3f0ab95d0891d7d2dc73a8ba8cf4c44f.ppt
- Количество слайдов: 44
 A Tool To Manage & View Whole Genome Shotgun (WGS) Sequence Assemblies René Warren January 8 th, 2004
A Tool To Manage & View Whole Genome Shotgun (WGS) Sequence Assemblies René Warren January 8 th, 2004
 WGS Sequence Projects @ GSC Rhodococcus sp. RHA 1 -Abundant aerobic soil bacteria with potential use for bioremediation -Capable of degrading polychlorinated biphenyls (PCBs) and other aromatic compounds -Produces a number of secondary metabolites that may represent a pharmacological interest Cryptococcus neoformans WM 276 -Cryptococcus is a ubiquitous human pathogen -Leading cause of infectious meningitis -Australian environmental strain WM 276; Sister strain to serotype B that caused the outbreak on Vancouver Island infecting >50 immuno-competent people
WGS Sequence Projects @ GSC Rhodococcus sp. RHA 1 -Abundant aerobic soil bacteria with potential use for bioremediation -Capable of degrading polychlorinated biphenyls (PCBs) and other aromatic compounds -Produces a number of secondary metabolites that may represent a pharmacological interest Cryptococcus neoformans WM 276 -Cryptococcus is a ubiquitous human pathogen -Leading cause of infectious meningitis -Australian environmental strain WM 276; Sister strain to serotype B that caused the outbreak on Vancouver Island infecting >50 immuno-competent people
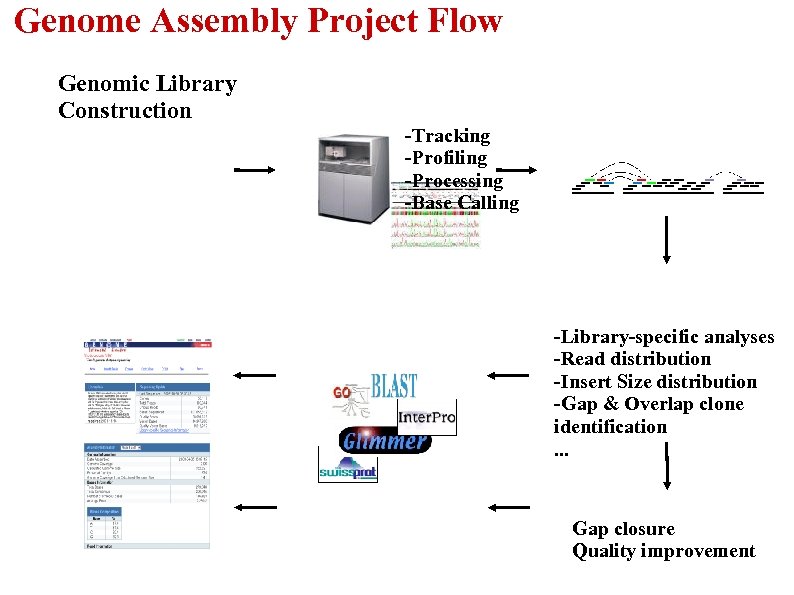 Genome Assembly Project Flow Genomic Library Construction -Tracking -Profiling -Processing -Base Calling -Library-specific analyses -Read distribution -Insert Size distribution -Gap & Overlap clone identification. . . Gap closure Quality improvement
Genome Assembly Project Flow Genomic Library Construction -Tracking -Profiling -Processing -Base Calling -Library-specific analyses -Read distribution -Insert Size distribution -Gap & Overlap clone identification. . . Gap closure Quality improvement
