9b0967a5e1d0d94ca96bdcdb854159dc.ppt
- Количество слайдов: 26
 A SAGE Approach to Discovery of Genes Involved in Autophagic Cell Death
A SAGE Approach to Discovery of Genes Involved in Autophagic Cell Death
 Acknowledgements Genome Sciences Centre Victor Ling Marco Marra Functional Genomics Suganthi Chittaranjan Doug Freeman Carrie Anderson Shaun Coughlin Sequencing Genome Sciences Centre Sequencing Team Bioinformatics Steven Jones Erin Garland Richard Varhol Scott Zuyderduyn SAGE team University of Maryland Biotech Institute Eric Baehrecke
Acknowledgements Genome Sciences Centre Victor Ling Marco Marra Functional Genomics Suganthi Chittaranjan Doug Freeman Carrie Anderson Shaun Coughlin Sequencing Genome Sciences Centre Sequencing Team Bioinformatics Steven Jones Erin Garland Richard Varhol Scott Zuyderduyn SAGE team University of Maryland Biotech Institute Eric Baehrecke
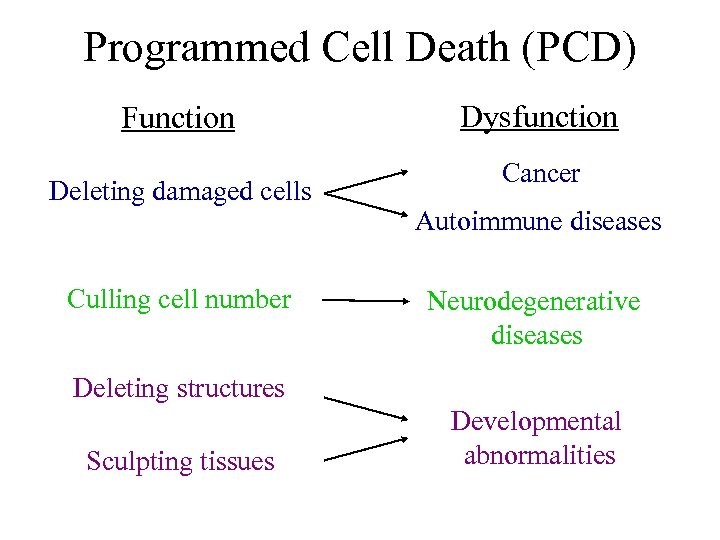 Programmed Cell Death (PCD) Function Deleting damaged cells Culling cell number Dysfunction Cancer Autoimmune diseases Neurodegenerative diseases Deleting structures Sculpting tissues Developmental abnormalities
Programmed Cell Death (PCD) Function Deleting damaged cells Culling cell number Dysfunction Cancer Autoimmune diseases Neurodegenerative diseases Deleting structures Sculpting tissues Developmental abnormalities
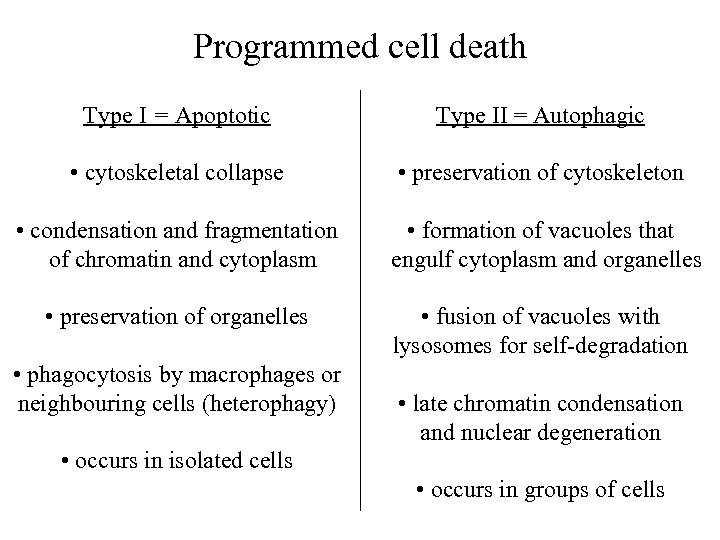 Programmed cell death Type I = Apoptotic Type II = Autophagic • cytoskeletal collapse • preservation of cytoskeleton • formation of vacuoles that engulf cytoplasm and organelles • condensation and fragmentation of chromatin and cytoplasm • preservation of organelles • fusion of vacuoles with lysosomes for self-degradation • phagocytosis by macrophages or neighbouring cells (heterophagy) • late chromatin condensation and nuclear degeneration • occurs in isolated cells • occurs in groups of cells
Programmed cell death Type I = Apoptotic Type II = Autophagic • cytoskeletal collapse • preservation of cytoskeleton • formation of vacuoles that engulf cytoplasm and organelles • condensation and fragmentation of chromatin and cytoplasm • preservation of organelles • fusion of vacuoles with lysosomes for self-degradation • phagocytosis by macrophages or neighbouring cells (heterophagy) • late chromatin condensation and nuclear degeneration • occurs in isolated cells • occurs in groups of cells
