Alibekova Alina spectrometry applications in proteomics. Recent innovations


Alibekova Alina spectrometry applications in proteomics

Recent innovations in spectrometry have significantly improved its application in the study of protein structure and function The value of mass spectrometry to biologists has been established by its effectiveness in characterizing the structure and dynamics of proteins at the femtomole to picomole level. Mass spectrometry can be used to characterize function-critical posttranslational modifications, including phosphorylation and glycosylation, to determine the numbers and positions of disulfide bonds in proteins, and to investigate macromolecular complexes, such as protein-ligand, protein-protein, and protein-DNA interactions .
Proteomics, which is generally defined as the characterization of the complete protein complement of a cell, tissue, or organism (a proteome), has moved to the forefront of "big science" now that the complete genomes of several organisms--including man--have been sequenced. But why is proteomics important? Simply put, proteins define the function of cells, tissues, and even organisms. Every cell in the human body contains an equivalent set of genes, but not every cell expresses the same ones. Only B cells, for example, express immunoglobulins. In addition, each gene can give rise to multiple proteins, either through alternative splicing or post-translational modifications. Finally, there is no direct correlation between mRNA abundance and steady-state protein levels. For all of these reasons, the field of proteomics is booming.
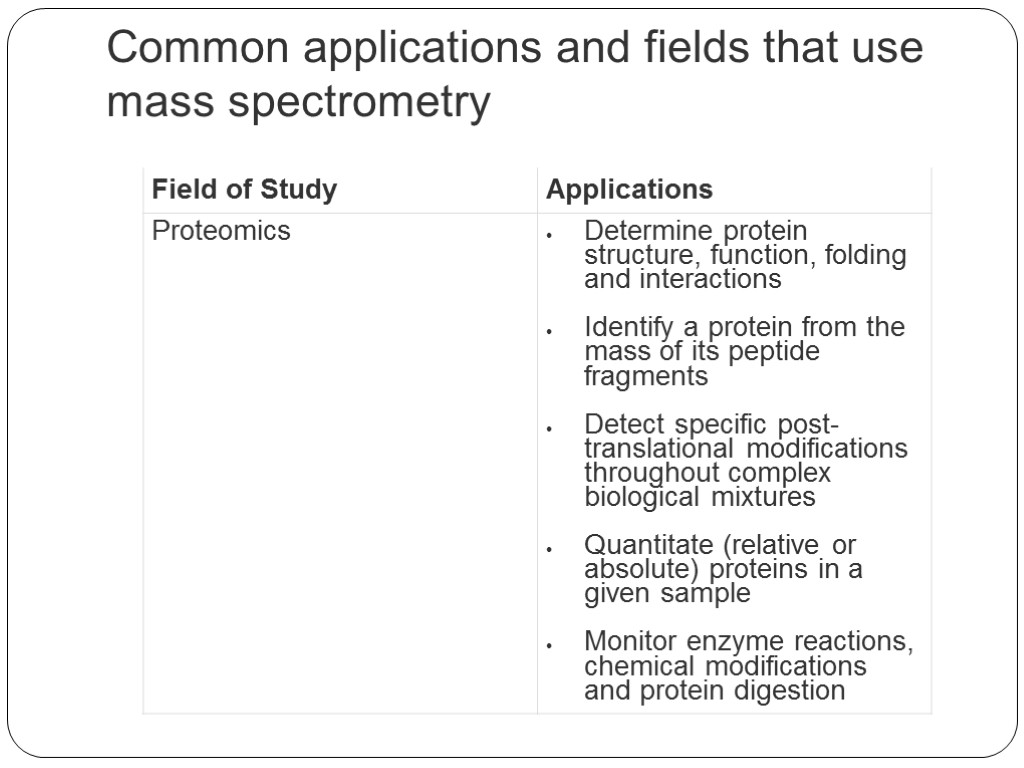
Common applications and fields that use mass spectrometry
Gas chromatography (GC) and liquid chromatography (LC) are common methods of pre-MS separation that are used when analyzing complex gas or liquid samples by MS, respectively. LC-MS is typically applied to the analysis of thermally unstable and nonvolatile molecules (e.g., sensitive biological fluids), while GC-MS is used for the analysis of volatile compounds such as petrochemicals. LC-MS and GC-MS also use different methods for ionization of the compound as it is introduced into the mass spectrometer. With LC-MS, electrospray ionization (EI) is commonly applied, resulting in the production of aerosolized ions. With GC-MS, the sample may be ionized directly or indirectly via EI.