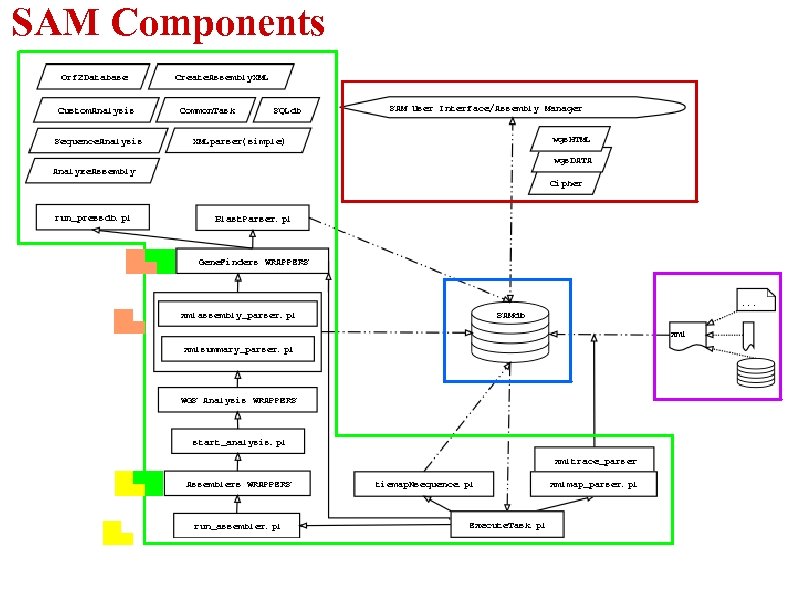
 What Is SAM & Why Using It? -Generic platform for manipulating WGS assembly data and data analysis, regardless of input type -Execute, View, Analyze, Manipulate WGS assemblies -Provide an automated platform for gene prediction and annotation -Provide a convenient/easy way to store & access WGS assembly data -Eases the use of external programs through web interface -Central repository for storing assembly, analysis, annotation & finishing information
What Is SAM & Why Using It? -Generic platform for manipulating WGS assembly data and data analysis, regardless of input type -Execute, View, Analyze, Manipulate WGS assemblies -Provide an automated platform for gene prediction and annotation -Provide a convenient/easy way to store & access WGS assembly data -Eases the use of external programs through web interface -Central repository for storing assembly, analysis, annotation & finishing information
 SAM Components
SAM Components
 SAM Components
SAM Components
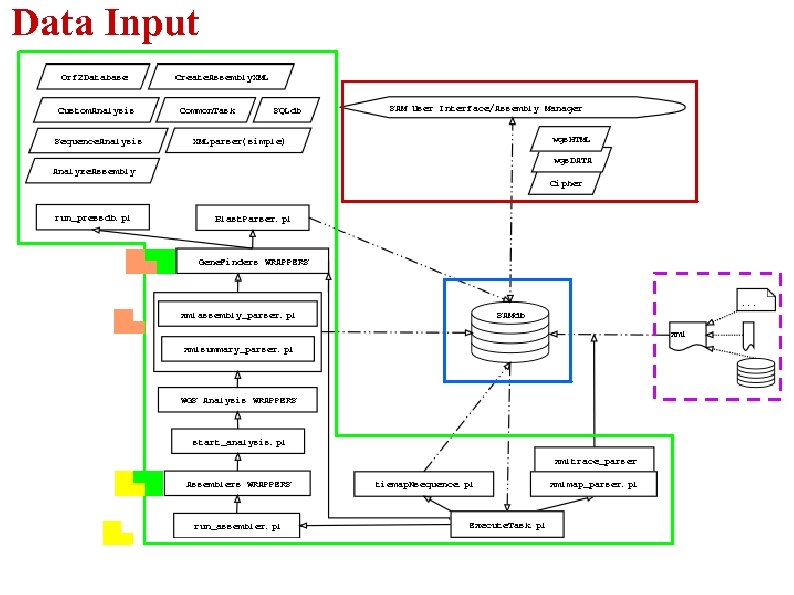 Data Input
Data Input
 Data Input -Two Types of Input: -Trace Archive Data -Fingerprint Map Data -To account for different data sources, XML files are created -Sequence db on lims-dbm and fingerprint clone flat file are our current sources -Additional programs must be written to account for the different sources -Those programs are NOT an integral part of the system -XML makes system generic and flexible
Data Input -Two Types of Input: -Trace Archive Data -Fingerprint Map Data -To account for different data sources, XML files are created -Sequence db on lims-dbm and fingerprint clone flat file are our current sources -Additional programs must be written to account for the different sources -Those programs are NOT an integral part of the system -XML makes system generic and flexible
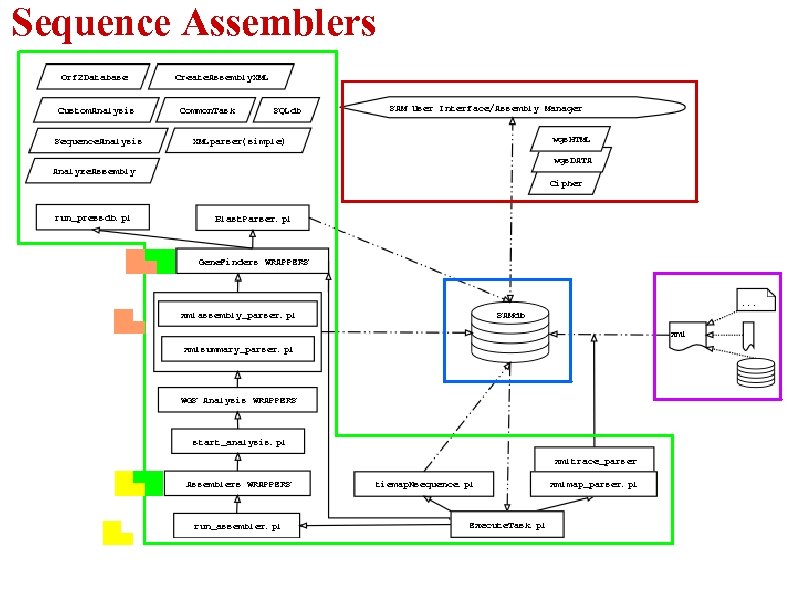 Sequence Assemblers