 Autophagic cell death in normal physiology • Dictyostelium sorocarp formation • insect metamorphosis • intersegmental muscle, gut, salivary glands • mammalian embryogenesis • regression of interdigital webs, sexual anlagen • mammalian adulthood • intestine, mammary gland post-weaning, ovarian atretic follicles
Autophagic cell death in normal physiology • Dictyostelium sorocarp formation • insect metamorphosis • intersegmental muscle, gut, salivary glands • mammalian embryogenesis • regression of interdigital webs, sexual anlagen • mammalian adulthood • intestine, mammary gland post-weaning, ovarian atretic follicles
 Autophagic cell death in disease • human neurodegenerative diseases (Alzheimer and Parkinson) • cardiomyocyte degeneration • spontaneous regression of human neuroblastoma • tamoxifen-treated mammary carcinoma cells (MCF-7) • bcl-2 antisense treatment of human leukemic HL 60 cells • beclin-1 (apg 6) promotes autophagy and inhibits tumorigenesis; expressed at decreased levels in human breast carcinoma
Autophagic cell death in disease • human neurodegenerative diseases (Alzheimer and Parkinson) • cardiomyocyte degeneration • spontaneous regression of human neuroblastoma • tamoxifen-treated mammary carcinoma cells (MCF-7) • bcl-2 antisense treatment of human leukemic HL 60 cells • beclin-1 (apg 6) promotes autophagy and inhibits tumorigenesis; expressed at decreased levels in human breast carcinoma
 Questions What is the relationship between Autophagic Cell Death and Cancer? • common mechanism in breast and other cancers? • gene mutation What is therapeutic potential of autophagic cell death in cancer? • solid tumours • apoptotic-resistant tumours
Questions What is the relationship between Autophagic Cell Death and Cancer? • common mechanism in breast and other cancers? • gene mutation What is therapeutic potential of autophagic cell death in cancer? • solid tumours • apoptotic-resistant tumours
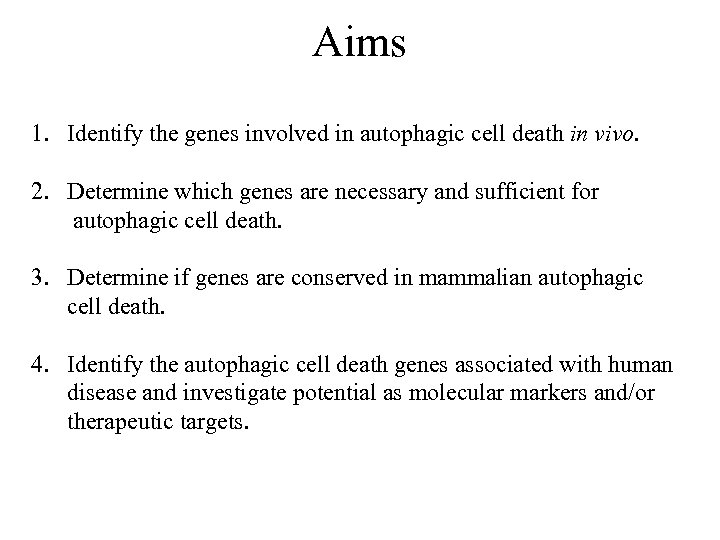 Aims 1. Identify the genes involved in autophagic cell death in vivo. 2. Determine which genes are necessary and sufficient for autophagic cell death. 3. Determine if genes are conserved in mammalian autophagic cell death. 4. Identify the autophagic cell death genes associated with human disease and investigate potential as molecular markers and/or therapeutic targets.
