b80598224cd6c97996334d066cc10aec.ppt
- Количество слайдов: 21
Accurate Multiplex Polony Sequencing of an Evolved Bacterial Genome Jay Shendure, Gregory J. Porreca, Nikos B. Reppas, Xiaoxia Lin, John P. Mc. Cutcheon. Abraham M. Rosenbaum, Michael D. Wang, Kun Zhang, Robi D. Mitra, George M. Church Science 2005 vol 309, pg 1728 -32

Authors - BA 1973: Duke University - Ph. D 1984 : Harvard University - Professor of Genetics at Harvard Medical School - Director of the Harvard/MIT DOE Genomes-to-Life Center - Director of the Harvard/MIT/Wash. U George M. Church NHGRI CEGS Jay Shendure - MD-Ph. D Student in the Church Lab
Background - Sanger Sequencing • Developed By Fredrick Sanger • Uses dd. NTPs to block DNA synthesis • DNA fragments run in gel and analyzed

Sequence analysis - Then • Use radioactively labeled d. NTPs • Run samples in four columns • Expose your film • Manually determine sequence www. carnegieinstitution. org
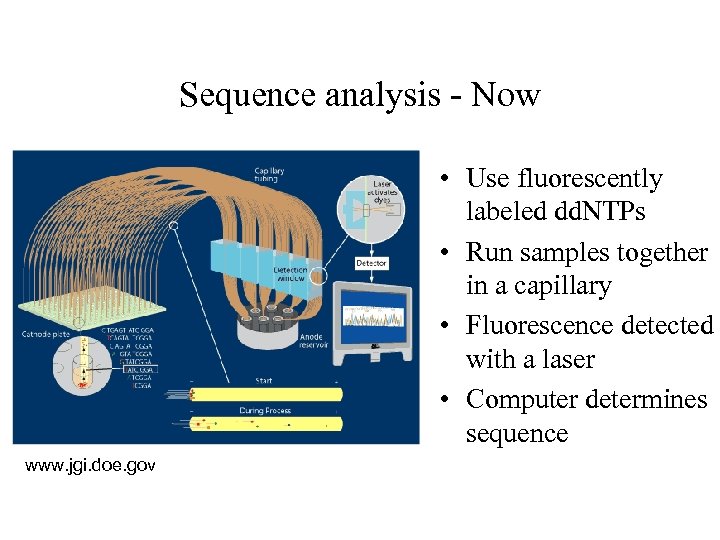
Sequence analysis - Now • Use fluorescently labeled dd. NTPs • Run samples together in a capillary • Fluorescence detected with a laser • Computer determines sequence www. jgi. doe. gov

