes are alternatively spliced
Alternative splicing in metazoans Human splicing statistics Estimating rates of alternative splicing in mammals and invertebrates. NATURE GENETICS VOLUME 36 | NUMBER 9 | SEPTEMBER 2004 The Alternative Splicing Gallery (ASG): bridging the gap between genome and transcriptome Nucleic Acids Research, 2004, Vol. 32, No. 13 • Alternative splicing is well characterized in animals • In humans, the vast majority of genes have multiple spliceforms • Estimates of up to 80% of human genes are alternatively spliced
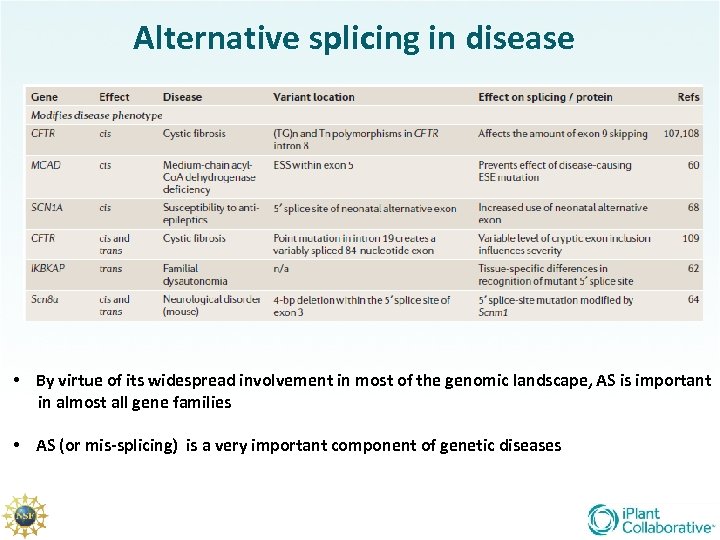 Alternative splicing in disease • By virtue of its widespread involvement in most of the genomic landscape, AS is important in almost all gene families • AS (or mis-splicing) is a very important component of genetic diseases
Alternative splicing in disease • By virtue of its widespread involvement in most of the genomic landscape, AS is important in almost all gene families • AS (or mis-splicing) is a very important component of genetic diseases
 Mechanisms of splicing
Mechanisms of splicing
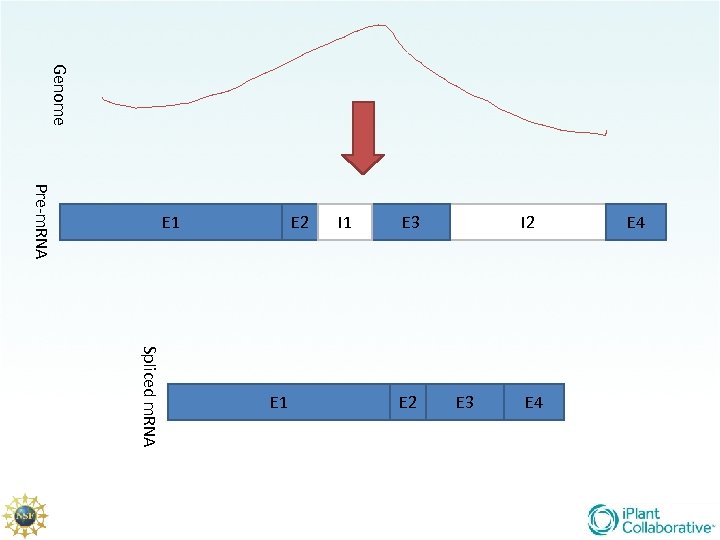 Genome Pre-m. RNA E 4 E 3 E 2 Spliced m. RNA E 1 E 4 I 2 E 3 I 1 E 2 E 1
Genome Pre-m. RNA E 4 E 3 E 2 Spliced m. RNA E 1 E 4 I 2 E 3 I 1 E 2 E 1