Aims 1. Identify the genes involved in autophagic cell death in vivo. 2. Determine which genes are necessary and sufficient for autophagic cell death. 3. Determine if genes are conserved in mammalian autophagic cell death. 4. Identify the autophagic cell death genes associated with human disease and investigate potential as molecular markers and/or therapeutic targets.
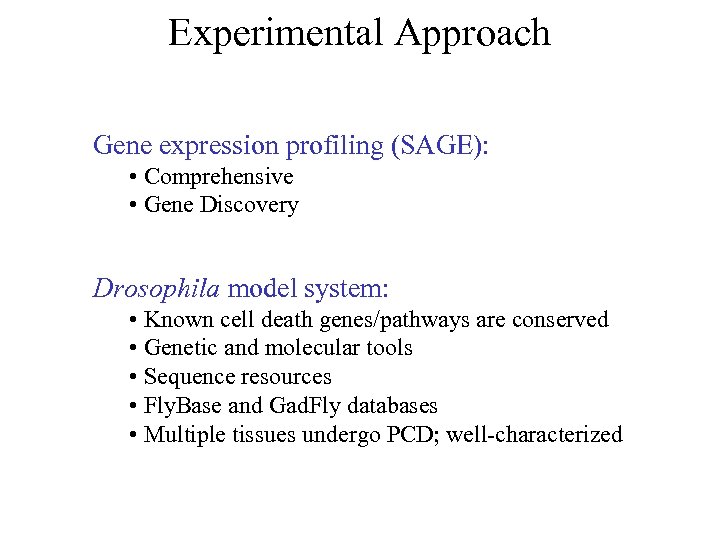 Experimental Approach Gene expression profiling (SAGE): • Comprehensive • Gene Discovery Drosophila model system: • Known cell death genes/pathways are conserved • Genetic and molecular tools • Sequence resources • Fly. Base and Gad. Fly databases • Multiple tissues undergo PCD; well-characterized
Experimental Approach Gene expression profiling (SAGE): • Comprehensive • Gene Discovery Drosophila model system: • Known cell death genes/pathways are conserved • Genetic and molecular tools • Sequence resources • Fly. Base and Gad. Fly databases • Multiple tissues undergo PCD; well-characterized
 Drosophila salivary gland PCD • autophagic • stage-specific • synchronous (Jiang et al. , 1997) • known cell death genes are regulated transcriptionally RT-PCR analysis Reverse transcription hr (APF, 18°C) diap 2 rpr hid - + - + -+ - + 16 18 20 22 23 24
Drosophila salivary gland PCD • autophagic • stage-specific • synchronous (Jiang et al. , 1997) • known cell death genes are regulated transcriptionally RT-PCR analysis Reverse transcription hr (APF, 18°C) diap 2 rpr hid - + - + -+ - + 16 18 20 22 23 24
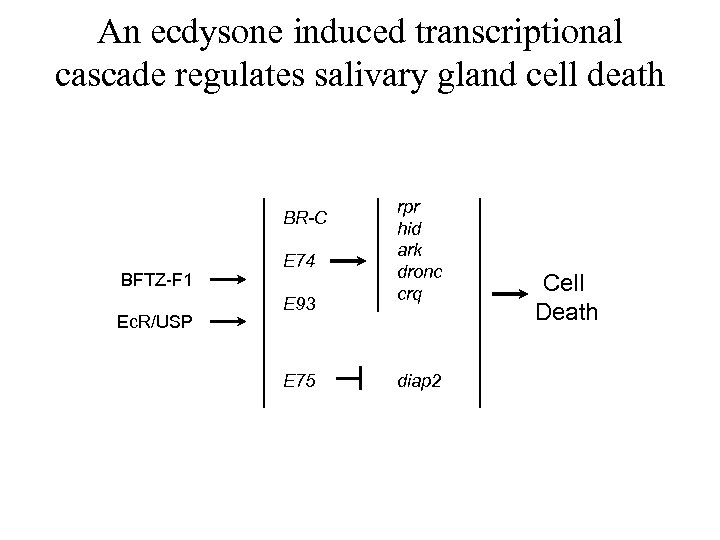 An ecdysone induced transcriptional cascade regulates salivary gland cell death BR-C BFTZ-F 1 Ec. R/USP E 74 E 93 E 75 rpr hid ark dronc crq diap 2 Cell Death
An ecdysone induced transcriptional cascade regulates salivary gland cell death BR-C BFTZ-F 1 Ec. R/USP E 74 E 93 E 75 rpr hid ark dronc crq diap 2 Cell Death
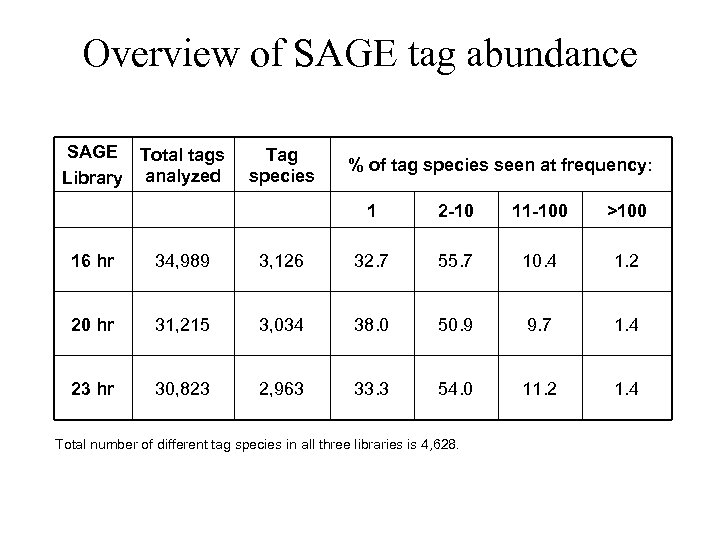 Overview of SAGE tag abundance SAGE Total tags Library analyzed Tag species % of tag species seen at frequency: 1 2 -10 11 -100 >100 16 hr 34, 989 3, 126 32. 7 55. 7 10. 4 1. 2 20 hr 31, 215 3, 034 38. 0 50. 9 9. 7 1. 4 23 hr 30, 823 2, 963 33. 3 54. 0 11. 2 1. 4 Total number of different tag species in all three libraries is 4, 628.
Overview of SAGE tag abundance SAGE Total tags Library analyzed Tag species % of tag species seen at frequency: 1 2 -10 11 -100 >100 16 hr 34, 989 3, 126 32. 7 55. 7 10. 4 1. 2 20 hr 31, 215 3, 034 38. 0 50. 9 9. 7 1. 4 23 hr 30, 823 2, 963 33. 3 54. 0 11. 2 1. 4 Total number of different tag species in all three libraries is 4, 628.