Sequence Assemblers
 Sequence Assemblers -Concurrently use 2 public Assemblers: Phrap and Arachne -System is designed to easily “Plug in” any new assemblers Proper wrapper must be written for each new added programs Phrap ●Allows use of entire read (not just Arachne ●Trims terminal regions of low quality and trimmed high quality part) eliminate reads with overall low quality ●Construct ● contig sequence as a Eliminate duplicate reads(exact and highly mosaic of the highest quality parts of similar) the reads ●Build contigs by merging read pairs that don't cross a repeat boundary ●Supercontig creation by linking pairs of contigs sharing >=2 forward-reverse links (2
Sequence Assemblers -Concurrently use 2 public Assemblers: Phrap and Arachne -System is designed to easily “Plug in” any new assemblers Proper wrapper must be written for each new added programs Phrap ●Allows use of entire read (not just Arachne ●Trims terminal regions of low quality and trimmed high quality part) eliminate reads with overall low quality ●Construct ● contig sequence as a Eliminate duplicate reads(exact and highly mosaic of the highest quality parts of similar) the reads ●Build contigs by merging read pairs that don't cross a repeat boundary ●Supercontig creation by linking pairs of contigs sharing >=2 forward-reverse links (2
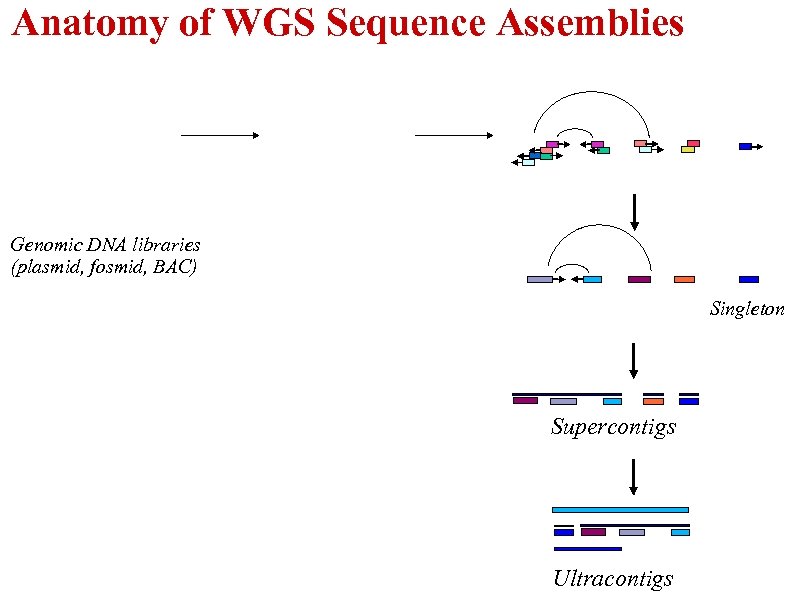 Anatomy of WGS Sequence Assemblies Genomic DNA libraries (plasmid, fosmid, BAC) Singleton Supercontigs Ultracontigs
Anatomy of WGS Sequence Assemblies Genomic DNA libraries (plasmid, fosmid, BAC) Singleton Supercontigs Ultracontigs
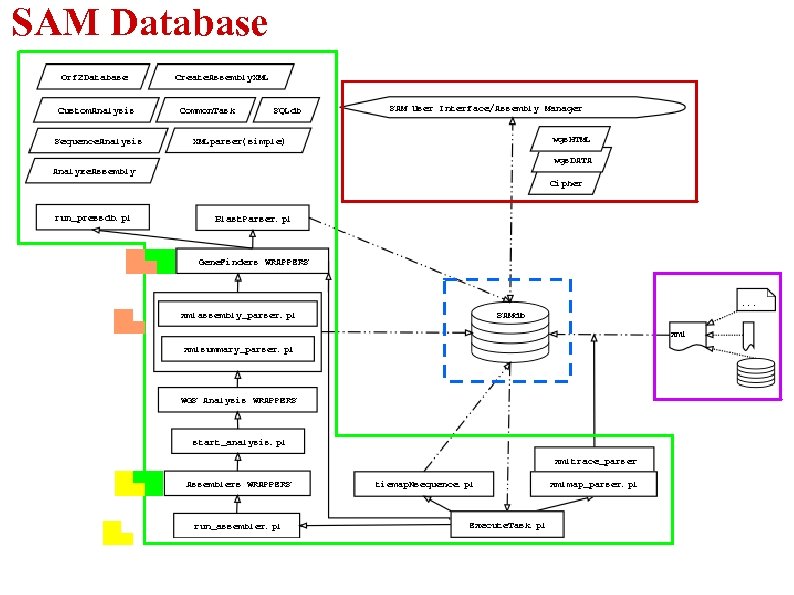 SAM Database
SAM Database
 SAM Database -My. SQL database -37 tables -Info stored as follows:
SAM Database -My. SQL database -37 tables -Info stored as follows:
 SAM Database -Modules (PERL) written for managing DB connection / allow basic read/write interactions with the database -Credentials needed to read/write from/to SAMdb is provided: -at command line -from web (fairly secure using PERL modules that freezes, compresses, encrypts and encodes data)
SAM Database -Modules (PERL) written for managing DB connection / allow basic read/write interactions with the database -Credentials needed to read/write from/to SAMdb is provided: -at command line -from web (fairly secure using PERL modules that freezes, compresses, encrypts and encodes data)
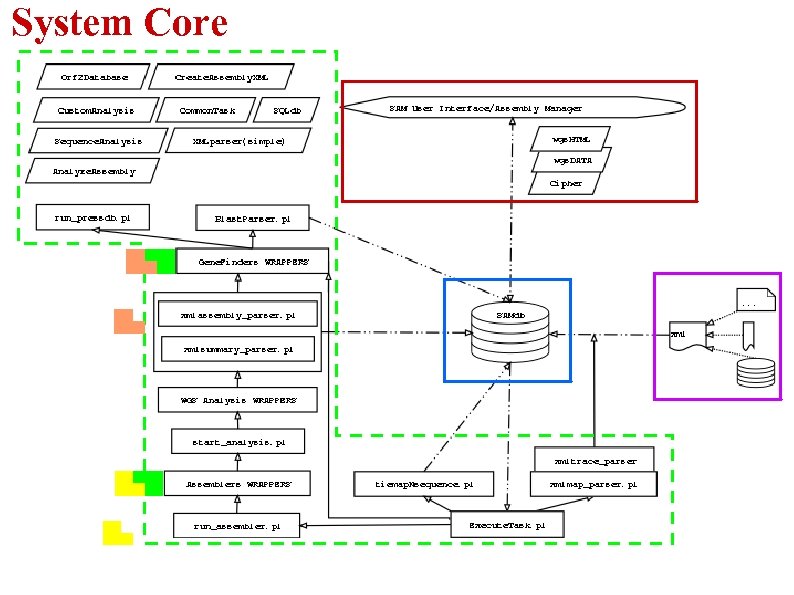 System Core
System Core
 System Core -10 PERL scripts -9 Wrappers (PERL/Shell Script) -11 modules -System is designed to use any assembly program and any gene finders as they become available -External program parameters, location, input, output directories are all located in the database (Config table) -Typically, configuration information is initially retrieved from SAMdb by head program and passed on. -Individual libraries perform set of related task: DB Interaction (basic) XML creation/parsing HTML creation Assembly Parsing Assembly Analysis Gap/Lib/Overlap Encryption Data Manip(WEB) DB dumps
System Core -10 PERL scripts -9 Wrappers (PERL/Shell Script) -11 modules -System is designed to use any assembly program and any gene finders as they become available -External program parameters, location, input, output directories are all located in the database (Config table) -Typically, configuration information is initially retrieved from SAMdb by head program and passed on. -Individual libraries perform set of related task: DB Interaction (basic) XML creation/parsing HTML creation Assembly Parsing Assembly Analysis Gap/Lib/Overlap Encryption Data Manip(WEB) DB dumps
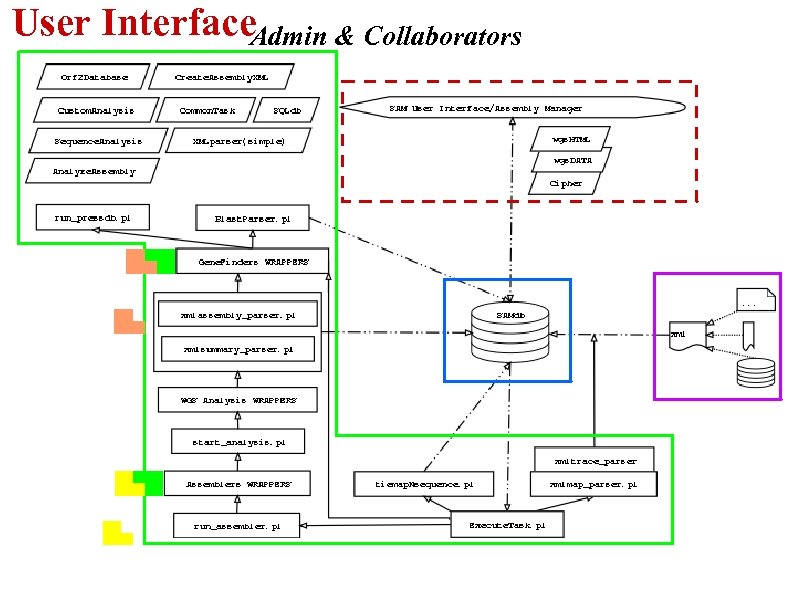 User Interface. Admin & Collaborators
User Interface. Admin & Collaborators
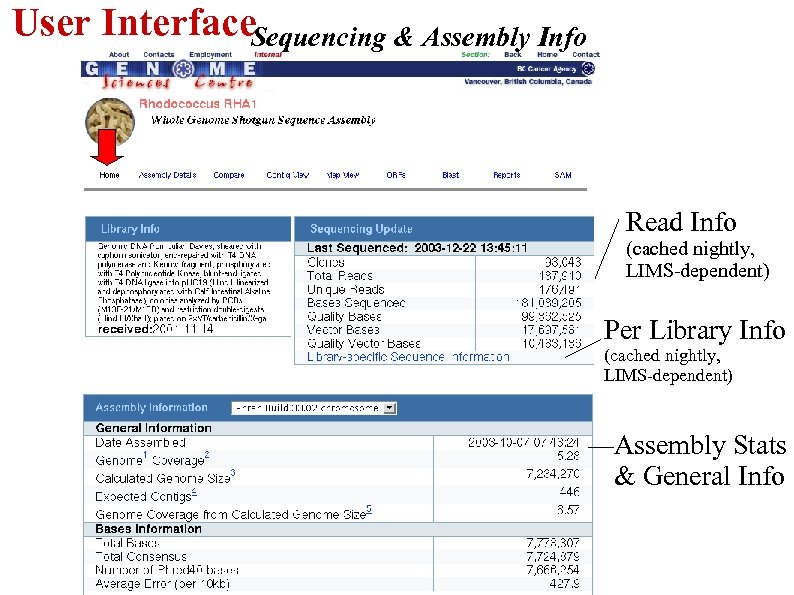 User Interface. Sequencing & Assembly Info Read Info (cached nightly, LIMS-dependent) Per Library Info (cached nightly, LIMS-dependent) Assembly Stats & General Info