 Alternative splicing of Ru. Bis. Co was one of the first examples of AS in plants “The data presented here demonstrate the existence of alternative splicing in plant systems, but the physiological significance of synthesizing two forms of rubisco activase remains unclear. However, this process may have important implications in photosynthesis. if these polypeptides were functionally equivalent enzymes in the chloroplast, there would be no need for the production of both polypeptides, and alternative splicing of the rubisco activase m. RNA would likely become a dispensable process. ” The majority of AS events have not been functionally characterized
Alternative splicing of Ru. Bis. Co was one of the first examples of AS in plants “The data presented here demonstrate the existence of alternative splicing in plant systems, but the physiological significance of synthesizing two forms of rubisco activase remains unclear. However, this process may have important implications in photosynthesis. if these polypeptides were functionally equivalent enzymes in the chloroplast, there would be no need for the production of both polypeptides, and alternative splicing of the rubisco activase m. RNA would likely become a dispensable process. ” The majority of AS events have not been functionally characterized
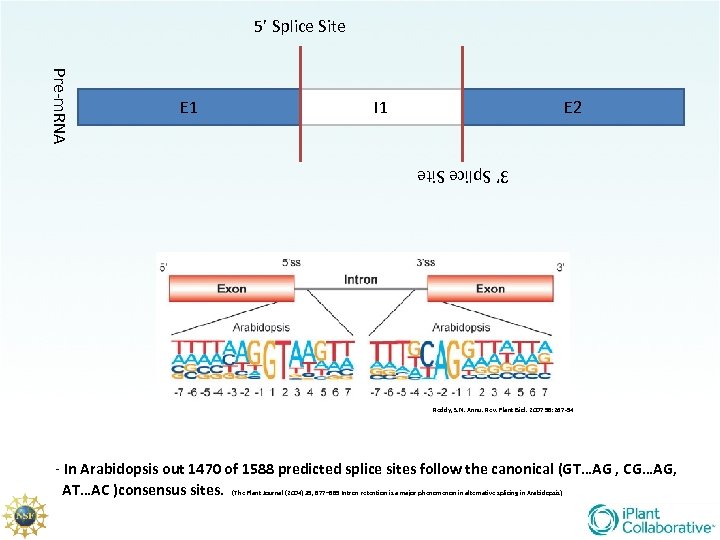 5’ Splice Site Pre-m. RNA E 1 I 1 E 2 3’ Splice Site Reddy, S. N. Annu. Rev. Plant Biol. 2007 58: 267 -94 - In Arabidopsis out 1470 of 1588 predicted splice sites follow the canonical (GT…AG , CG…AG, AT…AC )consensus sites. (The Plant Journal (2004) 39, 877– 885 Intron retention is a major phenomenon in alternative splicing in Arabidopsis)
