b118600e04636dc9696df3970c86f204.ppt
- Количество слайдов: 45
Alignments and alignment reliability The first critical step in sequence analysis – the know how Eyal Privman and Osnat Penn Tel Aviv University COST Training School Rehovot, 2010
What are alignments good for? n To compare sequences n Find homology n Similar sequence similar function n To learn about sequence evolution n Mismatch = point mutation n Gap = indel (insertion or deletion) n Reconstruct phylogenetic tree n Infer selection forces, e. g. , detecting positive selection
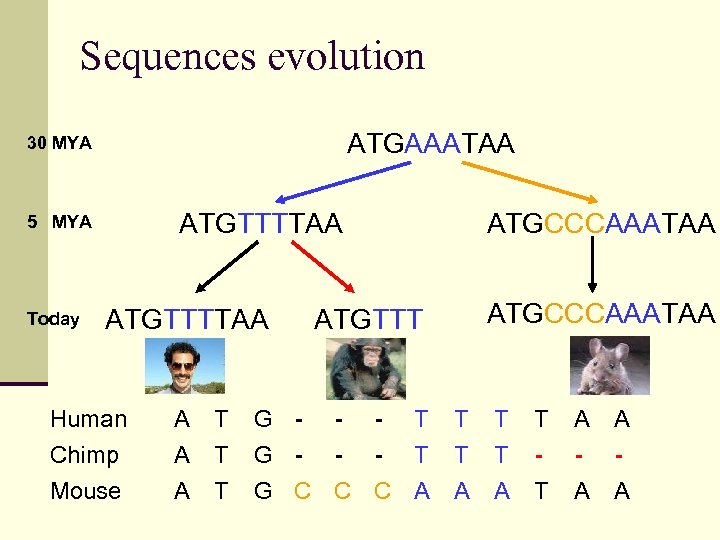
Sequences evolution ATGAAATAA 30 MYA ATGTTTTAA 5 MYA Today ATGTTTTAA Human Chimp Mouse A A A T T T ATGCCCAAATAA ATGTTT G - - - T G C C C A T T A A
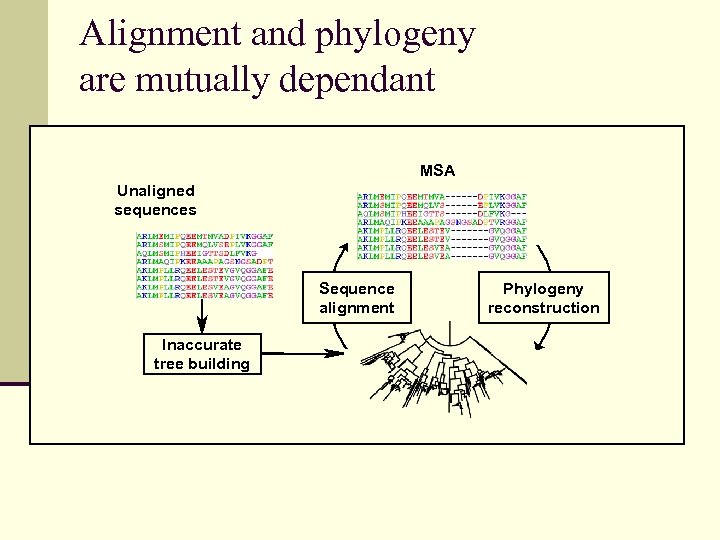
Alignment and phylogeny are mutually dependant MSA Unaligned sequences Sequence alignment Inaccurate tree building Phylogeny reconstruction
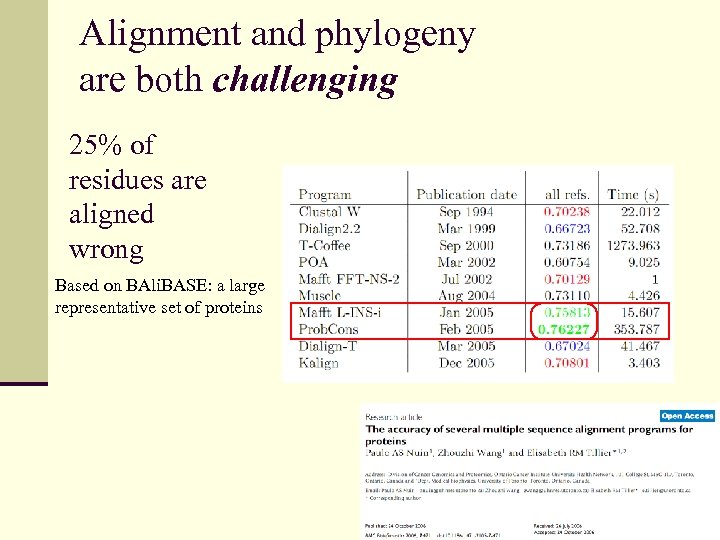
Alignment and phylogeny are both challenging 25% of residues are aligned wrong Based on BAli. BASE: a large representative set of proteins
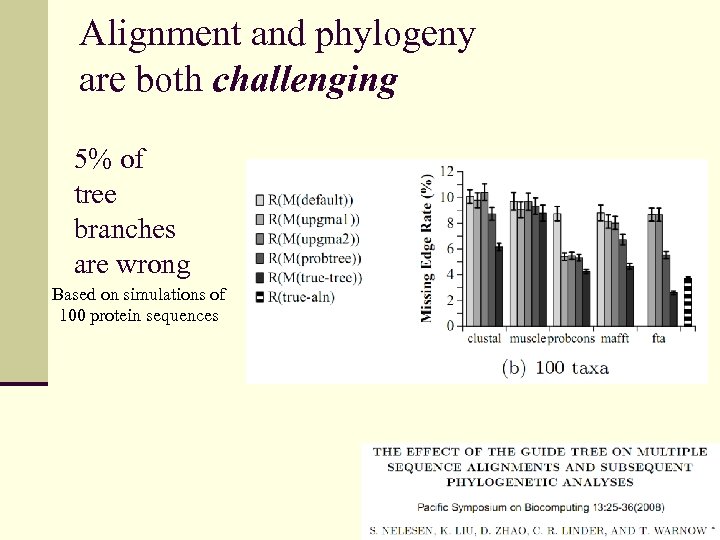