User Interface. Sequencing & Assembly Info Read Info (cached nightly, LIMS-dependent) Per Library Info (cached nightly, LIMS-dependent) Assembly Stats & General Info
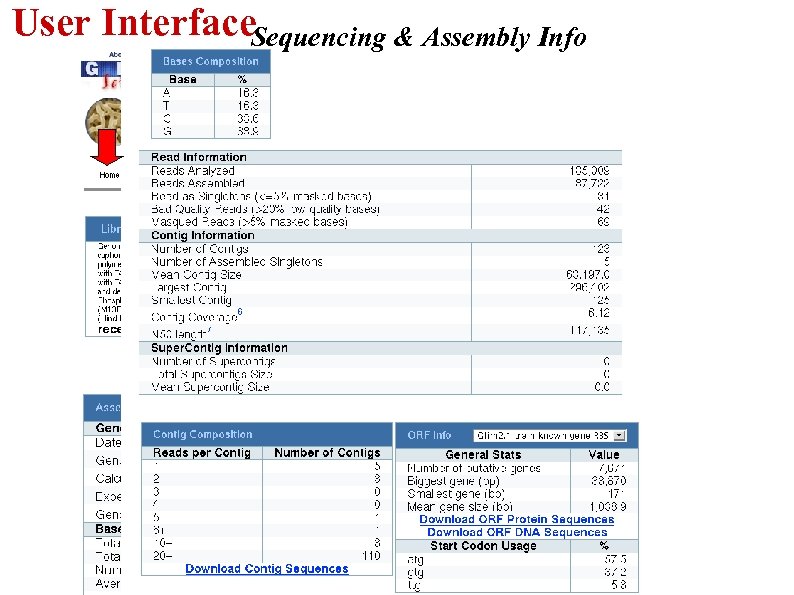 User Interface. Sequencing & Assembly Info
User Interface. Sequencing & Assembly Info
 User Interface Assembly Analyses Assembly Analysis Pages: Access pre-computed data From SAMdb
User Interface Assembly Analyses Assembly Analysis Pages: Access pre-computed data From SAMdb
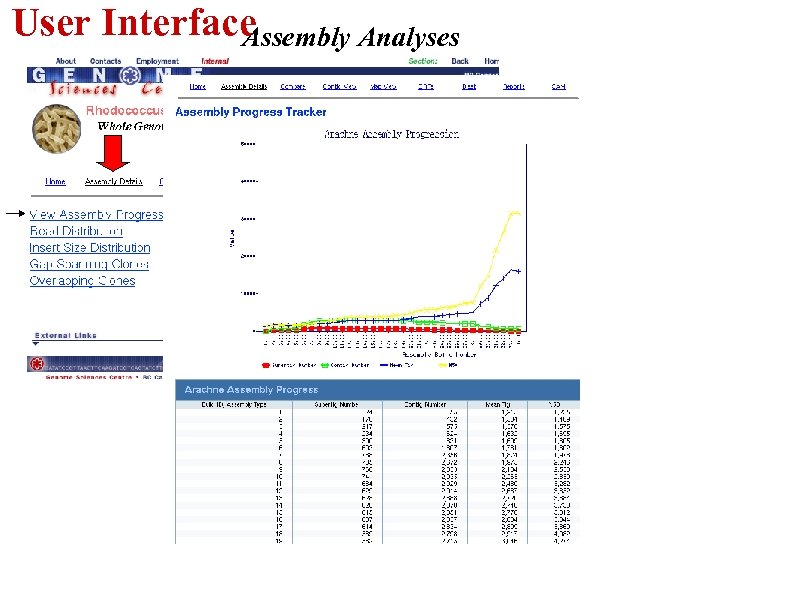 User Interface Assembly Analyses
User Interface Assembly Analyses
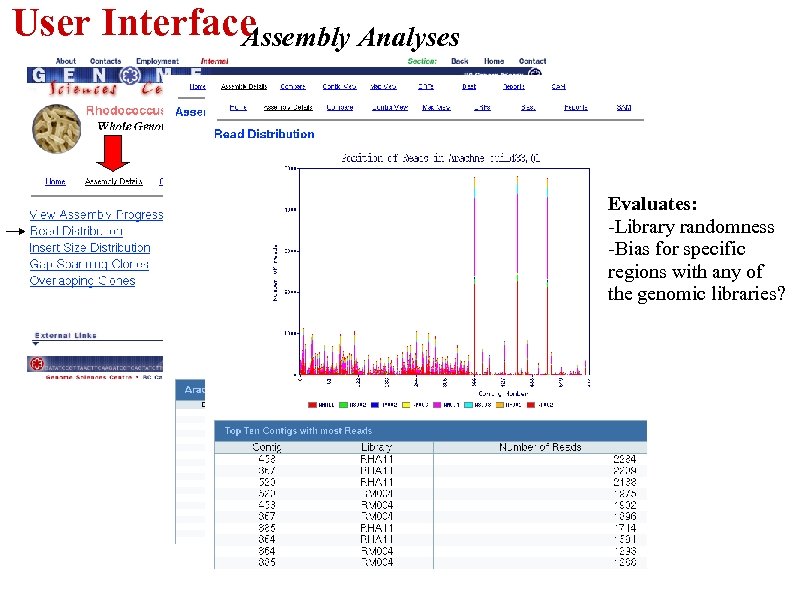 User Interface Assembly Analyses Evaluates: -Library randomness -Bias for specific regions with any of the genomic libraries?
User Interface Assembly Analyses Evaluates: -Library randomness -Bias for specific regions with any of the genomic libraries?
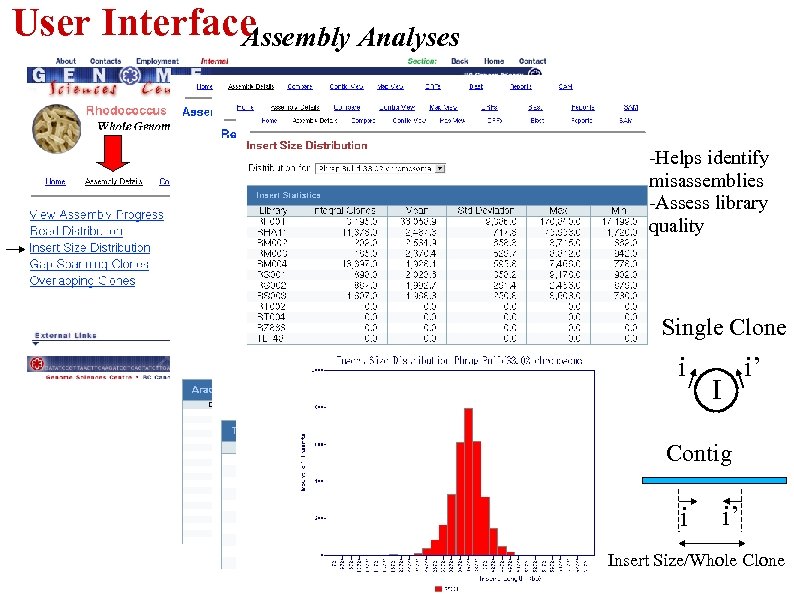 User Interface Assembly Analyses -Helps identify misassemblies -Assess library quality Single Clone i i’ I Contig i i’ Insert Size/Whole Clone
User Interface Assembly Analyses -Helps identify misassemblies -Assess library quality Single Clone i i’ I Contig i i’ Insert Size/Whole Clone
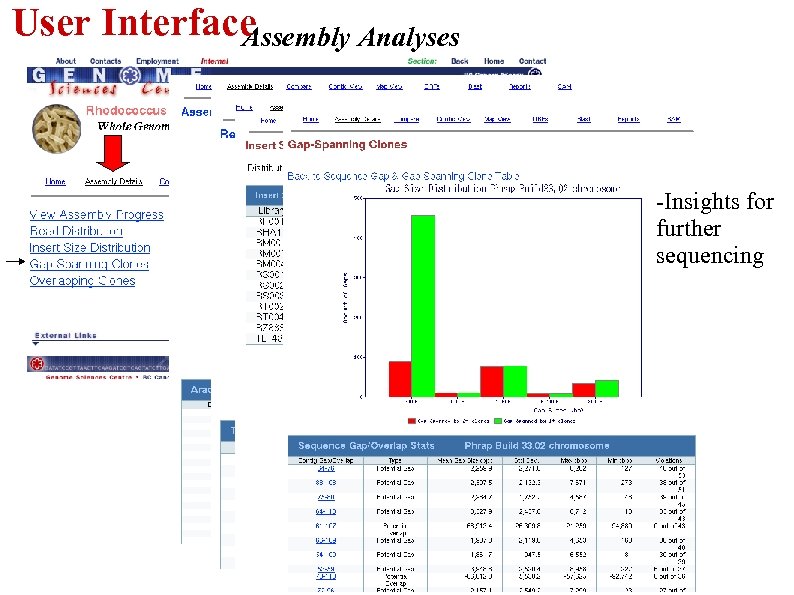 User Interface Assembly Analyses -Insights for further sequencing
User Interface Assembly Analyses -Insights for further sequencing
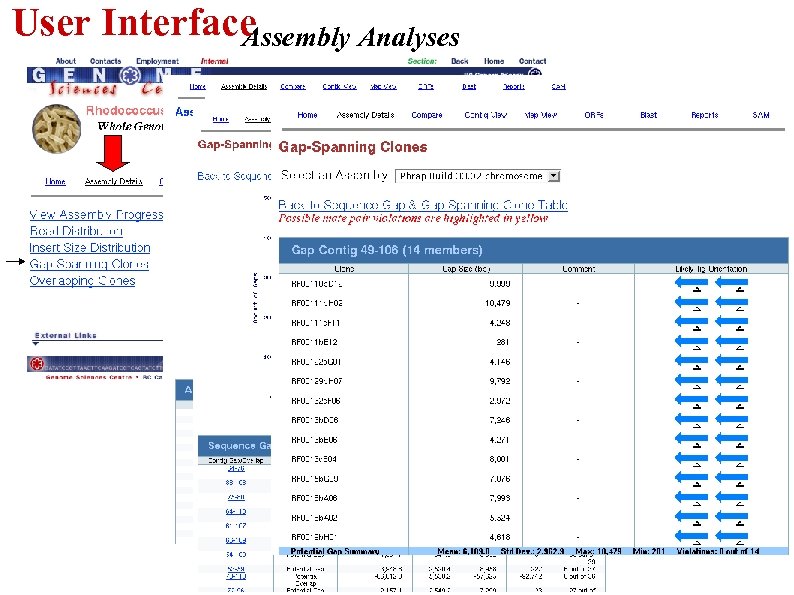 User Interface Assembly Analyses
User Interface Assembly Analyses
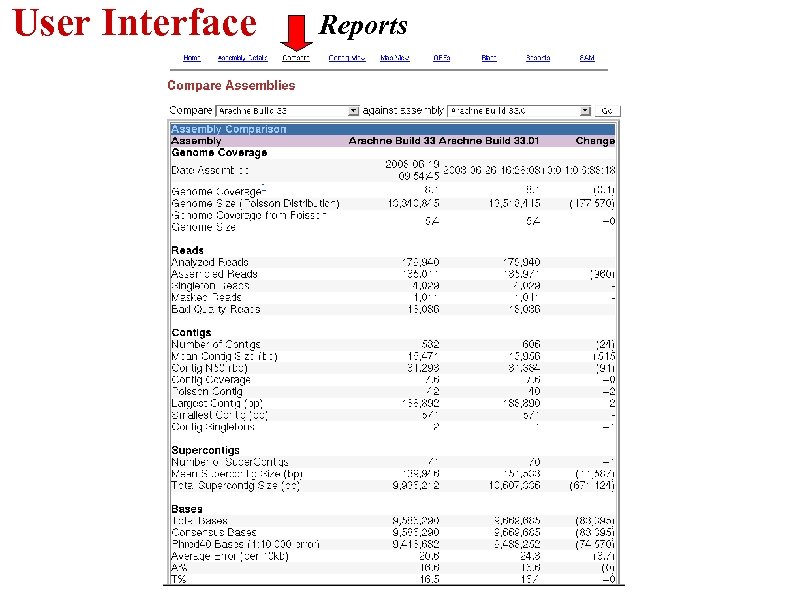 User Interface Reports
User Interface Reports
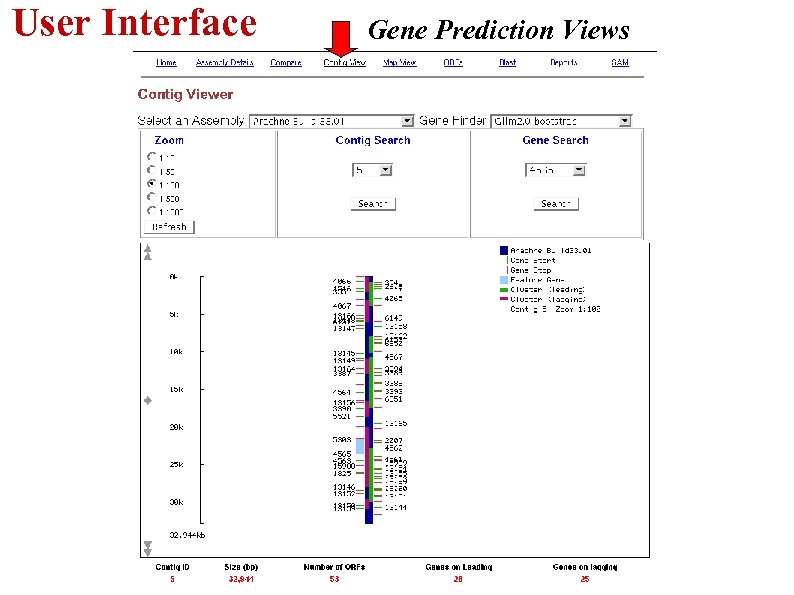 User Interface Gene Prediction Views
User Interface Gene Prediction Views
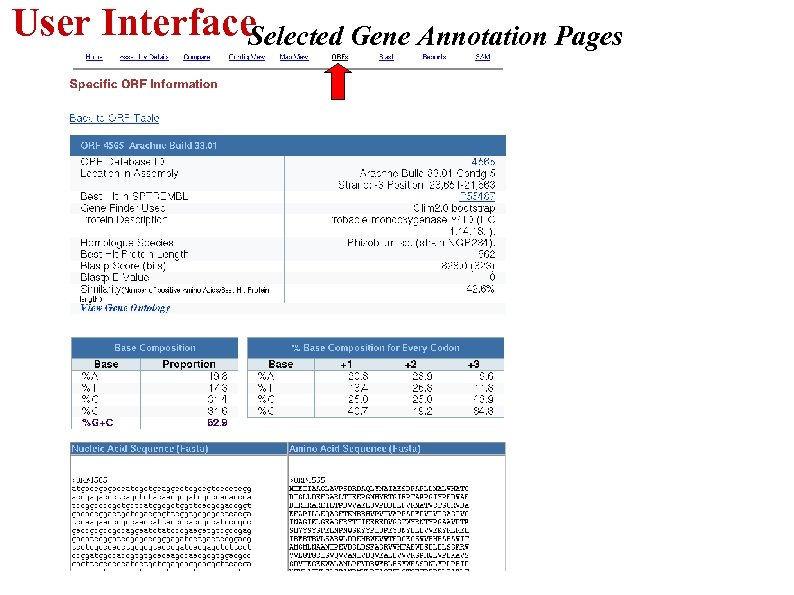 User Interface Selected Gene Annotation Pages
User Interface Selected Gene Annotation Pages