5’ Splice Site Pre-m. RNA E 1 I 1 E 2 3’ Splice Site Reddy, S. N. Annu. Rev. Plant Biol. 2007 58: 267 -94 - In Arabidopsis out 1470 of 1588 predicted splice sites follow the canonical (GT…AG , CG…AG, AT…AC )consensus sites. (The Plant Journal (2004) 39, 877– 885 Intron retention is a major phenomenon in alternative splicing in Arabidopsis)
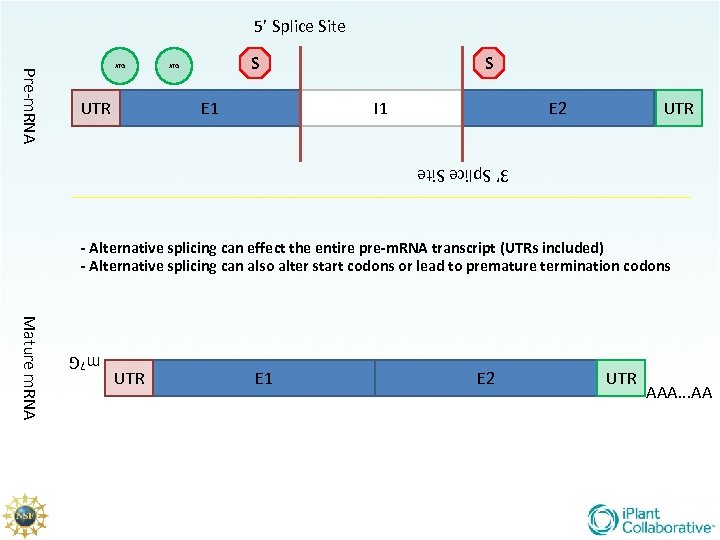 5’ Splice Site Pre-m. RNA ATG UTR S ATG E 1 S I 1 E 2 UTR 3’ Splice Site - Alternative splicing can effect the entire pre-m. RNA transcript (UTRs included) - Alternative splicing can also alter start codons or lead to premature termination codons m 7 G Mature m. RNA UTR E 1 E 2 UTR AAA. . . AA
5’ Splice Site Pre-m. RNA ATG UTR S ATG E 1 S I 1 E 2 UTR 3’ Splice Site - Alternative splicing can effect the entire pre-m. RNA transcript (UTRs included) - Alternative splicing can also alter start codons or lead to premature termination codons m 7 G Mature m. RNA UTR E 1 E 2 UTR AAA. . . AA
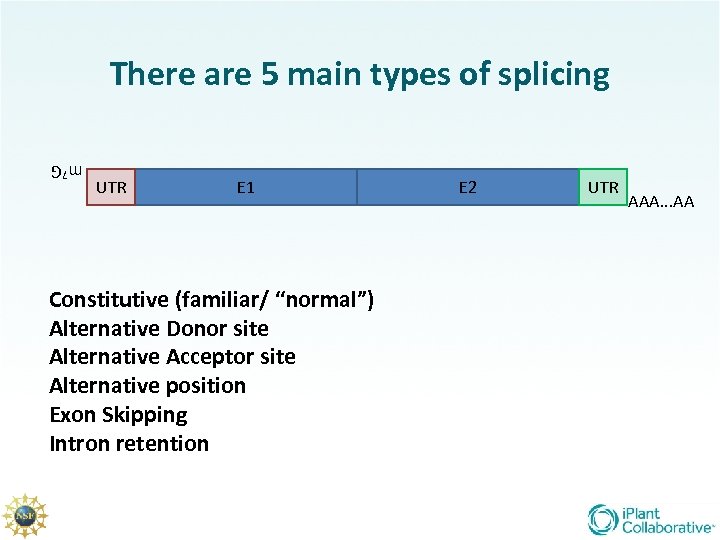 There are 5 main types of splicing UTR E 1 m 7 G Constitutive (familiar/ “normal”) Alternative Donor site Alternative Acceptor site Alternative position Exon Skipping Intron retention E 2 UTR AAA. . . AA
There are 5 main types of splicing UTR E 1 m 7 G Constitutive (familiar/ “normal”) Alternative Donor site Alternative Acceptor site Alternative position Exon Skipping Intron retention E 2 UTR AAA. . . AA