Alignment and phylogeny are both challenging 5% of tree branches are wrong Based on simulations of 100 protein sequences
Making an alignment n For 2 sequences : use exact methods. n For more sequences: n Exact methods are not feasible (too slow) n We use heuristic methods
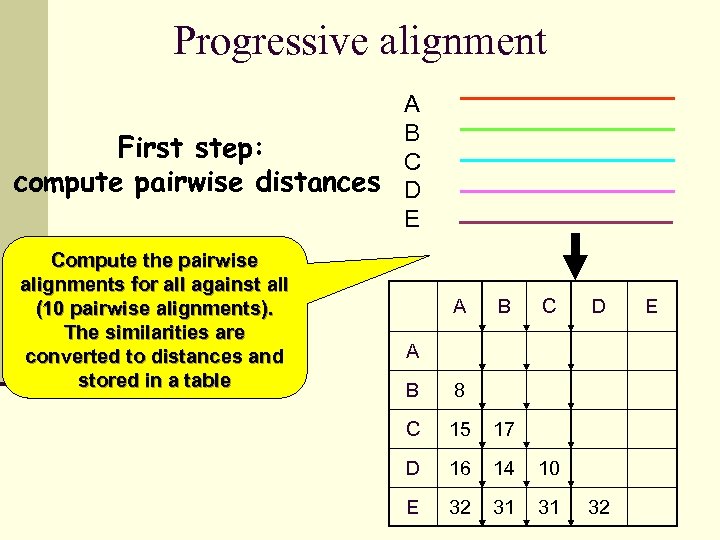
Progressive alignment A B First step: C compute pairwise distances D E Compute the pairwise alignments for all against all (10 pairwise alignments). The similarities are converted to distances and stored in a table A B C D A B 8 C 15 17 D 16 14 10 E 32 31 31 32 E
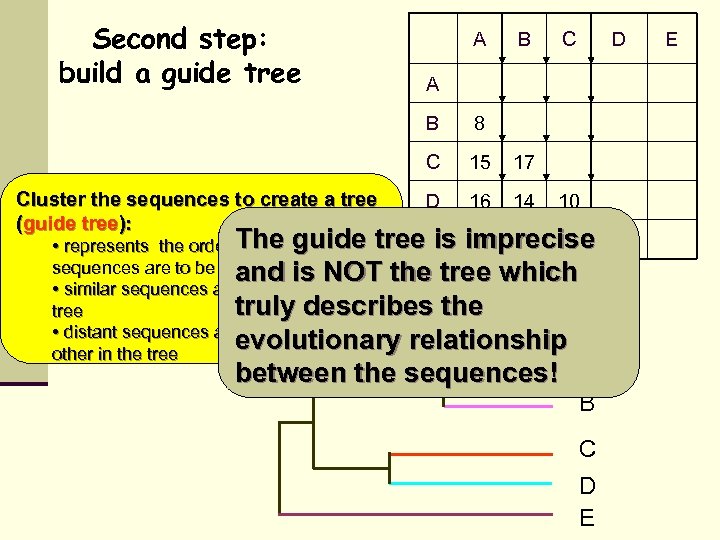
Second step: build a guide tree A B D A B 8 C Cluster the sequences to create a tree (guide tree): C 15 17 D 16 14 10 The guide is 32 31 31 • represents the order in which pairs of tree E imprecise 32 sequences are to be aligned is NOT the tree which and • similar sequences are neighbors in the truly describes the tree • distant sequences are distant from each evolutionary relationship A other in the tree between the sequences! B C D E E
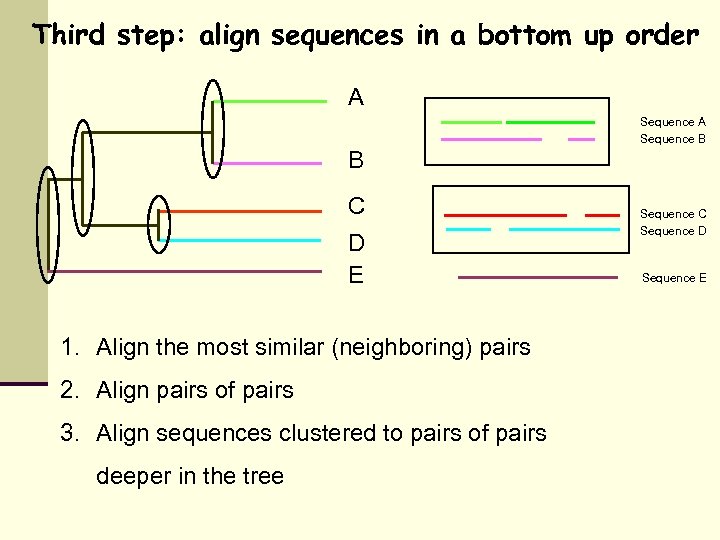