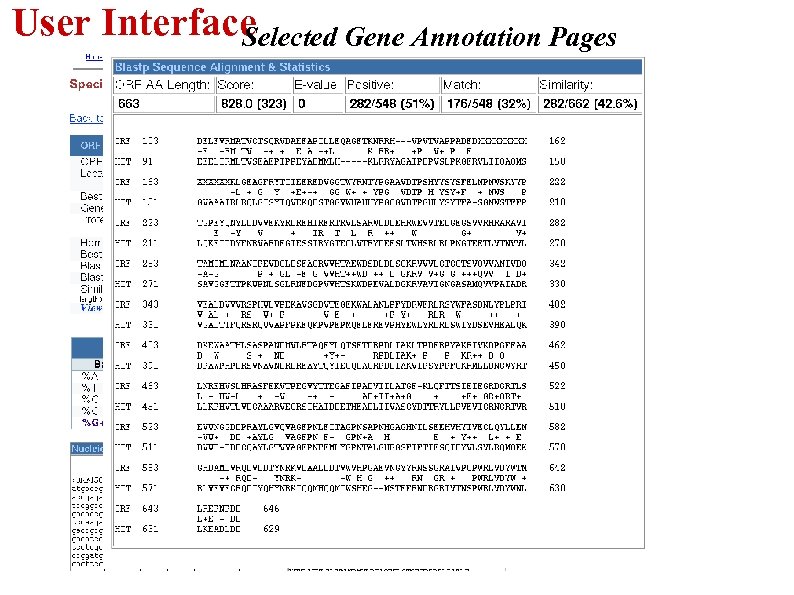 User Interface Selected Gene Annotation Pages
User Interface Selected Gene Annotation Pages
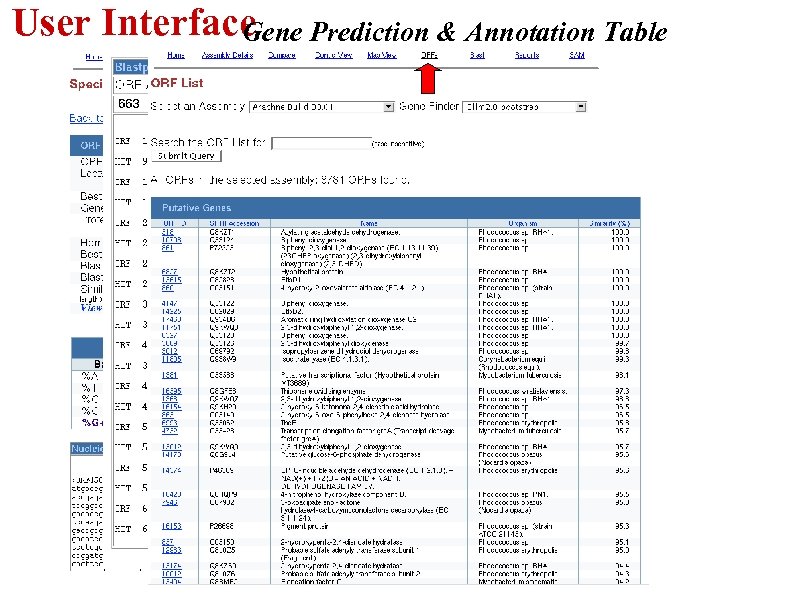 User Interface ene Prediction & Annotation Table G
User Interface ene Prediction & Annotation Table G
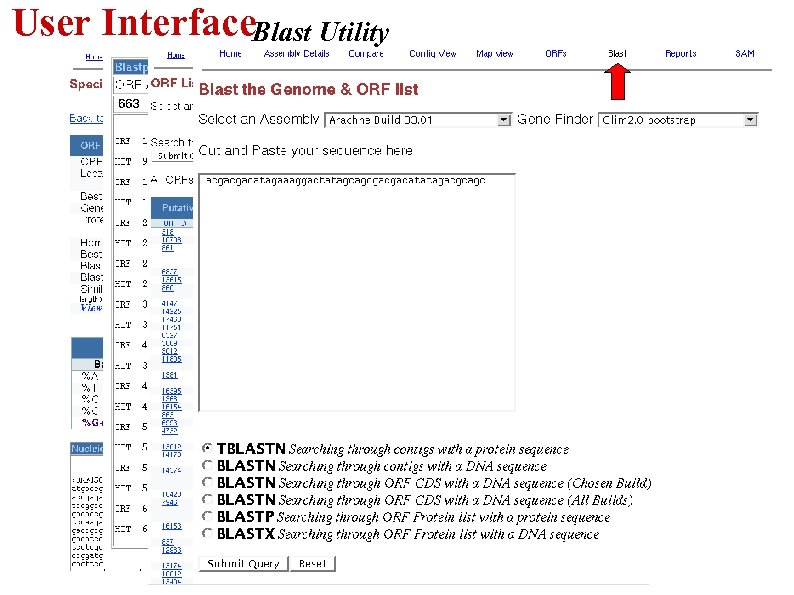 User Interface. Blast Utility
User Interface. Blast Utility
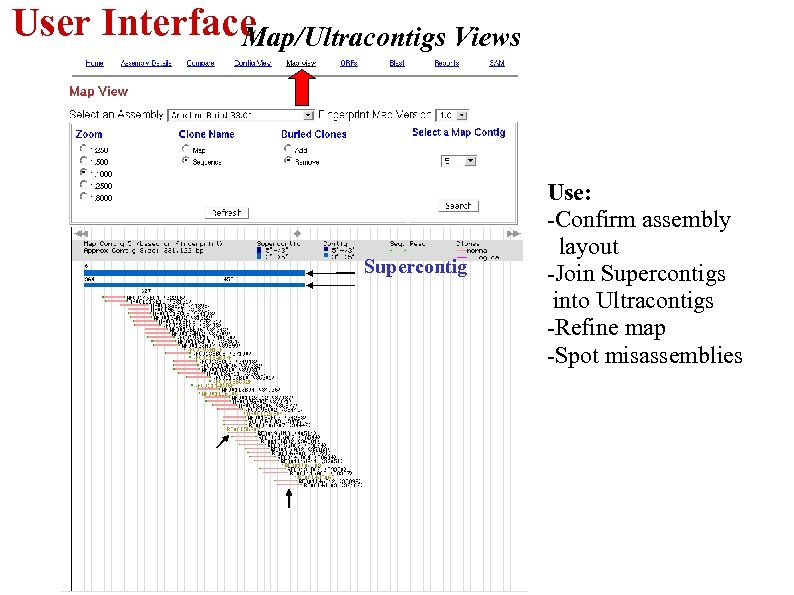 User Interface Map/Ultracontigs Views Supercontig Use: -Confirm assembly layout -Join Supercontigs into Ultracontigs -Refine map -Spot misassemblies
User Interface Map/Ultracontigs Views Supercontig Use: -Confirm assembly layout -Join Supercontigs into Ultracontigs -Refine map -Spot misassemblies
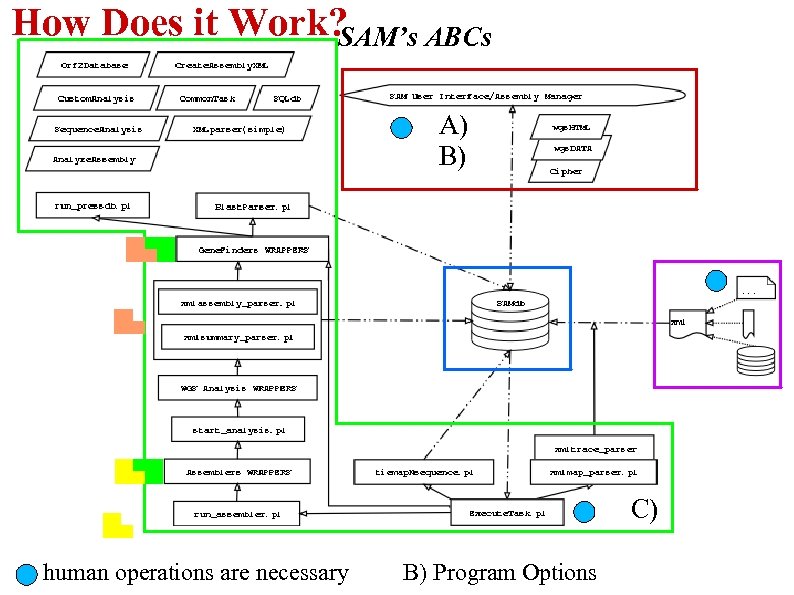 How Does it Work? SAM’s ABCs A) B) C) human operations are necessary B) Program Options
How Does it Work? SAM’s ABCs A) B) C) human operations are necessary B) Program Options
 Executing a Task A) Admin user simply login using My. SQL database credentials
Executing a Task A) Admin user simply login using My. SQL database credentials
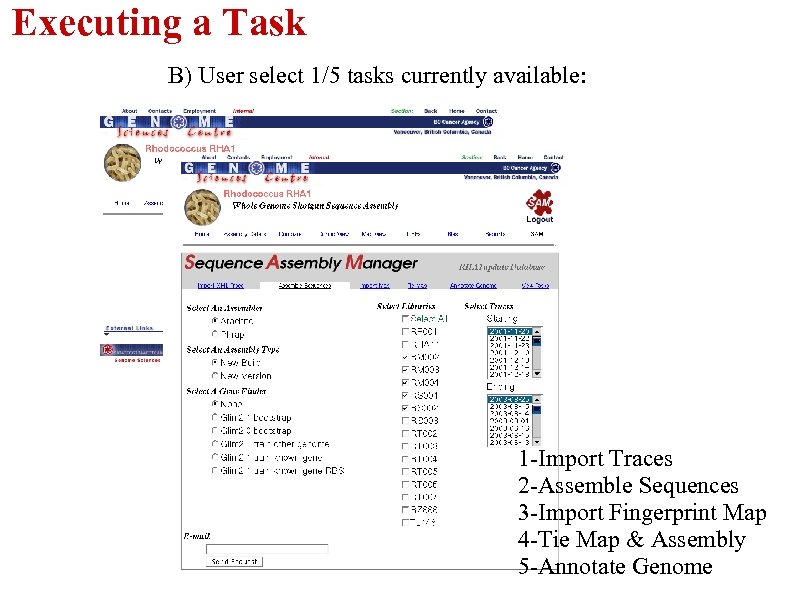 Executing a Task B) User select 1/5 tasks currently available: 1 -Import Traces 2 -Assemble Sequences 3 -Import Fingerprint Map 4 -Tie Map & Assembly 5 -Annotate Genome
Executing a Task B) User select 1/5 tasks currently available: 1 -Import Traces 2 -Assemble Sequences 3 -Import Fingerprint Map 4 -Tie Map & Assembly 5 -Annotate Genome
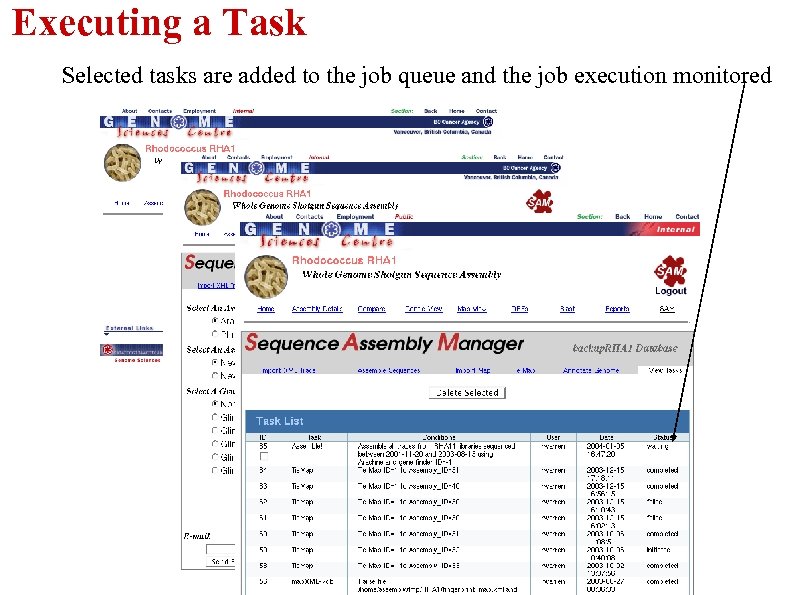 Executing a Task Selected tasks are added to the job queue and the job execution monitored 3) Task
Executing a Task Selected tasks are added to the job queue and the job execution monitored 3) Task