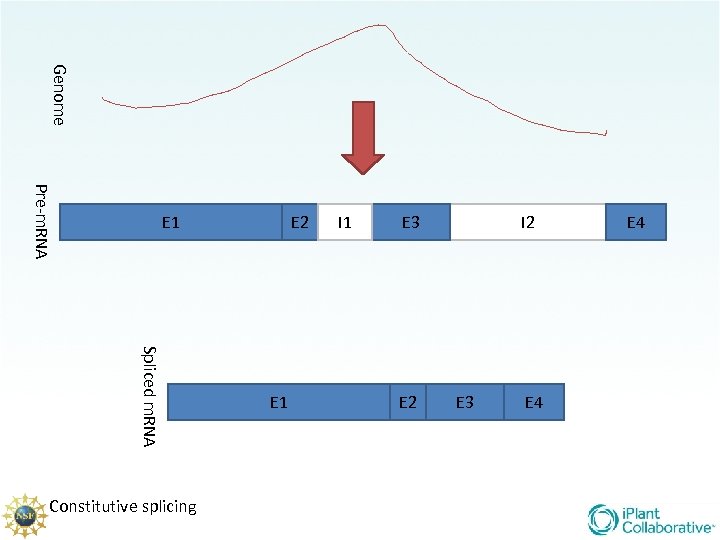 Genome Pre-m. RNA E 1 Spliced m. RNA Constitutive splicing E 2 E 1 I 1 E 3 E 2 I 2 E 3 E 4
Genome Pre-m. RNA E 1 Spliced m. RNA Constitutive splicing E 2 E 1 I 1 E 3 E 2 I 2 E 3 E 4
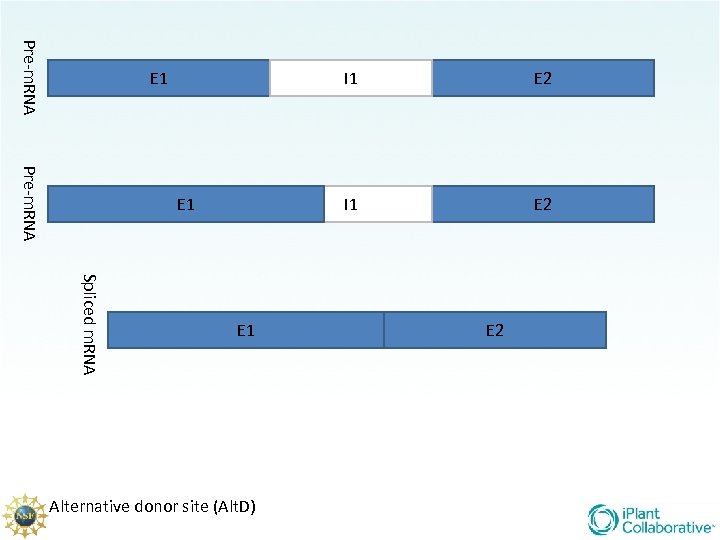 Pre-m. RNA E 1 I 1 E 2 Spliced m. RNA E 1 Alternative donor site (Alt. D) E 2
Pre-m. RNA E 1 I 1 E 2 Spliced m. RNA E 1 Alternative donor site (Alt. D) E 2
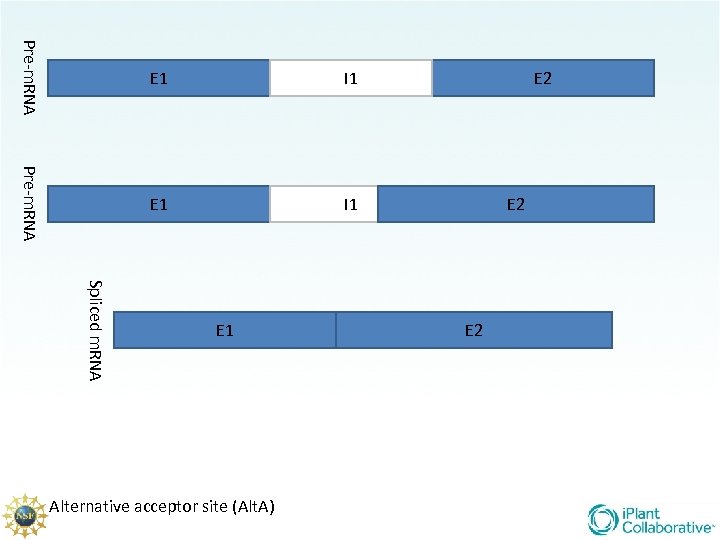 Pre-m. RNA E 1 I 1 E 2 E 2 Spliced m. RNA E 1 Alternative acceptor site (Alt. A) E 2
Pre-m. RNA E 1 I 1 E 2 E 2 Spliced m. RNA E 1 Alternative acceptor site (Alt. A) E 2
 Pre-m. RNA E 1 I 1 E 2 Spliced m. RNA Alternative Position (Alt. P) E 1 E 2
Pre-m. RNA E 1 I 1 E 2 Spliced m. RNA Alternative Position (Alt. P) E 1 E 2
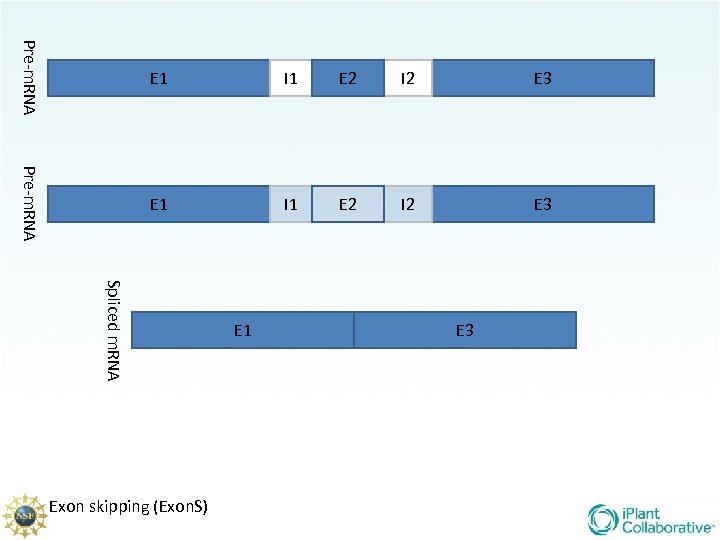 Pre-m. RNA E 1 I 1 E 2 I 2 E 3 Spliced m. RNA Exon skipping (Exon. S) E 1 E 3
Pre-m. RNA E 1 I 1 E 2 I 2 E 3 Spliced m. RNA Exon skipping (Exon. S) E 1 E 3
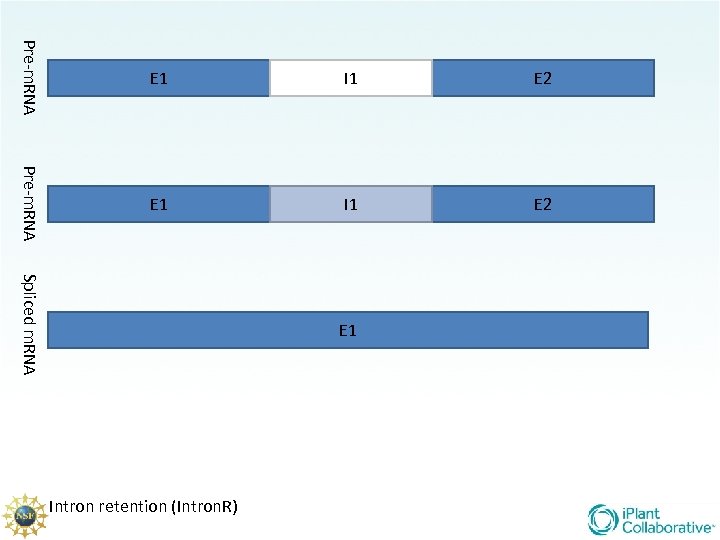 Pre-m. RNA E 1 I 1 E 2 Spliced m. RNA E 1 Intron retention (Intron. R)
Pre-m. RNA E 1 I 1 E 2 Spliced m. RNA E 1 Intron retention (Intron. R)
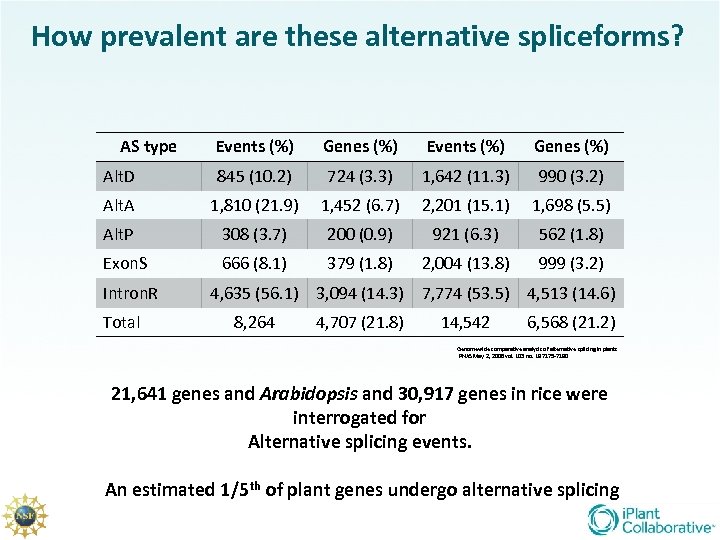 How prevalent are these alternative spliceforms? AS type Events (%) Genes (%) Events (%) Genes (%) Alt. D 845 (10. 2) 724 (3. 3) 1, 642 (11. 3) 990 (3. 2) Alt. A 1, 810 (21. 9) 1, 452 (6. 7) 2, 201 (15. 1) 1, 698 (5. 5) Alt. P 308 (3. 7) 200 (0. 9) 921 (6. 3) 562 (1. 8) Exon. S 666 (8. 1) 379 (1. 