Third step: align sequences in a bottom up order A Sequence B B C D E 1. Align the most similar (neighboring) pairs 2. Align pairs of pairs 3. Align sequences clustered to pairs of pairs deeper in the tree Sequence C Sequence D Sequence E
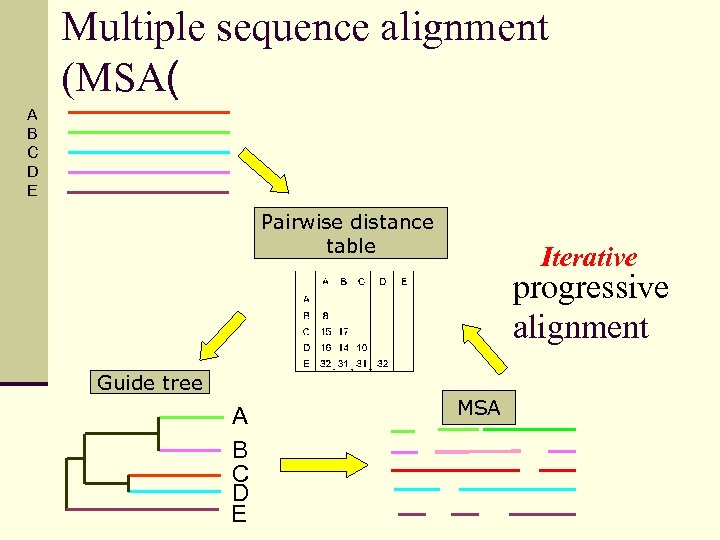
Multiple sequence alignment (MSA( A B C D E Pairwise distance table Iterative progressive alignment Guide tree A B C D E MSA

Multiple sequence alignment (MSA( Several advanced MSA programs are available. Today we will use two: n MAFFT – fastest and one of the most accurate n PRANK – distinct from all other MSA programs because of its correct treatment of insertions/deletions
Nucleic Acids Research, 2002, Vol. 30, No. 14 3059 -3066 © 2002 Oxford University Press MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform Kazutaka Katoh, Kazuharu Misawa 1, Kei-ichi Kuma and Takashi Miyata* n Web server & download: http: //align. bmr. kyushu-u. ac. jp/mafft/online/server/ n Efficiency-tuned variants quick & dirty or slow but accurate
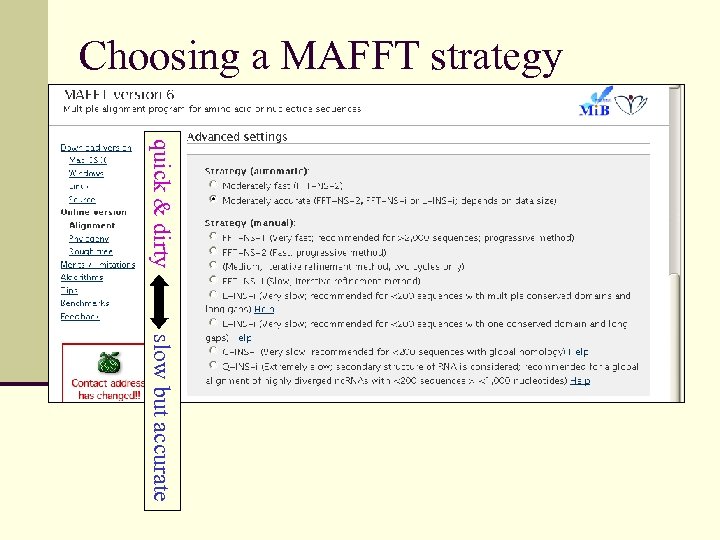
Choosing a MAFFT strategy quick & dirty slow but accurate
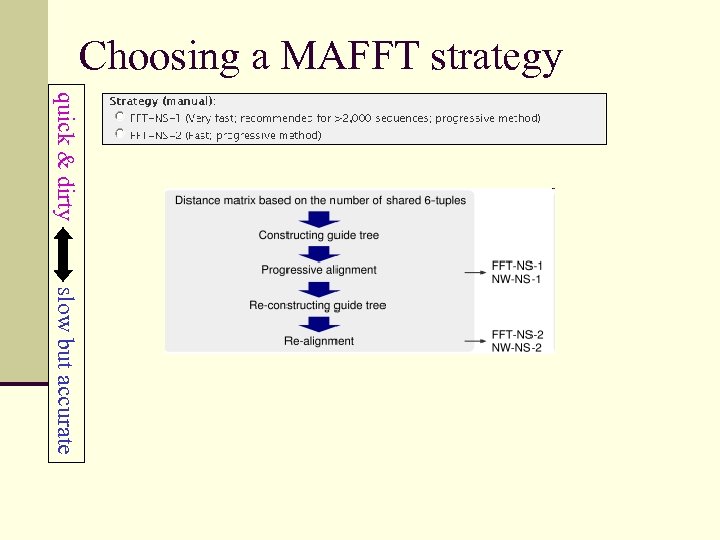