 Executing a Task C) Job is executed by simple command line (could go in cron) *Off course, all parameters must be set properly in the database and appropriate config files required by external programs must exist in the proper format in order for SAM to work properly Debugging -Limited. -A few error traps have been set in order to catch major problems -User must be familiar with Assemblers in order to troubleshoot -Admin user gets notified by email of success/failure when job is done/crashed
Executing a Task C) Job is executed by simple command line (could go in cron) *Off course, all parameters must be set properly in the database and appropriate config files required by external programs must exist in the proper format in order for SAM to work properly Debugging -Limited. -A few error traps have been set in order to catch major problems -User must be familiar with Assemblers in order to troubleshoot -Admin user gets notified by email of success/failure when job is done/crashed
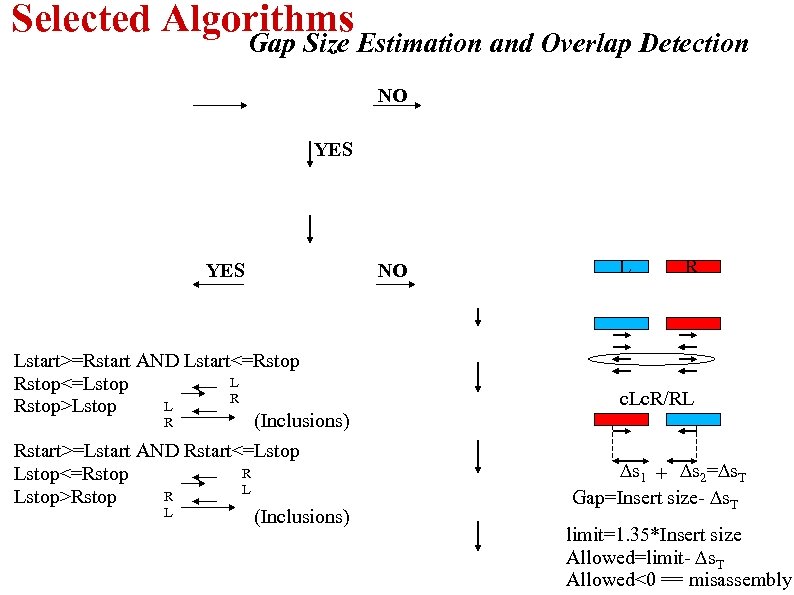 Selected Algorithms Gap Size Estimation and Overlap Detection NO YES NO L R Lstart>=Rstart AND Lstart<=Rstop L Rstop<=Lstop R L Rstop>Lstop R (Inclusions) c. Lc. R/RL Rstart>=Lstart AND Rstart<=Lstop R Lstop<=Rstop L R Lstop>Rstop L (Inclusions) ∆s 1 + ∆s 2=∆s. T Gap=Insert size- ∆s. T limit=1. 35*Insert size Allowed=limit- ∆s. T Allowed<0 == misassembly
Selected Algorithms Gap Size Estimation and Overlap Detection NO YES NO L R Lstart>=Rstart AND Lstart<=Rstop L Rstop<=Lstop R L Rstop>Lstop R (Inclusions) c. Lc. R/RL Rstart>=Lstart AND Rstart<=Lstop R Lstop<=Rstop L R Lstop>Rstop L (Inclusions) ∆s 1 + ∆s 2=∆s. T Gap=Insert size- ∆s. T limit=1. 35*Insert size Allowed=limit- ∆s. T Allowed<0 == misassembly
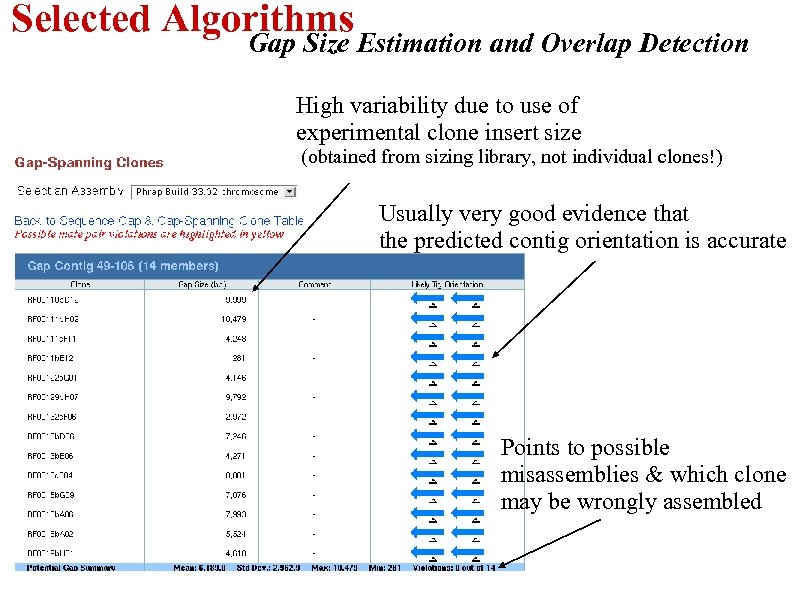 Selected Algorithms Gap Size Estimation and Overlap Detection High variability due to use of experimental clone insert size (obtained from sizing library, not individual clones!) Usually very good evidence that the predicted contig orientation is accurate Points to possible misassemblies & which clone may be wrongly assembled