Electrospray ionization is the primary ion source used in liquid chromatography-mass spectrometry because it is a liquid-gas interface capable of coupling liquid chomatography with mass spectrometry.
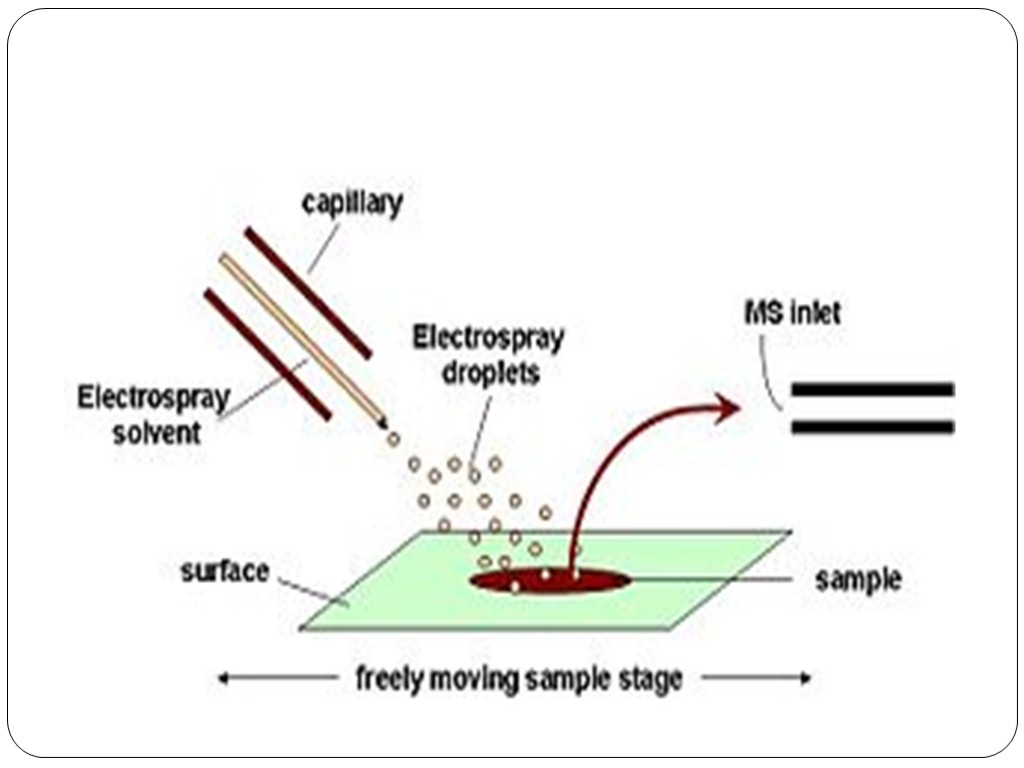
In ambient ionization, the formation of ions occurs outside the mass spectrometer without sample preparation. Electrospray is used for ion formation is a number of ambient ion sources. Desorption electrospray ionization (DESI) is an ambient ionization technique in which a solvent electrospray is directed at a sample. The electrospray is attracted to the surface by applying a voltage to the sample. Sample compounds are extracted into the solvent which is again aerosolized as highly charged droplets that evaporate to form highly charged ions. After ionization, the ions enter the atmospheric pressure interface of the mass spectrometer. DESI allows for ambient ionization of samples at atmospheric pressure, with little sample preparation. Extractive electrospray ionization is an spray-type, ambient ionization method that uses two merged sprays, one of which is generated by electrospray. Laser-based electrospray-based ambient ionization is a two-step process in which a pulsed laser is used to desorb or ablate material from a sample and the plume of material interacts with an electrospray to create ions. For ambient ionization, the sample material is deposited on a target near the electrospray. The laser desorbs or ablates material from the sample which is ejected from the surface and into the electrospray which produces highly charged ions. Examples are electrospray laser desorption ionization, matrix-assisted laser desorption electrospray ionization, and laser ablation electrospray ionization