Choosing a MAFFT strategy quick & dirty slow but accurate
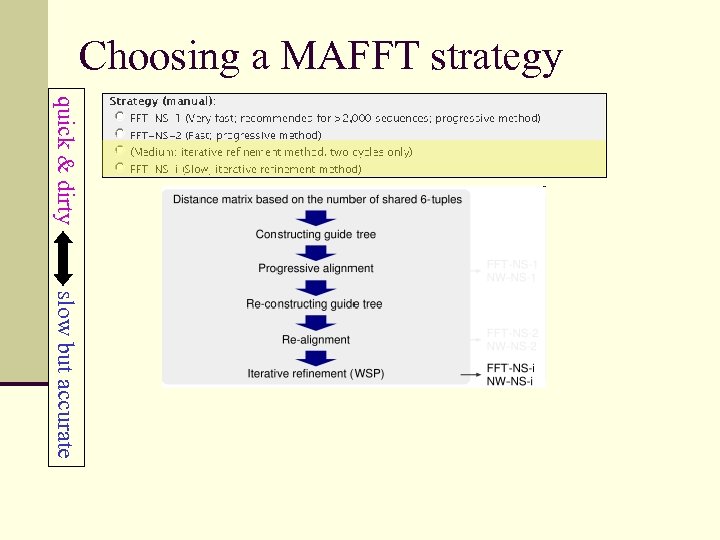
Choosing a MAFFT strategy quick & dirty slow but accurate
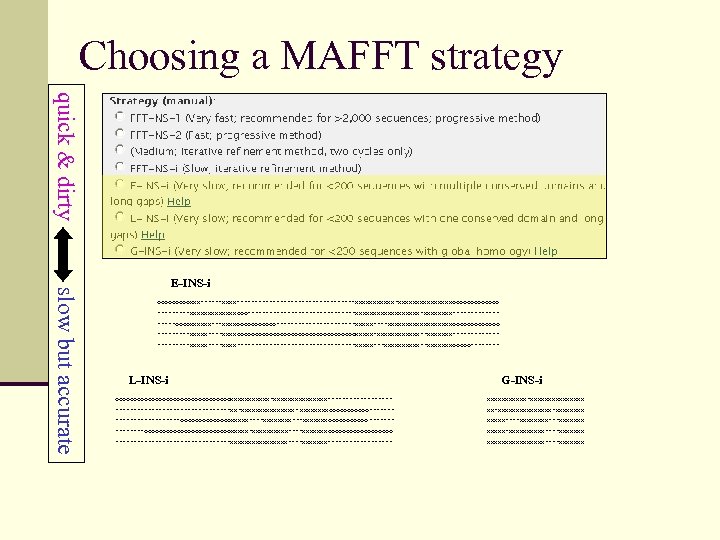
Choosing a MAFFT strategy quick & dirty slow but accurate E-INS-i ooooo. XXX------XXXX-----------------XXXXXXXXXXXXXXXooooooo -----XXXXXXXooo---------------XXXXXXXXX-XXXX---------oooo. XXXXXX---XXXXoooooo-----------XXXXX----XXXXXXXXXooooooo -----XXXXX----XXXXooooooooooooooooo. XXXXX-XXXXXX--XXXXXXX-----------XXXXX-------------------XXXXX---XXXXX--XXXXXXXooooo---- L-INS-i G-INS-i oooooooooooooooo. XXXXXX-XXXXXXXX--------- XXXXXX-XXXXXXXX ----------------XX-XXXXXXXX- XXXXoooooo------- XX-XXXXXXXX-XXXX ---------ooooooo. XXXXX----XXXX---XXXXXXXoooooo ------- XXXXX----XXXX---XXXXXXX ----oooooooooooo. XXXXX-XXXXX----XXXXXXXooooooooo XXXXX-XXXXX----XXXXXXX ----------------XXXXXXXX----XXXXXXX--------- XXXXXXXX----XXXXXXX
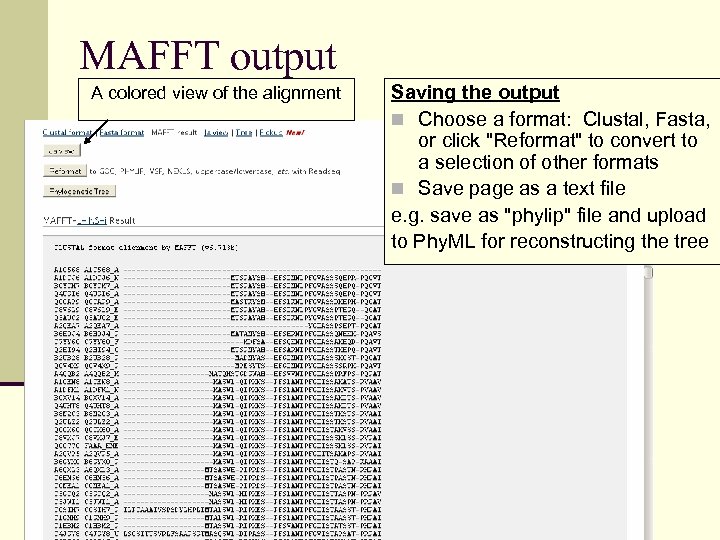
MAFFT output A colored view of the alignment Saving the output n Choose a format: Clustal, Fasta, or click "Reformat" to convert to a selection of other formats n Save page as a text file e. g. save as "phylip" file and upload to Phy. ML for reconstructing the tree