Selected Algorithms Gap Size Estimation and Overlap Detection High variability due to use of experimental clone insert size (obtained from sizing library, not individual clones!) Usually very good evidence that the predicted contig orientation is accurate Points to possible misassemblies & which clone may be wrongly assembled
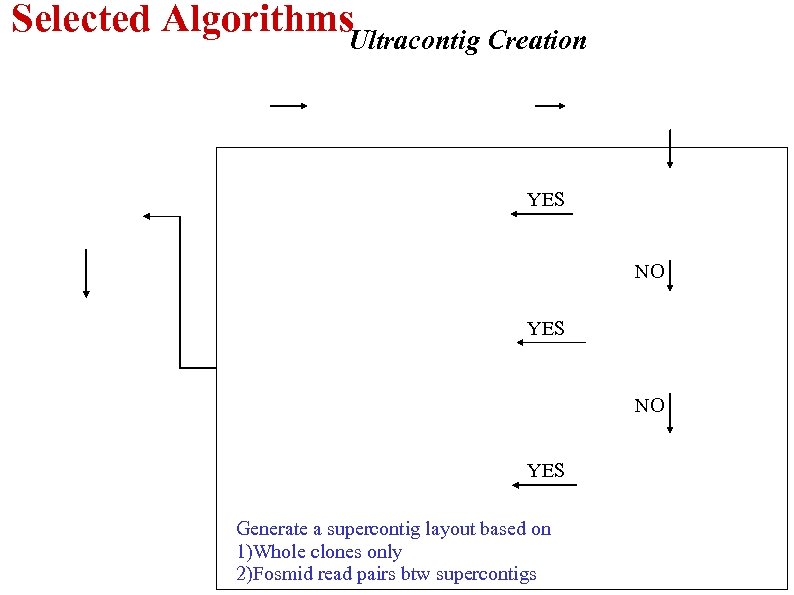 Selected Algorithms. Ultracontig Creation YES NO YES Generate a supercontig layout based on 1)Whole clones only 2)Fosmid read pairs btw supercontigs
Selected Algorithms. Ultracontig Creation YES NO YES Generate a supercontig layout based on 1)Whole clones only 2)Fosmid read pairs btw supercontigs
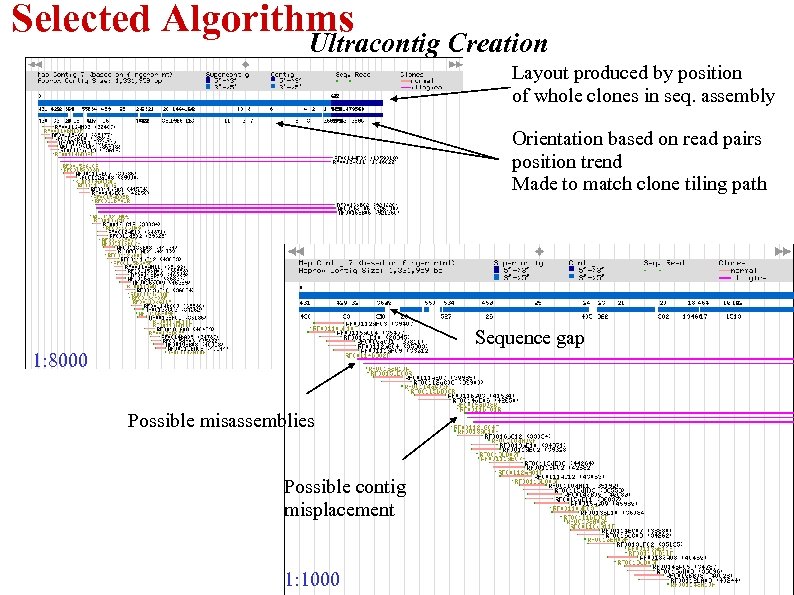 Selected Algorithms Ultracontig Creation Layout produced by position of whole clones in seq. assembly Orientation based on read pairs position trend Made to match clone tiling path Sequence gap 1: 8000 Possible misassemblies Possible contig misplacement 1: 1000
Selected Algorithms Ultracontig Creation Layout produced by position of whole clones in seq. assembly Orientation based on read pairs position trend Made to match clone tiling path Sequence gap 1: 8000 Possible misassemblies Possible contig misplacement 1: 1000
 Future Directions -Switch to OOPerl/ -Create API for database interaction -Break down modules -Improve error trapping -Have the system create on-the-fly configuration files needed by any given assemblers -Make system LIMS-independent
Future Directions -Switch to OOPerl/ -Create API for database interaction -Break down modules -Improve error trapping -Have the system create on-the-fly configuration files needed by any given assemblers -Make system LIMS-independent
 Bioinformatics René Warren Anca Petrescu Yaron Butterfield Mapping GSC mapping group Heesun Shin Jacquie Schein
Bioinformatics René Warren Anca Petrescu Yaron Butterfield Mapping GSC mapping group Heesun Shin Jacquie Schein