High performance liquid chromatography (HPLC) is the most common separation method to study biological samples by MS or MS/MS (termed LC-MS or LC-MS/MS, respectively), because the majority of biological samples are liquid and nonvolatile. LC columns have small diameters (e.g., 75 μm; nanoHPLC) and low flow rates (e.g., 200 nL/min), which are ideal for minute samples (2). Additionally, "in-line" liquid chromatography (LC linked directly to MS) provides a high-throughput approach to sample analysis, enabling multiple analytes to elute through the column at different rates to be immediately analyzed by MS. For example, 1-5 peptides in a complex biological mixture can be sequenced per second by in-line LC-MS/MS
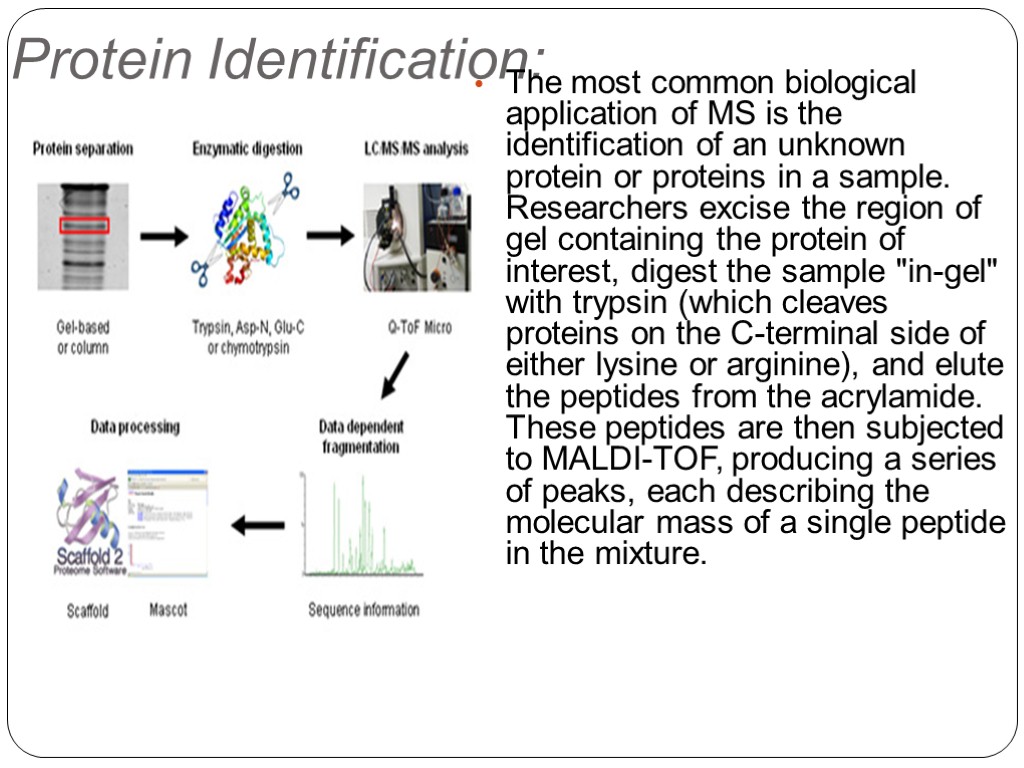
Protein Identification: The most common biological application of MS is the identification of an unknown protein or proteins in a sample. Researchers excise the region of gel containing the protein of interest, digest the sample "in-gel" with trypsin (which cleaves proteins on the C-terminal side of either lysine or arginine), and elute the peptides from the acrylamide. These peptides are then subjected to MALDI-TOF, producing a series of peaks, each describing the molecular mass of a single peptide in the mixture.

Peptide Sequencing: Researchers introduce the desired peptide into the machine, fragment it along the peptide backbone via collision-induced dissociation; mass analyze the resulting fragments. Two or more series of fragments are produced, depending on where the peptide bonds break. Fragments from one series will vary in mass from each other by the weight of a single amino acid, enabling the researcher to deduce the peptide sequence. For example, in a tryptic digestion, the C-terminal residue in the peptide chain is either lysine (145) or arginine (173). The mass of next larger fragment should then be the sum of 145 or 173, plus the mass of the next N-terminal residue. Ideally, peptide sequencing approaches will yield a "ladder" of peptide fragments, although in practice this is not always observed because only those fragments that include the charged ion can be analyzed.
spectrometry_applic.inproteomics.ppt
- Количество слайдов: 12