Phy. ML: tree reconstruction The most widely used maximum likelihood (ML) program n Web server & download: http: //www. atgc-montpellier. fr/phyml/
PRANK
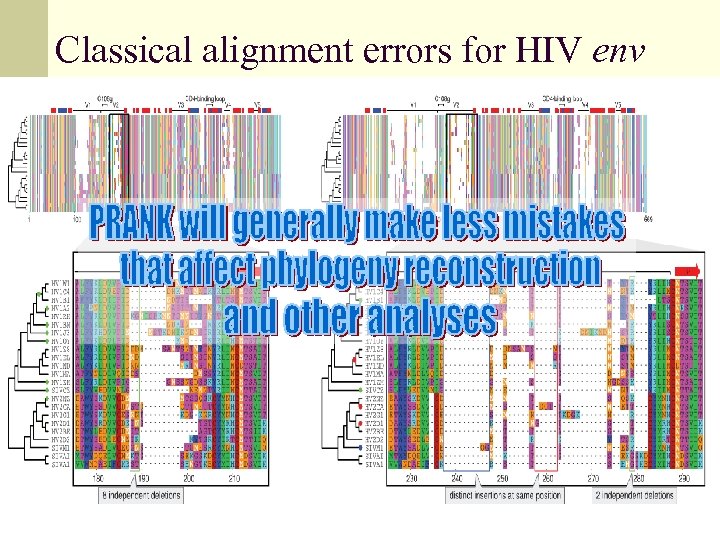
Classical alignment errors for HIV env
PRANK n Web server: http: //www. ebi. ac. uk/goldman-srv/web. PRANK/
PRANK output If you need a different format – copy the results to the READSEQ sequence converter: http: //www-bimas. cit. nih. gov/molbio/readseq/
1. Download and save the sequences file from Osnat's homepage (you can google “Osnat Penn" and look for the workshop materials under "Teaching"). Save the file as "trim 5 a. AA. fas" (File “Save page as”). This file contains 20 protein sequences in FASTA format. 2. Run PRANK web-server to create a protein alignment: a. In the “Default alignment” section browse for “trim 5 a. AA. fas”. b. Run (press the “Start alignment“ button). 3. While you wait: copy the sequences into the MAFFT web server and run the "automatic" "moderately accurate" strategy – which strategy did MAFFT choose for you? Click on the "Fasta format“ link, and save as “trim 5 a. AA. mafft. aln“ (File “Save page as”) and try the "Jalview" button. 