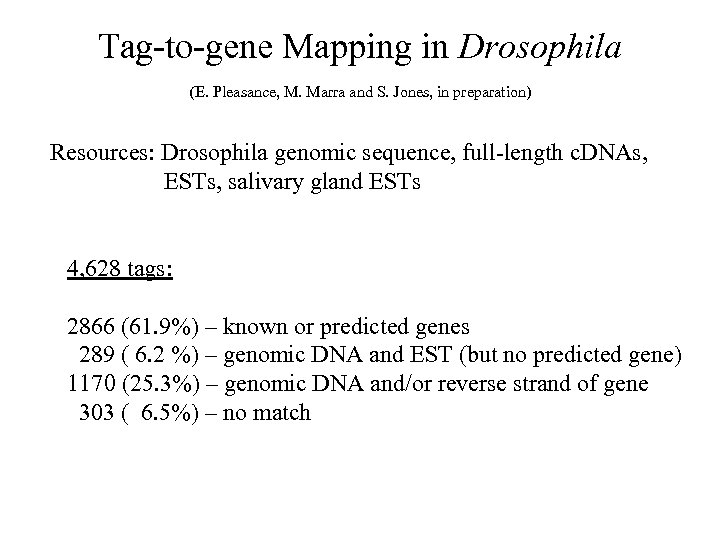 Tag-to-gene Mapping in Drosophila (E. Pleasance, M. Marra and S. Jones, in preparation) Resources: Drosophila genomic sequence, full-length c. DNAs, ESTs, salivary gland ESTs 4, 628 tags: 2866 (61. 9%) – known or predicted genes 289 ( 6. 2 %) – genomic DNA and EST (but no predicted gene) 1170 (25. 3%) – genomic DNA and/or reverse strand of gene 303 ( 6. 5%) – no match
Tag-to-gene Mapping in Drosophila (E. Pleasance, M. Marra and S. Jones, in preparation) Resources: Drosophila genomic sequence, full-length c. DNAs, ESTs, salivary gland ESTs 4, 628 tags: 2866 (61. 9%) – known or predicted genes 289 ( 6. 2 %) – genomic DNA and EST (but no predicted gene) 1170 (25. 3%) – genomic DNA and/or reverse strand of gene 303 ( 6. 5%) – no match
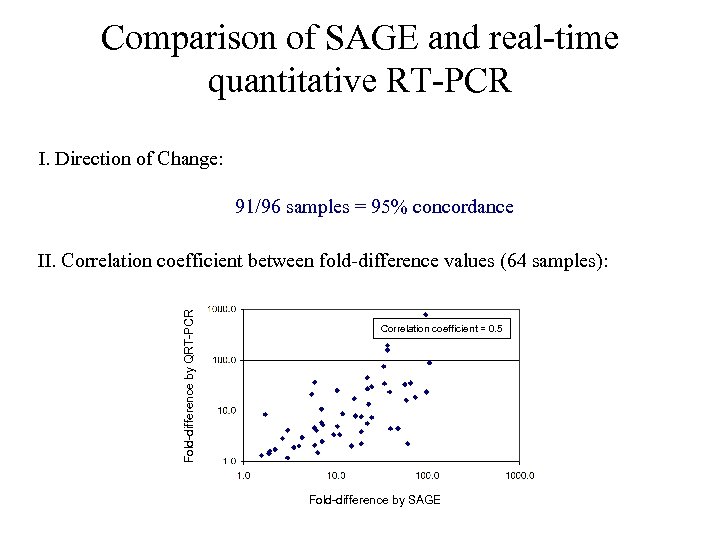 Comparison of SAGE and real-time quantitative RT-PCR I. Direction of Change: 91/96 samples = 95% concordance Fold-difference by QRT-PCR II. Correlation coefficient between fold-difference values (64 samples): Correlation coefficient = 0. 5 Fold-difference by SAGE
Comparison of SAGE and real-time quantitative RT-PCR I. Direction of Change: 91/96 samples = 95% concordance Fold-difference by QRT-PCR II. Correlation coefficient between fold-difference values (64 samples): Correlation coefficient = 0. 5 Fold-difference by SAGE
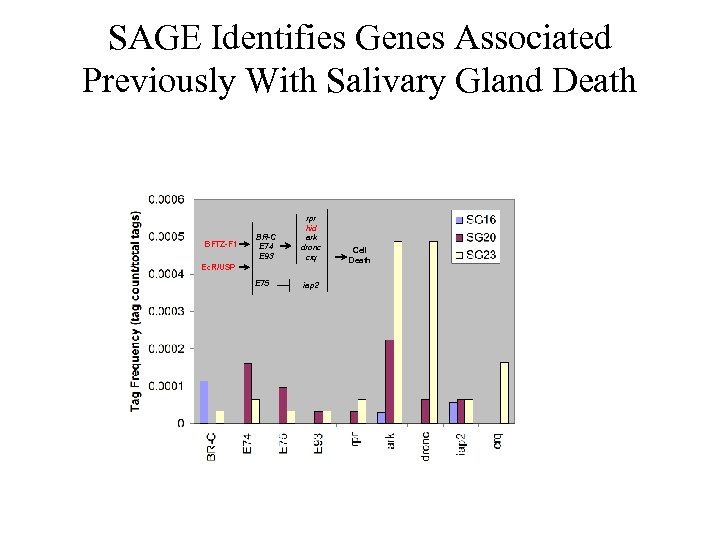 SAGE Identifies Genes Associated Previously With Salivary Gland Death BFTZ-F 1 BR-C E 74 E 93 rpr hid ark dronc crq Ec. R/USP E 75 iap 2 Cell Death
SAGE Identifies Genes Associated Previously With Salivary Gland Death BFTZ-F 1 BR-C E 74 E 93 rpr hid ark dronc crq Ec. R/USP E 75 iap 2 Cell Death
 1244 genes are expressed differentially prior to salivary gland PCD 512 genes have associated biological annotations (Flybase Gene Ontology) 732 genes have unknown functions 377 of these were unpredicted
1244 genes are expressed differentially prior to salivary gland PCD 512 genes have associated biological annotations (Flybase Gene Ontology) 732 genes have unknown functions 377 of these were unpredicted
 Secondary screening of differentially expressed genes I. Data Mining II. Gene expression in salivary gland cell death-defective mutant (E 93) III. Gene expression in other dying tissues IV. Loss-of-function and gain-of-function mutant analyses