8) 2, 004 (13. 8) 999 (3. 2) Intron. R Total 4, 635 (56. 1) 3, 094 (14. 3) 7, 774 (53. 5) 4, 513 (14. 6) 8, 264 4, 707 (21. 8) 14, 542 6, 568 (21. 2) Genomewide comparative analysis of alternative splicing in plants PNAS May 2, 2006 vol. 103 no. 18 7175 -7180 21, 641 genes and Arabidopsis and 30, 917 genes in rice were interrogated for Alternative splicing events. An estimated 1/5 th of plant genes undergo alternative splicing
How prevalent are these alternative spliceforms? AS type Events (%) Genes (%) Events (%) Genes (%) Alt. D 845 (10. 2) 724 (3. 3) 1, 642 (11. 3) 990 (3. 2) Alt. A 1, 810 (21. 9) 1, 452 (6. 7) 2, 201 (15. 1) 1, 698 (5. 5) Alt. P 308 (3. 7) 200 (0. 9) 921 (6. 3) 562 (1. 8) Exon. S 666 (8. 1) 379 (1. 8) 2, 004 (13. 8) 999 (3. 2) Intron. R Total 4, 635 (56. 1) 3, 094 (14. 3) 7, 774 (53. 5) 4, 513 (14. 6) 8, 264 4, 707 (21. 8) 14, 542 6, 568 (21. 2) Genomewide comparative analysis of alternative splicing in plants PNAS May 2, 2006 vol. 103 no. 18 7175 -7180 21, 641 genes and Arabidopsis and 30, 917 genes in rice were interrogated for Alternative splicing events. An estimated 1/5 th of plant genes undergo alternative splicing
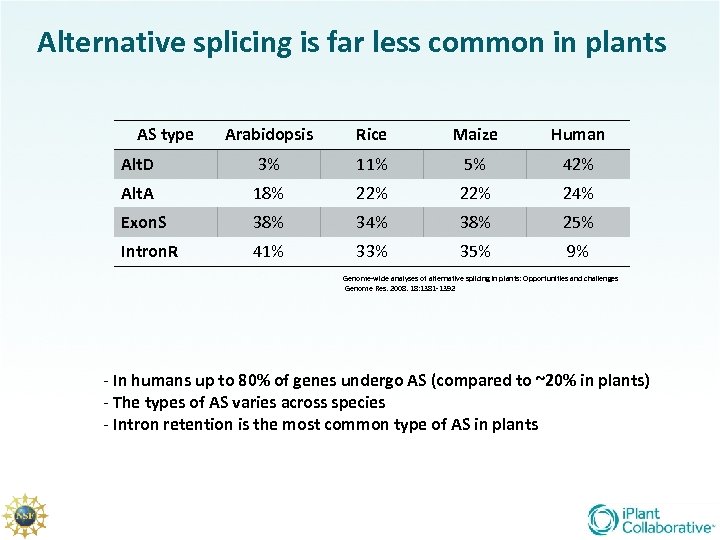 Alternative splicing is far less common in plants AS type Arabidopsis Rice Maize Human Alt. D 3% 11% 5% 42% Alt. A 18% 22% 24% Exon. S 38% 34% 38% 25% Intron. R 41% 33% 35% 9% Genome-wide analyses of alternative splicing in plants: Opportunities and challenges Genome Res. 2008. 18: 1381 -1392 - In humans up to 80% of genes undergo AS (compared to ~20% in plants) - The types of AS varies across species - Intron retention is the most common type of AS in plants