4. When PRANK finishes click on the “Show Fasta file” button, and save the MSA by the name “trim 5 a. AA. prank. aln“.
Sources of alignment errors Progressive alignment algorithms are greedy heuristics v Co-optimal solutions Heads-or-Tails (Ho. T) scores (Landan & Graur 2007) v Guide-tree errors GUIDANCE scores (Penn, Privman et al. MBE 2010)
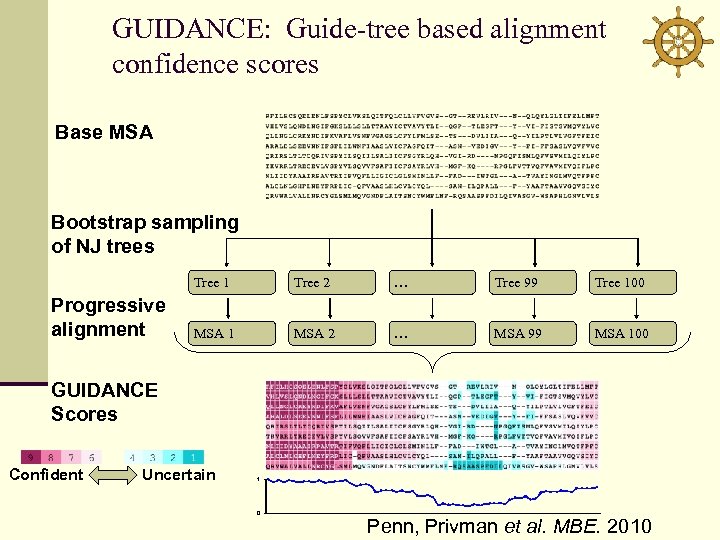
GUIDANCE: Guide-tree based alignment confidence scores Base MSA Bootstrap sampling of NJ trees Tree 1 Progressive alignment Tree 2 … Tree 99 Tree 100 MSA 1 MSA 2 … MSA 99 MSA 100 GUIDANCE Scores Confident Uncertain 1 0 Penn, Privman et al. MBE. 2010
http: //guidance. tau. ac. il
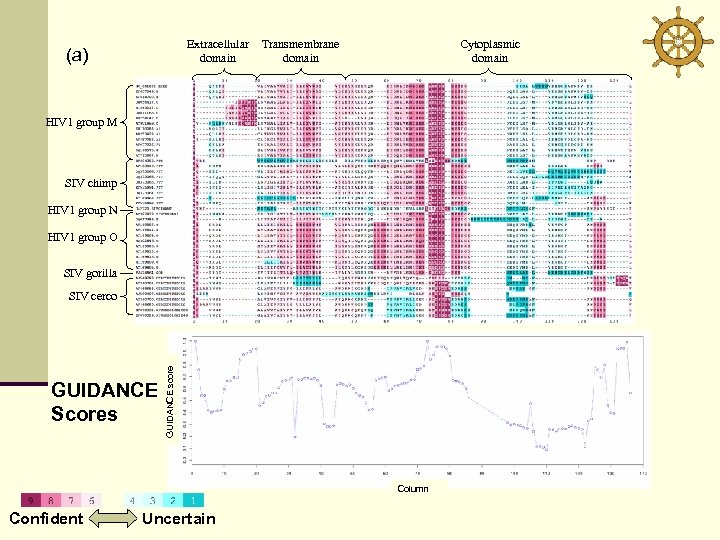
Extracellular domain (a) Transmembrane domain Cytoplasmic domain HIV 1 group M SIV chimp HIV 1 group N HIV 1 group O SIV gorilla GUIDANCE Scores GUIDANCE score SIV cerco Column Confident Uncertain
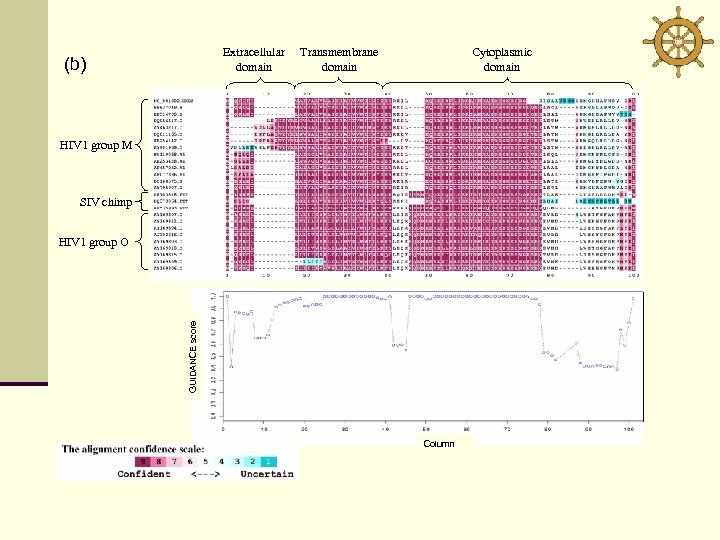
Extracellular domain (b) Transmembrane domain Cytoplasmic domain HIV 1 group M SIV chimp GUIDANCE score HIV 1 group O Column