Secondary screening of differentially expressed genes I. Data Mining II. Gene expression in salivary gland cell death-defective mutant (E 93) III. Gene expression in other dying tissues IV. Loss-of-function and gain-of-function mutant analyses
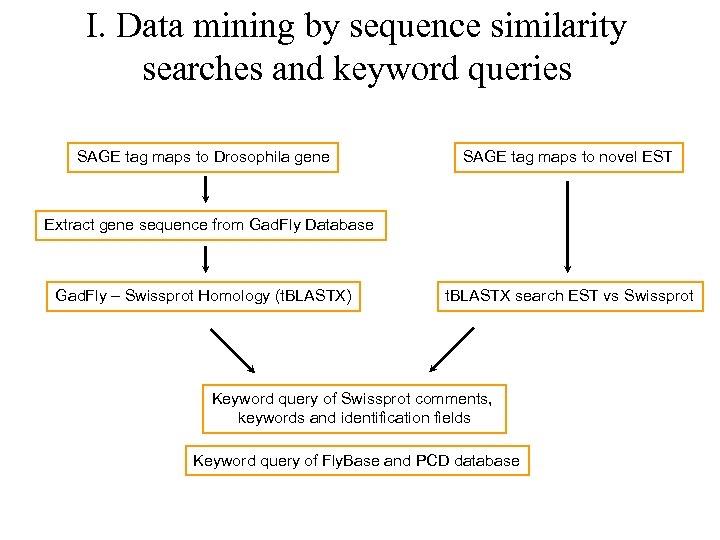 I. Data mining by sequence similarity searches and keyword queries SAGE tag maps to Drosophila gene SAGE tag maps to novel EST Extract gene sequence from Gad. Fly Database Gad. Fly – Swissprot Homology (t. BLASTX) t. BLASTX search EST vs Swissprot Keyword query of Swissprot comments, keywords and identification fields Keyword query of Fly. Base and PCD database
I. Data mining by sequence similarity searches and keyword queries SAGE tag maps to Drosophila gene SAGE tag maps to novel EST Extract gene sequence from Gad. Fly Database Gad. Fly – Swissprot Homology (t. BLASTX) t. BLASTX search EST vs Swissprot Keyword query of Swissprot comments, keywords and identification fields Keyword query of Fly. Base and PCD database
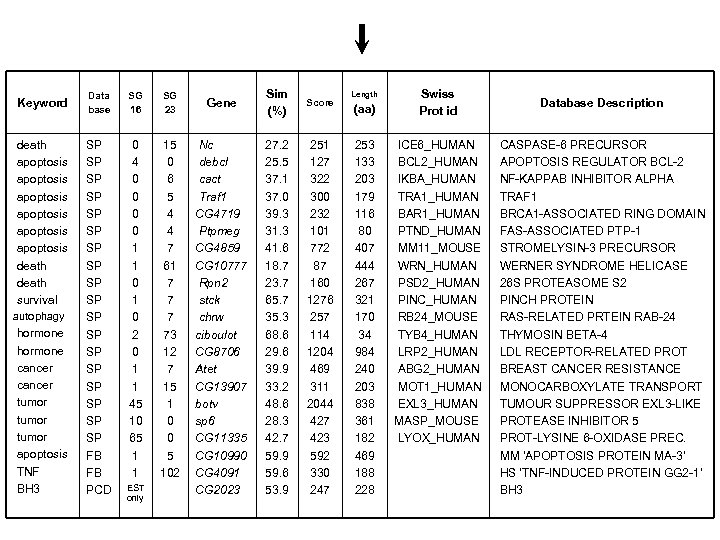 Keyword death apoptosis apoptosis death survival autophagy hormone cancer tumor apoptosis TNF BH 3 Data base SG 16 SG 23 SP SP SP SP SP FB FB PCD 0 4 0 0 1 1 0 2 0 1 1 45 10 65 1 1 15 0 6 5 4 4 7 61 7 73 12 7 15 1 0 0 5 102 EST only Gene Sim (%) Score Nc debcl cact Traf 1 CG 4719 Ptpmeg CG 4859 CG 10777 Rpn 2 stck chrw ciboulot CG 8706 Atet CG 13907 botv sp 6 CG 11335 CG 10990 CG 4091 CG 2023 27. 2 25. 5 37. 1 37. 0 39. 3 31. 3 41. 6 18. 7 23. 7 65. 7 35. 3 68. 6 29. 6 39. 9 33. 2 48. 6 28. 3 42. 7 59. 9 59. 6 53. 9 251 127 322 300 232 101 772 87 160 1276 257 114 1204 469 311 2044 427 423 592 330 247 (aa) Swiss Prot id Database Description 253 133 203 179 116 80 407 444 267 321 170 34 984 240 203 838 361 182 469 188 228 ICE 6_HUMAN BCL 2_HUMAN IKBA_HUMAN TRA 1_HUMAN BAR 1_HUMAN PTND_HUMAN MM 11_MOUSE WRN_HUMAN PSD 2_HUMAN PINC_HUMAN RB 24_MOUSE TYB 4_HUMAN LRP 2_HUMAN ABG 2_HUMAN MOT 1_HUMAN EXL 3_HUMAN MASP_MOUSE LYOX_HUMAN CASPASE-6 PRECURSOR APOPTOSIS REGULATOR BCL-2 NF-KAPPAB INHIBITOR ALPHA TRAF 1 BRCA 1 -ASSOCIATED RING DOMAIN FAS-ASSOCIATED PTP-1 STROMELYSIN-3 PRECURSOR WERNER SYNDROME HELICASE 26 S PROTEASOME S 2 PINCH PROTEIN RAS-RELATED PRTEIN RAB-24 THYMOSIN BETA-4 LDL RECEPTOR-RELATED PROT BREAST CANCER RESISTANCE MONOCARBOXYLATE TRANSPORT TUMOUR SUPPRESSOR EXL 3 -LIKE PROTEASE INHIBITOR 5 PROT-LYSINE 6 -OXIDASE PREC. MM 'APOPTOSIS PROTEIN MA-3' HS 'TNF-INDUCED PROTEIN GG 2 -1' BH 3 Length