The Problem • Electrophoretic methods may very well be reaching their limits • New sequencing method need to be developed if we are to achieve the goal of the $1000 genome
The Problem • Electrophoretic methods may very well be reaching their limits • New sequencing method need to be developed if we are to achieve the goal of the $1000 genome • The Solution: Use a cyclic array method
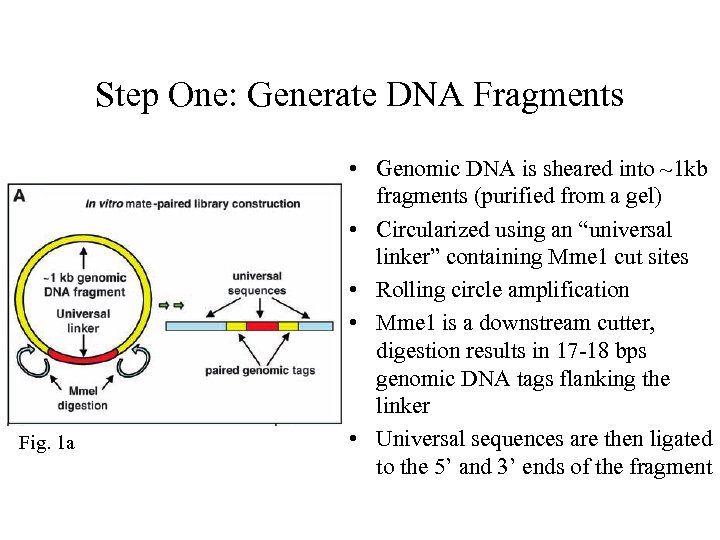
Step One: Generate DNA Fragments Fig. 1 a • Genomic DNA is sheared into ~1 kb fragments (purified from a gel) • Circularized using an “universal linker” containing Mme 1 cut sites • Rolling circle amplification • Mme 1 is a downstream cutter, digestion results in 17 -18 bps genomic DNA tags flanking the linker • Universal sequences are then ligated to the 5’ and 3’ ends of the fragment
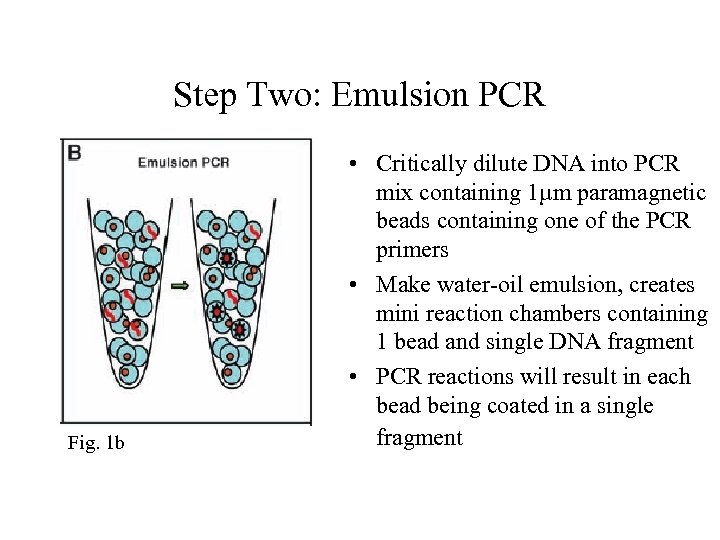