1. Run GUIDANCE web-server to calculate confidence scores for the MAFFT alignment: a. In the “Upload your sequence file” window browse for “trim 5 a. AA. fas”. b. Choose “Amino Acids” in the “Sequences Type” option. c. In order to speed the run, change the “Number of bootstrap repeats” in the “Advanced options” section to 30. Note that this is not recommended for real life. d. Run (press the “Submit“ button).

Detecting selection forces Positive selection
Empirical findings variation among genes: “Important” proteins evolve slower than “unimportant” ones unimportant
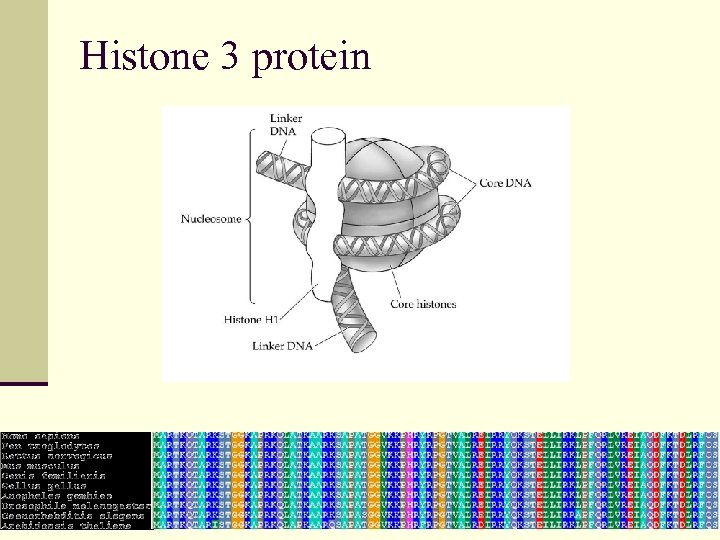
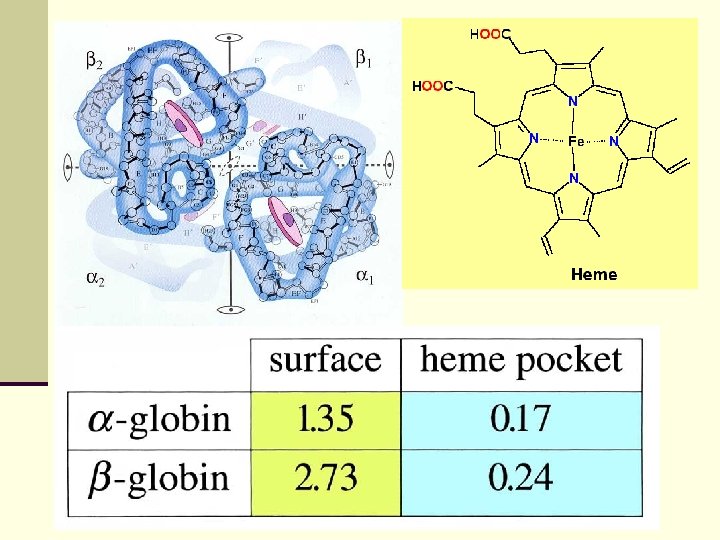
Histone 3 protein
Empirical findings variation among sites: Functional sites evolve slower than nonfunctional sites
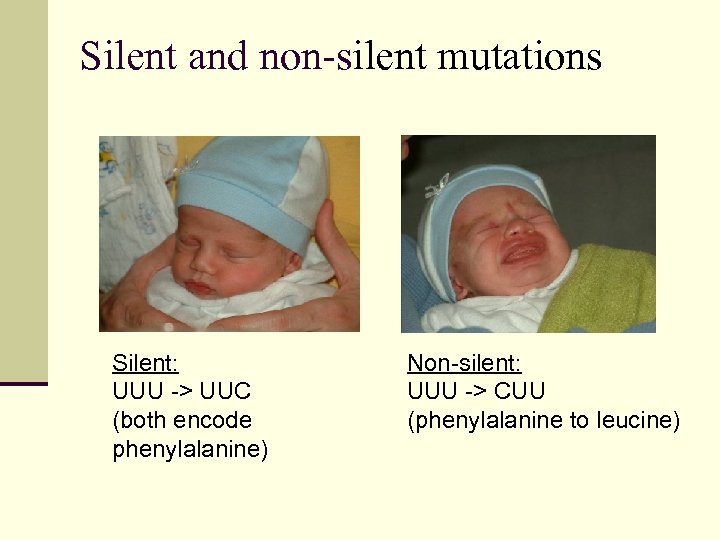
Silent and non-silent mutations Silent: UUU -> UUC (both encode phenylalanine) Non-silent: UUU -> CUU (phenylalanine to leucine)

For most proteins, the rate of silent substitutions is much higher than the non-silent rate This is called purifying selection = conservation