Keyword death apoptosis apoptosis death survival autophagy hormone cancer tumor apoptosis TNF BH 3 Data base SG 16 SG 23 SP SP SP SP SP FB FB PCD 0 4 0 0 1 1 0 2 0 1 1 45 10 65 1 1 15 0 6 5 4 4 7 61 7 73 12 7 15 1 0 0 5 102 EST only Gene Sim (%) Score Nc debcl cact Traf 1 CG 4719 Ptpmeg CG 4859 CG 10777 Rpn 2 stck chrw ciboulot CG 8706 Atet CG 13907 botv sp 6 CG 11335 CG 10990 CG 4091 CG 2023 27. 2 25. 5 37. 1 37. 0 39. 3 31. 3 41. 6 18. 7 23. 7 65. 7 35. 3 68. 6 29. 6 39. 9 33. 2 48. 6 28. 3 42. 7 59. 9 59. 6 53. 9 251 127 322 300 232 101 772 87 160 1276 257 114 1204 469 311 2044 427 423 592 330 247 (aa) Swiss Prot id Database Description 253 133 203 179 116 80 407 444 267 321 170 34 984 240 203 838 361 182 469 188 228 ICE 6_HUMAN BCL 2_HUMAN IKBA_HUMAN TRA 1_HUMAN BAR 1_HUMAN PTND_HUMAN MM 11_MOUSE WRN_HUMAN PSD 2_HUMAN PINC_HUMAN RB 24_MOUSE TYB 4_HUMAN LRP 2_HUMAN ABG 2_HUMAN MOT 1_HUMAN EXL 3_HUMAN MASP_MOUSE LYOX_HUMAN CASPASE-6 PRECURSOR APOPTOSIS REGULATOR BCL-2 NF-KAPPAB INHIBITOR ALPHA TRAF 1 BRCA 1 -ASSOCIATED RING DOMAIN FAS-ASSOCIATED PTP-1 STROMELYSIN-3 PRECURSOR WERNER SYNDROME HELICASE 26 S PROTEASOME S 2 PINCH PROTEIN RAS-RELATED PRTEIN RAB-24 THYMOSIN BETA-4 LDL RECEPTOR-RELATED PROT BREAST CANCER RESISTANCE MONOCARBOXYLATE TRANSPORT TUMOUR SUPPRESSOR EXL 3 -LIKE PROTEASE INHIBITOR 5 PROT-LYSINE 6 -OXIDASE PREC. MM 'APOPTOSIS PROTEIN MA-3' HS 'TNF-INDUCED PROTEIN GG 2 -1' BH 3 Length
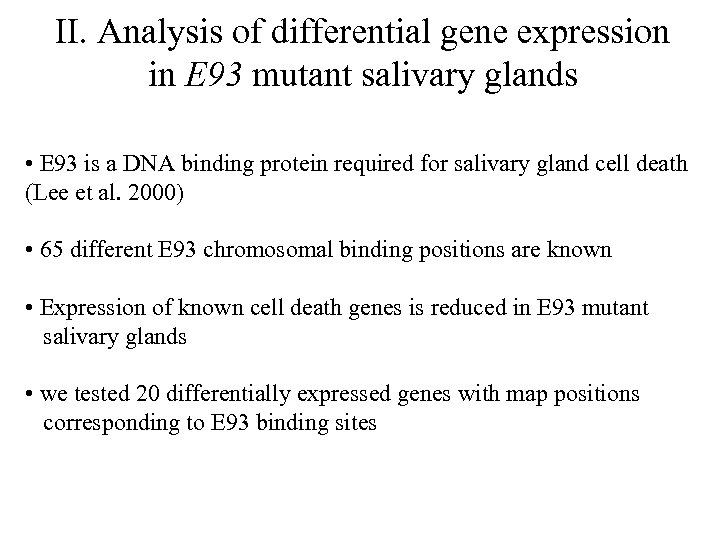 II. Analysis of differential gene expression in E 93 mutant salivary glands • E 93 is a DNA binding protein required for salivary gland cell death (Lee et al. 2000) • 65 different E 93 chromosomal binding positions are known • Expression of known cell death genes is reduced in E 93 mutant salivary glands • we tested 20 differentially expressed genes with map positions corresponding to E 93 binding sites
II. Analysis of differential gene expression in E 93 mutant salivary glands • E 93 is a DNA binding protein required for salivary gland cell death (Lee et al. 2000) • 65 different E 93 chromosomal binding positions are known • Expression of known cell death genes is reduced in E 93 mutant salivary glands • we tested 20 differentially expressed genes with map positions corresponding to E 93 binding sites