Step Two: Emulsion PCR Fig. 1 b • Critically dilute DNA into PCR mix containing 1µm paramagnetic beads containing one of the PCR primers • Make water-oil emulsion, creates mini reaction chambers containing 1 bead and single DNA fragment • PCR reactions will result in each bead being coated in a single fragment
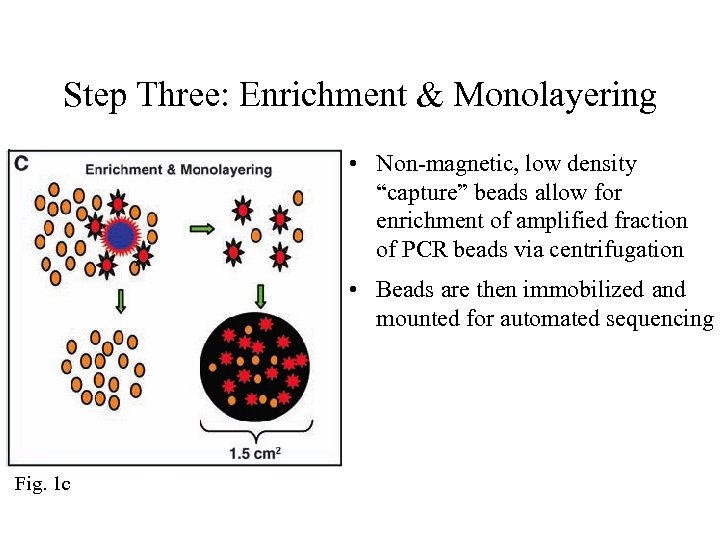
Step Three: Enrichment & Monolayering • Non-magnetic, low density “capture” beads allow for enrichment of amplified fraction of PCR beads via centrifugation • Beads are then immobilized and mounted for automated sequencing Fig. 1 c

Step Four: Cycles of Sequencing and Imaging • Sequencing is done in automated cycles • Computer algorithm identifies each bead and determines which color it is fluorescing each round Fig. 1 d
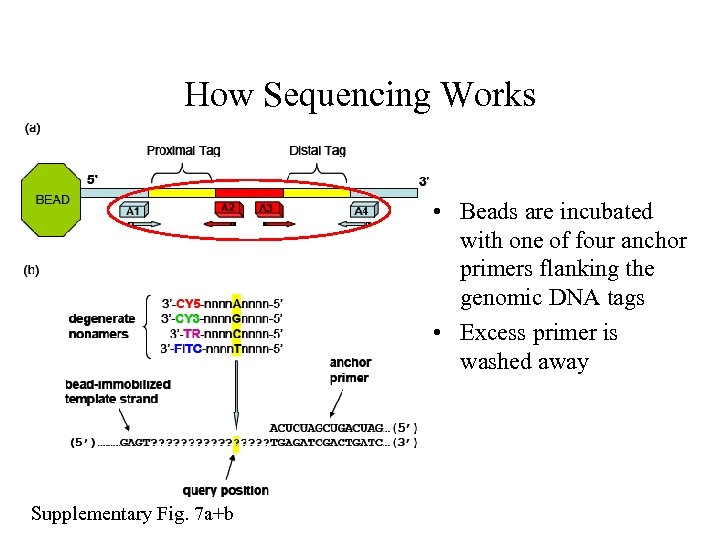
How Sequencing Works • Beads are incubated with one of four anchor primers flanking the genomic DNA tags • Excess primer is washed away Supplementary Fig. 7 a+b
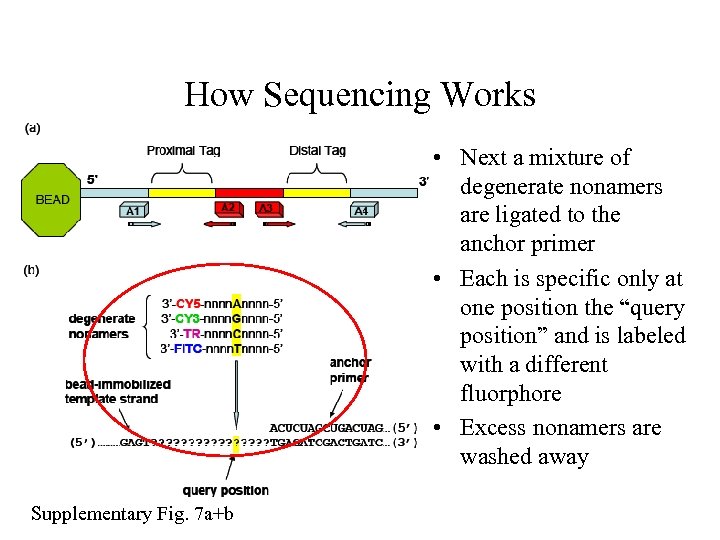
How Sequencing Works • Next a mixture of degenerate nonamers are ligated to the anchor primer • Each is specific only at one position the “query position” and is labeled with a different fluorphore • Excess nonamers are washed away Supplementary Fig. 7 a+b