Alternative splicing is far less common in plants AS type Arabidopsis Rice Maize Human Alt. D 3% 11% 5% 42% Alt. A 18% 22% 24% Exon. S 38% 34% 38% 25% Intron. R 41% 33% 35% 9% Genome-wide analyses of alternative splicing in plants: Opportunities and challenges Genome Res. 2008. 18: 1381 -1392 - In humans up to 80% of genes undergo AS (compared to ~20% in plants) - The types of AS varies across species - Intron retention is the most common type of AS in plants
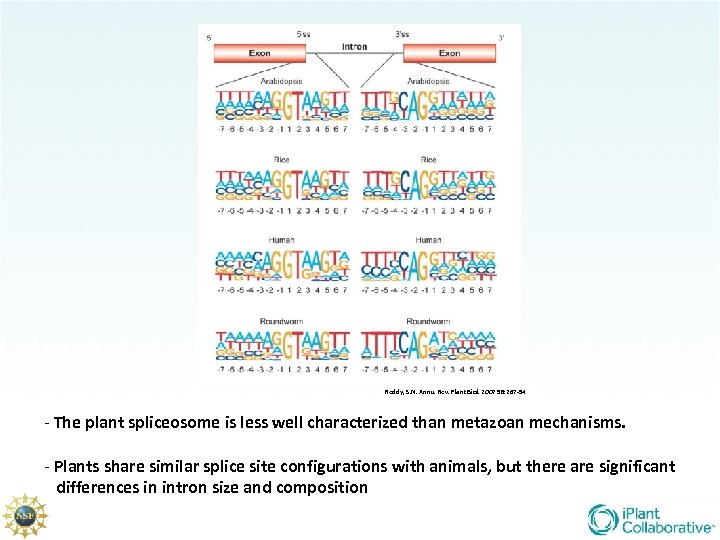 Reddy, S. N. Annu. Rev. Plant Biol. 2007 58: 267 -94 - The plant spliceosome is less well characterized than metazoan mechanisms. - Plants share similar splice site configurations with animals, but there are significant differences in intron size and composition
Reddy, S. N. Annu. Rev. Plant Biol. 2007 58: 267 -94 - The plant spliceosome is less well characterized than metazoan mechanisms. - Plants share similar splice site configurations with animals, but there are significant differences in intron size and composition
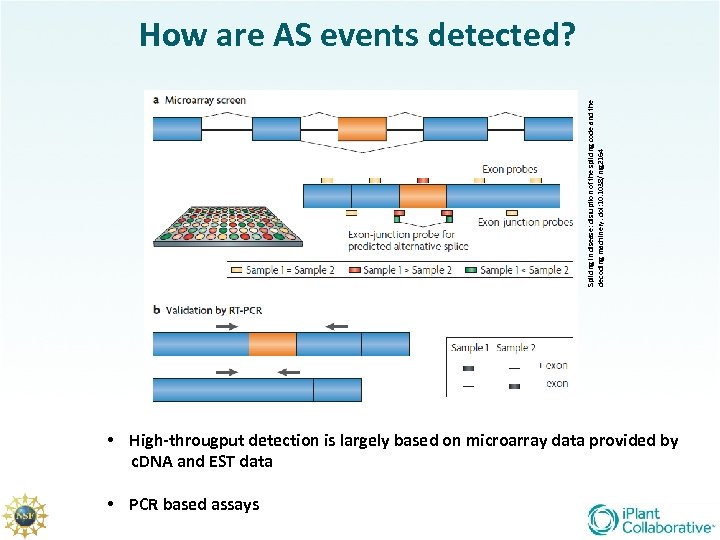 Splicing in disease: disruption of the splicing code and the decoding machinery. doi: 10. 1038/nrg 2164 How are AS events detected? • High-througput detection is largely based on microarray data provided by c. DNA and EST data • PCR based assays