There are rare cases where the non-silent rate is much higher than the silent rate This is called positive selection
Positive Selection Examples: n Pathogen proteins evading the host immune system n Proteins of the immune system detecting pathogen proteins n Pathogen proteins that are drug targets n Proteins that are products of gene duplication n Proteins involved in the reproductive system
http: //selecton. tau. ac. il

Selecton results
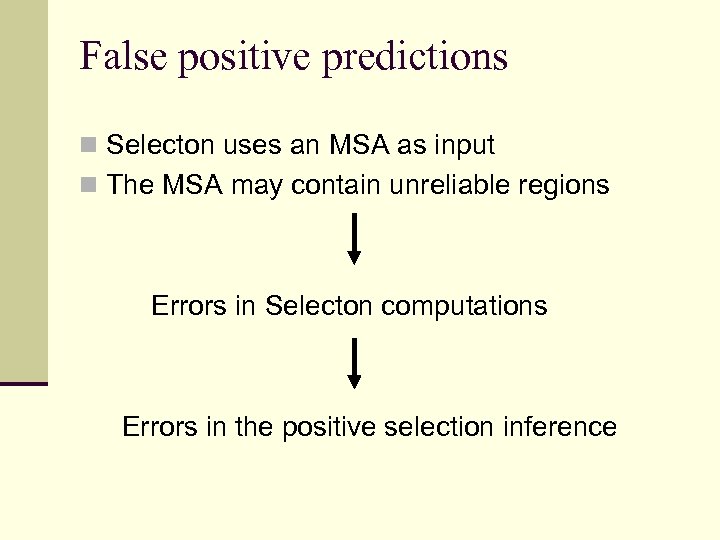
False positive predictions n Selecton uses an MSA as input n The MSA may contain unreliable regions Errors in Selecton computations Errors in the positive selection inference

1. Go to the GUIDANCE results of the last exercise. 2. Which columns are not well aligned? Are these sites also predicted to evolve under positive selection? See Selecton results in: http: //selecton. tau. ac. il/results/1268662868/colors. html
Summary n Different alignment programs may result different MSAs. n Alignment uncertainty may cause errors in downstream analyses such as positive selection analysis. n GUIDANCE can detect alignment errors.

Thanks for your attention!
b118600e04636dc9696df3970c86f204.ppt