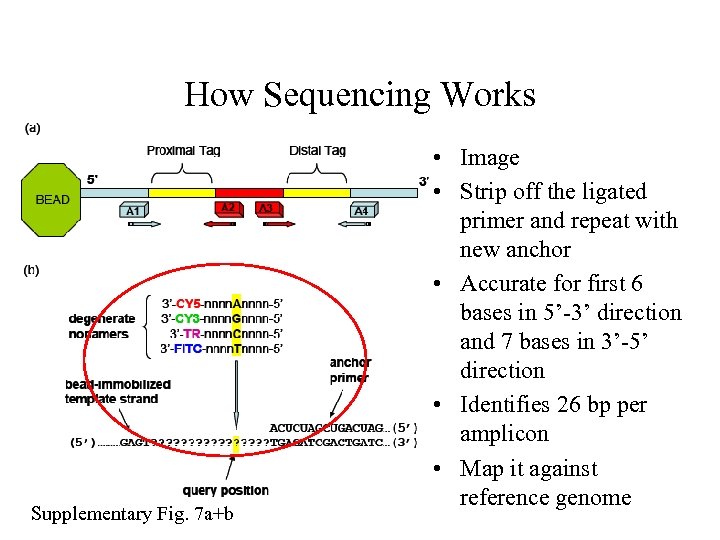
How Sequencing Works Supplementary Fig. 7 a+b • Image • Strip off the ligated primer and repeat with new anchor • Accurate for first 6 bases in 5’-3’ direction and 7 bases in 3’-5’ direction • Identifies 26 bp per amplicon • Map it against reference genome
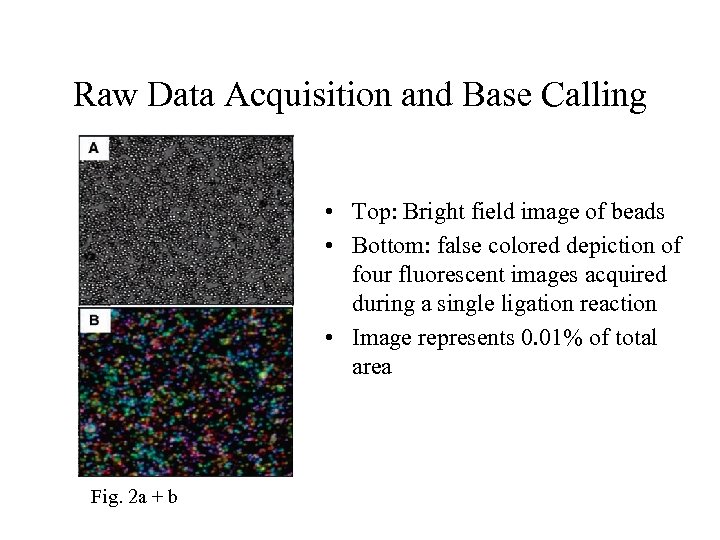
Raw Data Acquisition and Base Calling • Top: Bright field image of beads • Bottom: false colored depiction of four fluorescent images acquired during a single ligation reaction • Image represents 0. 01% of total area Fig. 2 a + b
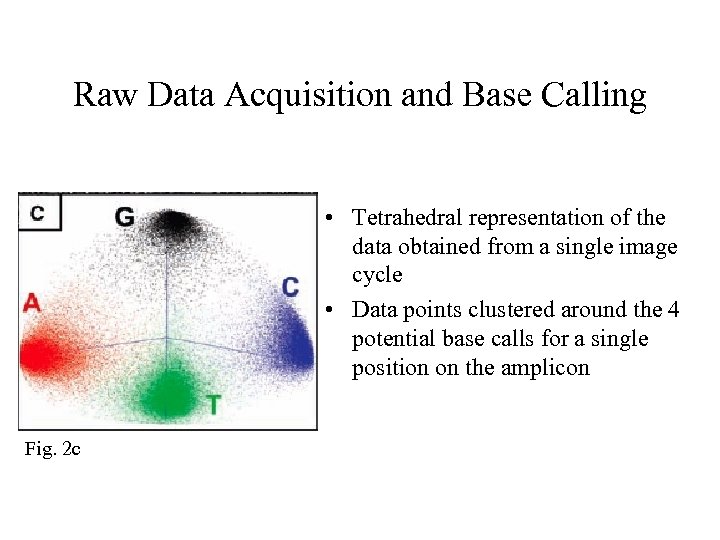
Raw Data Acquisition and Base Calling • Tetrahedral representation of the data obtained from a single image cycle • Data points clustered around the 4 potential base calls for a single position on the amplicon Fig. 2 c
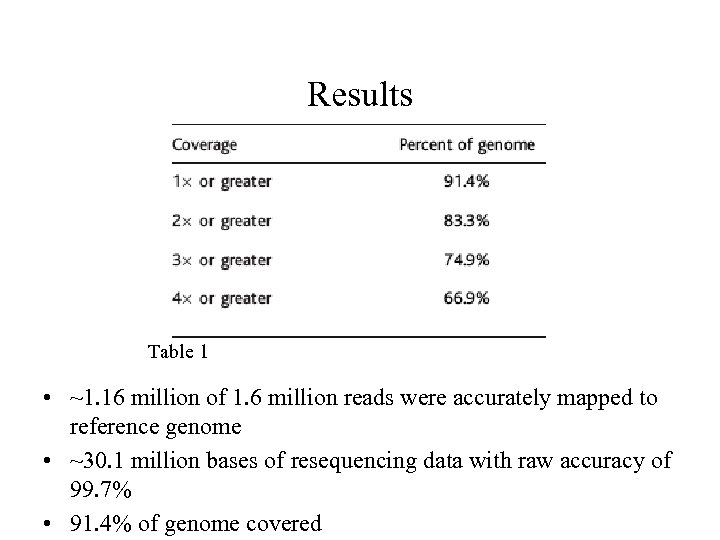
Results Table 1 • ~1. 16 million of 1. 6 million reads were accurately mapped to reference genome • ~30. 1 million bases of resequencing data with raw accuracy of 99. 7% • 91. 4% of genome covered