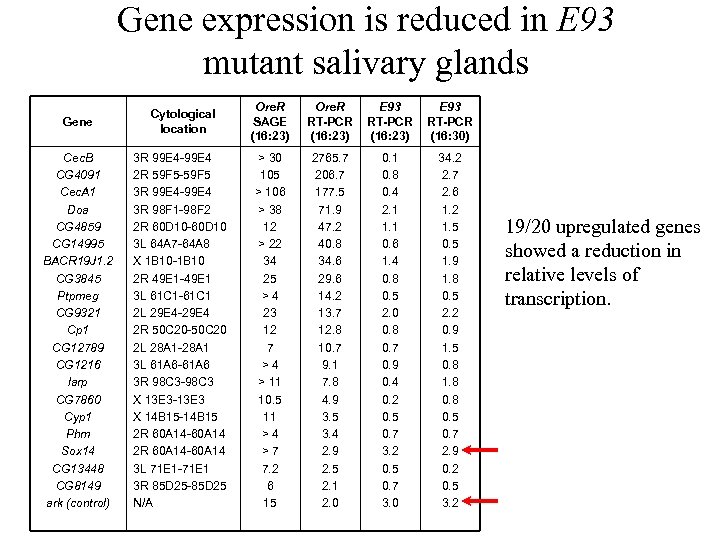 Gene expression is reduced in E 93 mutant salivary glands Gene Cytological location Ore. R SAGE (16: 23) Ore. R RT-PCR (16: 23) E 93 RT-PCR (16: 30) Cec. B CG 4091 Cec. A 1 Doa CG 4859 CG 14995 BACR 19 J 1. 2 CG 3845 Ptpmeg CG 9321 Cp 1 CG 12789 CG 1216 larp CG 7860 Cyp 1 Phm Sox 14 CG 13448 CG 8149 ark (control) 3 R 99 E 4 -99 E 4 2 R 59 F 5 -59 F 5 3 R 99 E 4 -99 E 4 3 R 98 F 1 -98 F 2 2 R 60 D 10 -60 D 10 3 L 64 A 7 -64 A 8 X 1 B 10 -1 B 10 2 R 49 E 1 -49 E 1 3 L 61 C 1 -61 C 1 2 L 29 E 4 -29 E 4 2 R 50 C 20 -50 C 20 2 L 28 A 1 -28 A 1 3 L 61 A 6 -61 A 6 3 R 98 C 3 -98 C 3 X 13 E 3 -13 E 3 X 14 B 15 -14 B 15 2 R 60 A 14 -60 A 14 3 L 71 E 1 -71 E 1 3 R 85 D 25 -85 D 25 N/A > 30 105 > 106 > 38 12 > 22 34 25 > 4 23 12 7 > 4 > 11 10. 5 11 > 4 > 7 7. 2 6 15 2765. 7 206. 7 177. 5 71. 9 47. 2 40. 8 34. 6 29. 6 14. 2 13. 7 12. 8 10. 7 9. 1 7. 8 4. 9 3. 5 3. 4 2. 9 2. 5 2. 1 2. 0 0. 1 0. 8 0. 4 2. 1 1. 1 0. 6 1. 4 0. 8 0. 5 2. 0 0. 8 0. 7 0. 9 0. 4 0. 2 0. 5 0. 7 3. 0 34. 2 2. 7 2. 6 1. 2 1. 5 0. 5 1. 9 1. 8 0. 5 2. 2 0. 9 1. 5 0. 8 1. 8 0. 5 0. 7 2. 9 0. 2 0. 5 3. 2 19/20 upregulated genes showed a reduction in relative levels of transcription.
Gene expression is reduced in E 93 mutant salivary glands Gene Cytological location Ore. R SAGE (16: 23) Ore. R RT-PCR (16: 23) E 93 RT-PCR (16: 30) Cec. B CG 4091 Cec. A 1 Doa CG 4859 CG 14995 BACR 19 J 1. 2 CG 3845 Ptpmeg CG 9321 Cp 1 CG 12789 CG 1216 larp CG 7860 Cyp 1 Phm Sox 14 CG 13448 CG 8149 ark (control) 3 R 99 E 4 -99 E 4 2 R 59 F 5 -59 F 5 3 R 99 E 4 -99 E 4 3 R 98 F 1 -98 F 2 2 R 60 D 10 -60 D 10 3 L 64 A 7 -64 A 8 X 1 B 10 -1 B 10 2 R 49 E 1 -49 E 1 3 L 61 C 1 -61 C 1 2 L 29 E 4 -29 E 4 2 R 50 C 20 -50 C 20 2 L 28 A 1 -28 A 1 3 L 61 A 6 -61 A 6 3 R 98 C 3 -98 C 3 X 13 E 3 -13 E 3 X 14 B 15 -14 B 15 2 R 60 A 14 -60 A 14 3 L 71 E 1 -71 E 1 3 R 85 D 25 -85 D 25 N/A > 30 105 > 106 > 38 12 > 22 34 25 > 4 23 12 7 > 4 > 11 10. 5 11 > 4 > 7 7. 2 6 15 2765. 7 206. 7 177. 5 71. 9 47. 2 40. 8 34. 6 29. 6 14. 2 13. 7 12. 8 10. 7 9. 1 7. 8 4. 9 3. 5 3. 4 2. 9 2. 5 2. 1 2. 0 0. 1 0. 8 0. 4 2. 1 1. 1 0. 6 1. 4 0. 8 0. 5 2. 0 0. 8 0. 7 0. 9 0. 4 0. 2 0. 5 0. 7 3. 0 34. 2 2. 7 2. 6 1. 2 1. 5 0. 5 1. 9 1. 8 0. 5 2. 2 0. 9 1. 5 0. 8 1. 8 0. 5 0. 7 2. 9 0. 2 0. 5 3. 2 19/20 upregulated genes showed a reduction in relative levels of transcription.