Splicing in disease: disruption of the splicing code and the decoding machinery. doi: 10. 1038/nrg 2164 How are AS events detected? • High-througput detection is largely based on microarray data provided by c. DNA and EST data • PCR based assays
 Biological importance of AS So far, AS has been implicated in a number of biologically important roles including: - Splicing - Transcriptions - Flowering regulation - Disease resistance - Enzymatic activity A database of AS genes is available at plantgdb. org/ASIP/
Biological importance of AS So far, AS has been implicated in a number of biologically important roles including: - Splicing - Transcriptions - Flowering regulation - Disease resistance - Enzymatic activity A database of AS genes is available at plantgdb. org/ASIP/
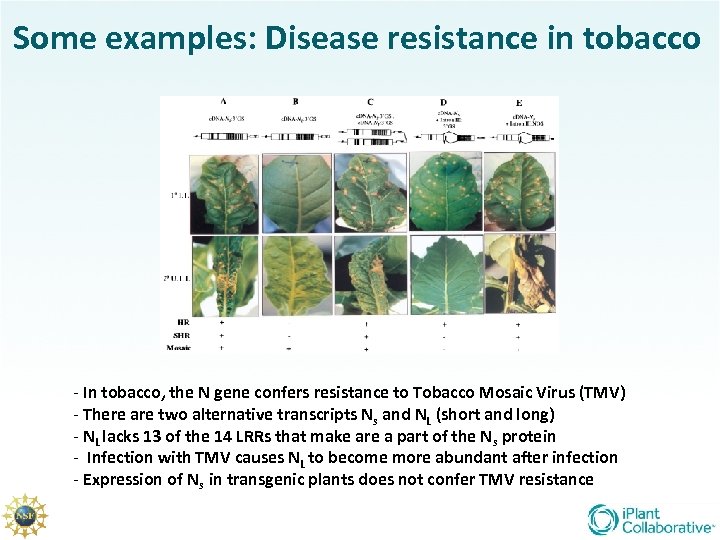 Some examples: Disease resistance in tobacco - In tobacco, the N gene confers resistance to Tobacco Mosaic Virus (TMV) - There are two alternative transcripts Ns and NL (short and long) - NL lacks 13 of the 14 LRRs that make are a part of the Ns protein - Infection with TMV causes NL to become more abundant after infection - Expression of Ns in transgenic plants does not confer TMV resistance
Some examples: Disease resistance in tobacco - In tobacco, the N gene confers resistance to Tobacco Mosaic Virus (TMV) - There are two alternative transcripts Ns and NL (short and long) - NL lacks 13 of the 14 LRRs that make are a part of the Ns protein - Infection with TMV causes NL to become more abundant after infection - Expression of Ns in transgenic plants does not confer TMV resistance