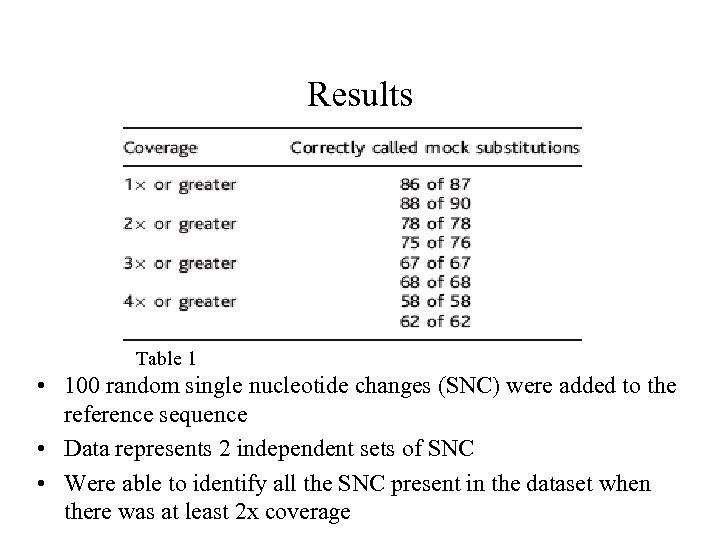
Results Table 1 • 100 random single nucleotide changes (SNC) were added to the reference sequence • Data represents 2 independent sets of SNC • Were able to identify all the SNC present in the dataset when there was at least 2 x coverage
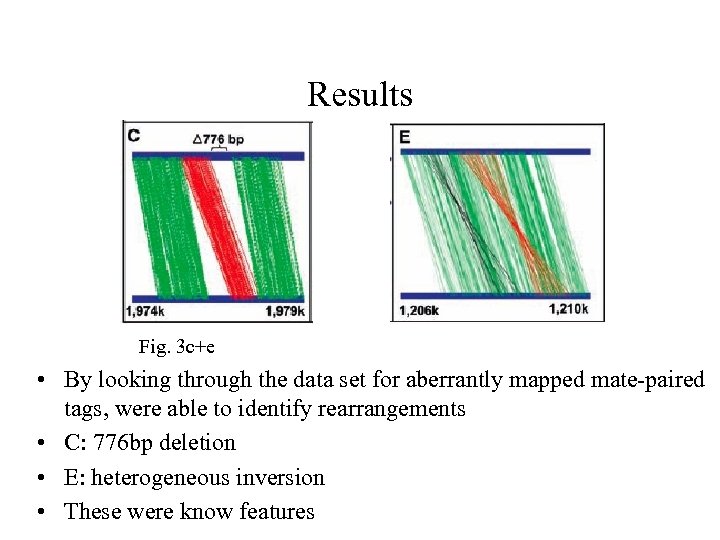
Results Fig. 3 c+e • By looking through the data set for aberrantly mapped mate-paired tags, were able to identify rearrangements • C: 776 bp deletion • E: heterogeneous inversion • These were know features
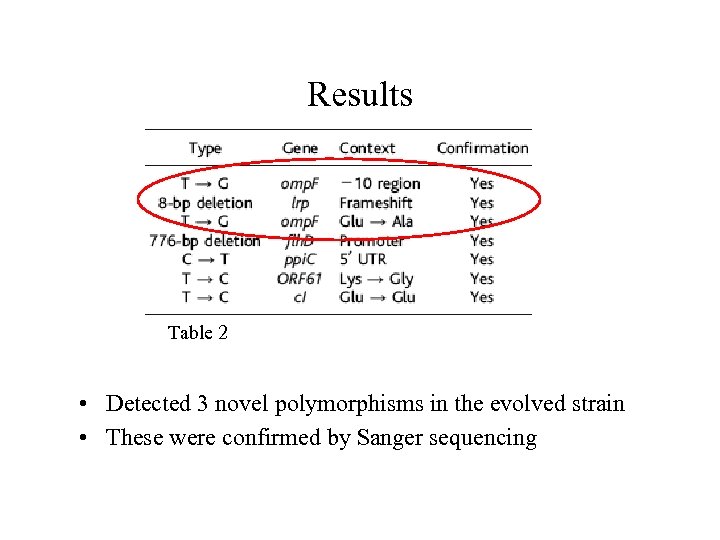
Results Table 2 • Detected 3 novel polymorphisms in the evolved strain • These were confirmed by Sanger sequencing

Conclusions • Current high-throughput centers sequence at a rate of 20 bases per instrument second at a cost of $1. 00 per kb • This method has an overall sequence rate of ~140 bp/s and a cost of $0. 08 per kb, in house enzyme production could drop this number even lower • Open source technology. • Reasonably cheap to set up equipment ~ $140, 000
b80598224cd6c97996334d066cc10aec.ppt