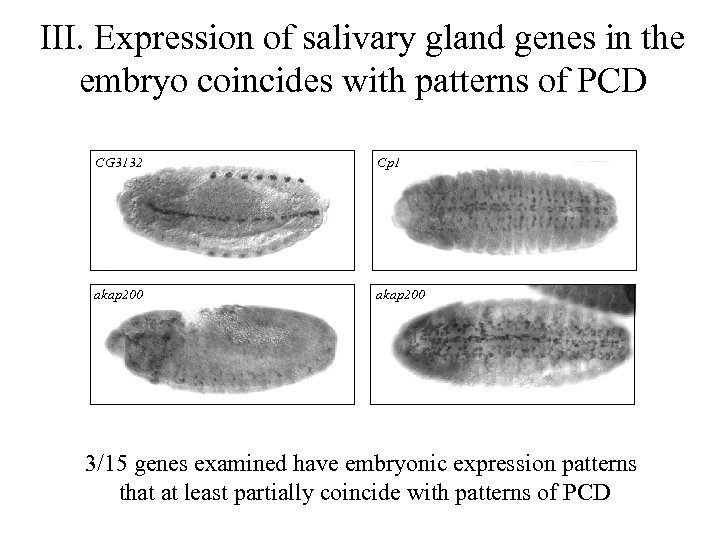 III. Expression of salivary gland genes in the embryo coincides with patterns of PCD CG 3132 Cp 1 akap 200 3/15 genes examined have embryonic expression patterns that at least partially coincide with patterns of PCD
III. Expression of salivary gland genes in the embryo coincides with patterns of PCD CG 3132 Cp 1 akap 200 3/15 genes examined have embryonic expression patterns that at least partially coincide with patterns of PCD
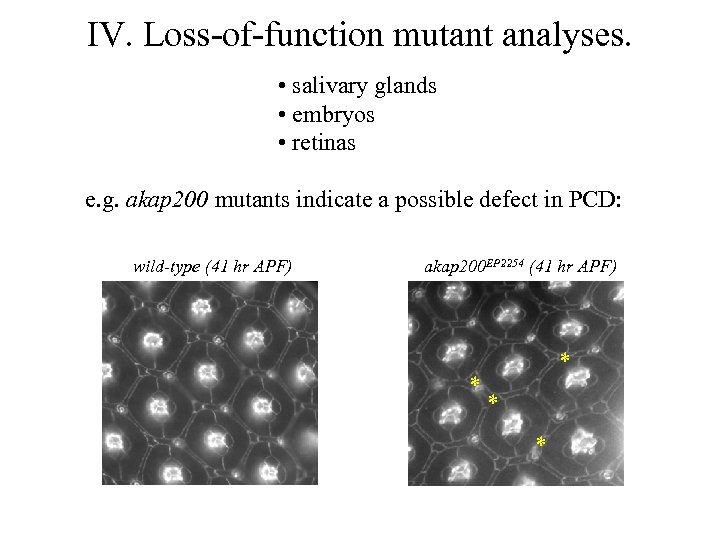 IV. Loss-of-function mutant analyses. • salivary glands • embryos • retinas e. g. akap 200 mutants indicate a possible defect in PCD: wild-type (41 hr APF) akap 200 EP 2254 (41 hr APF) * *
IV. Loss-of-function mutant analyses. • salivary glands • embryos • retinas e. g. akap 200 mutants indicate a possible defect in PCD: wild-type (41 hr APF) akap 200 EP 2254 (41 hr APF) * *
 Differentially expressed genes reveal molecular features associated with autophagic cell death • Autophagic cell death involves the induction of genes required for protein synthesis e. g. 6 different translation initiation factors • Novel transcription factors appear upregulated coordinately with known transcriptional regulators e. g. maf-S, CG 3350 • Components of multiple signal transduction pathways are involved e. g. TNF-a like pathway, akap 200, Doa
Differentially expressed genes reveal molecular features associated with autophagic cell death • Autophagic cell death involves the induction of genes required for protein synthesis e. g. 6 different translation initiation factors • Novel transcription factors appear upregulated coordinately with known transcriptional regulators e. g. maf-S, CG 3350 • Components of multiple signal transduction pathways are involved e. g. TNF-a like pathway, akap 200, Doa
 Differentially expressed genes and molecular features of autophagic cell death, continued. • apg-like genes can be regulated transcriptionally and this regulation is associated with autophagic cell death e. g. genes involved in two ubiquitin-like pathways CG 6194 (apg 4; novel cysteine protease) CG 5429 (apg 6/beclin-1) • Other autophagy-associated genes are likely involved e. g. lysosomal enzymes, rab-7 • Autophagic and apoptotic cell death appear to utilize at least some common pathways or pathway components
Differentially expressed genes and molecular features of autophagic cell death, continued. • apg-like genes can be regulated transcriptionally and this regulation is associated with autophagic cell death e. g. genes involved in two ubiquitin-like pathways CG 6194 (apg 4; novel cysteine protease) CG 5429 (apg 6/beclin-1) • Other autophagy-associated genes are likely involved e. g. lysosomal enzymes, rab-7 • Autophagic and apoptotic cell death appear to utilize at least some common pathways or pathway components
 Future Directions Autophagic cell death in cancer 1. Extent of role of autophagic cell death in different types of cancer • Gene expression comparisons btn wt, cancers and Dm SAGE (Erin) • Need to develop autophagic PCD specific markers Expt: expression profiling in tamoxifen-treated MCF-7 cells 2. Is it possible to induce autophagic PCD in tumours? • candidate genes to test: CG 6194 (a cysteine protease) (Is it possible to induce autophagic PCD in apoptotic-resistant tumours? ) 3. What molecules are necessary and/or sufficient for autophagic PCD? 1. Mutant analyses in Drosophila (existing mutants; RNAi) 2. GAL 4/UAS system to induce tissue-specific ectopic expression
Future Directions Autophagic cell death in cancer 1. Extent of role of autophagic cell death in different types of cancer • Gene expression comparisons btn wt, cancers and Dm SAGE (Erin) • Need to develop autophagic PCD specific markers Expt: expression profiling in tamoxifen-treated MCF-7 cells 2. Is it possible to induce autophagic PCD in tumours? • candidate genes to test: CG 6194 (a cysteine protease) (Is it possible to induce autophagic PCD in apoptotic-resistant tumours? ) 3. What molecules are necessary and/or sufficient for autophagic PCD? 1. Mutant analyses in Drosophila (existing mutants; RNAi) 2. GAL 4/UAS system to induce tissue-specific ectopic expression