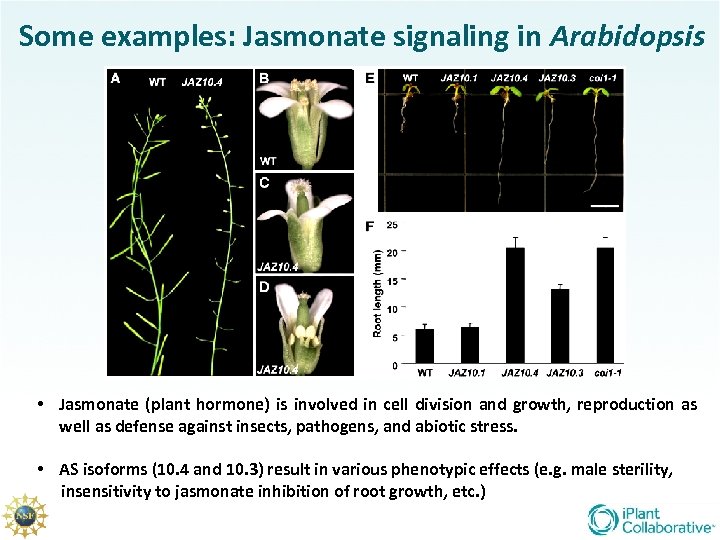 Some examples: Jasmonate signaling in Arabidopsis • Jasmonate (plant hormone) is involved in cell division and growth, reproduction as well as defense against insects, pathogens, and abiotic stress. • AS isoforms (10. 4 and 10. 3) result in various phenotypic effects (e. g. male sterility, insensitivity to jasmonate inhibition of root growth, etc. )
Some examples: Jasmonate signaling in Arabidopsis • Jasmonate (plant hormone) is involved in cell division and growth, reproduction as well as defense against insects, pathogens, and abiotic stress. • AS isoforms (10. 4 and 10. 3) result in various phenotypic effects (e. g. male sterility, insensitivity to jasmonate inhibition of root growth, etc. )
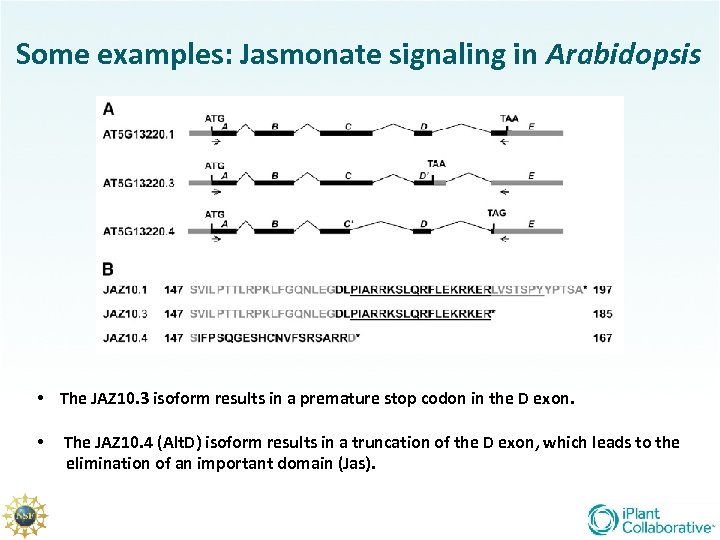 Some examples: Jasmonate signaling in Arabidopsis • The JAZ 10. 3 isoform results in a premature stop codon in the D exon. • The JAZ 10. 4 (Alt. D) isoform results in a truncation of the D exon, which leads to the elimination of an important domain (Jas).
Some examples: Jasmonate signaling in Arabidopsis • The JAZ 10. 3 isoform results in a premature stop codon in the D exon. • The JAZ 10. 4 (Alt. D) isoform results in a truncation of the D exon, which leads to the elimination of an important domain (Jas).


